Abstract
TRPV channels are important polymodal integrators of noxious stimuli mediating thermosensation and nociception. An ankyrin repeat domain (ARD), which is a common protein–protein recognition domain, is conserved in the N-terminal intracellular domain of all TRPV channels and predicted to contain three to four ankyrin repeats. Here we report the first structure from the TRPV channel subfamily, a 1.7 Å resolution crystal structure of the human TRPV2 ARD. Our crystal structure reveals a six ankyrin repeat stack with multiple insertions in each repeat generating several unique features compared with a canonical ARD. The surface typically used for ligand recognition, the ankyrin groove, contains extended loops with an exposed hydrophobic patch and a prominent kink resulting from a large rotational shift of the last two repeats. The TRPV2 ARD provides the first structural insight into a domain that coordinates nociceptive sensory transduction and is likely to be a prototype for other TRPV channel ARDs.
Keywords: TRPV channel, ankyrin repeat, crystal structure
Transient receptor potential (TRP) channels are biological sensors essential for processing numerous environmental stimuli (Clapham 2003). The TRPV subfamily is comprised of nonselective cation channels important for nociception, osmosensation, mechanosensation, maintenance of calcium homeostasis, and temperature sensing, with TRPV1 through 4 each exhibiting a distinct thermal activation threshold (Patapoutian et al. 2003). TRPV channels can be polymodally activated as demonstrated by TRPV1, which is activated by heat (≥43°C), acidification, vanilloid compounds such as capsaicin, the active ingredient of hot chili peppers, and several proinflammatory mediators (Caterina and Julius 2001). TRPV channels are also emerging as multifunctional receptors as shown by TRPV2, which in addition to responding to noxious thermal stimuli (≥52°C) (Caterina et al. 1999), neuropeptides (Boels et al. 2001), and growth factors (Kanzaki et al. 1999), is proposed to play a role in proinflammatory degranulation events in mast cells (Stokes et al. 2004) and mechanosensation in vascular smooth muscle (Muraki et al. 2003).
Although TRPV channels mediate a range of physiological and sensory responses, they share a common structural architecture. Their general topology consists of an intracellular N- and C-terminal domain and six transmembrane helices (TM1 to TM6) with TM5, TM6, and the connecting loop forming the cation-conducting pore (Voets et al. 2004). The channels assemble as tetramers (Kedei et al. 2001) and in some cases form heteromeric channels (Hoenderop et al. 2003).
The intracellular N-terminal domain of TRPV channels contains multiple ankyrin (ANK) repeats. This is a common motif utilized in protein–protein recognition, mediating a diverse range of functions, including ion transport, cell–cell signaling, transcription, cell-cycle regulation, cytoskeletal maintenance, development, and inflammation (Mosavi et al. 2004).
Little is known about the function of ANK repeats in TRPV channels, although a role in cell surface trafficking has been proposed. An interaction between the ANK repeats of TRPV1 and two vesicular proteins, Syt IX and Snapin, has been identified, suggesting that the ANK repeats may localize the channel in vesicles that participate in SNARE-dependent exocytosis in excitable cells (Morenilla-Palao et al. 2004). Recently, Fas-associated factor 1, which may play a role in channel clearance, was shown to bind the TRPV1 ANK repeats and reduce the sensitivity of the channel to agonists (Kim et al. 2006). Phosphorylation sites important for channel regulation have also been mapped to the ANK repeats of TRPV1 (Bhave et al. 2002; Zhang et al. 2005).
More is being learned about the roles of TRPV channels in sensory transduction, but the structure of a TRPV channel has not yet been elucidated and the molecular mechanisms by which external stimuli translate into channel gating are largely unknown. To gain further insight into TRPV channel function, we report the 1.7 Å resolution crystal structure of the ANK repeat domain (ARD) of human TRPV2, the first structure from the TRPV channel family.
Results and Discussion
Overall structure of the TRPV2 ARD
The crystal structure of the human TRPV2 N-terminal fragment reveals six ANK repeats (Fig. 1A). As with canonical ANK repeats, each TRPV2 ANK repeat is defined by two antiparallel helices (α1 and α2) followed by a loop region linking to a β-hairpin structure (Fig. 1B). The loop and β-hairpin region are missing in the last ANK repeat (ANK6), which may be due to the early termination of the crystallization construct. The helix-loop-helix motifs stack on top of one another with the β-hairpin structures projecting out almost perpendicular to the helices. Each ANK repeat is rotated counterclockwise relative to the previous repeat, generating an elongated ARD structure with a slight left-handed curve and approximate dimensions of 70 Å × 30 Å × 30 Å.
Figure 1.
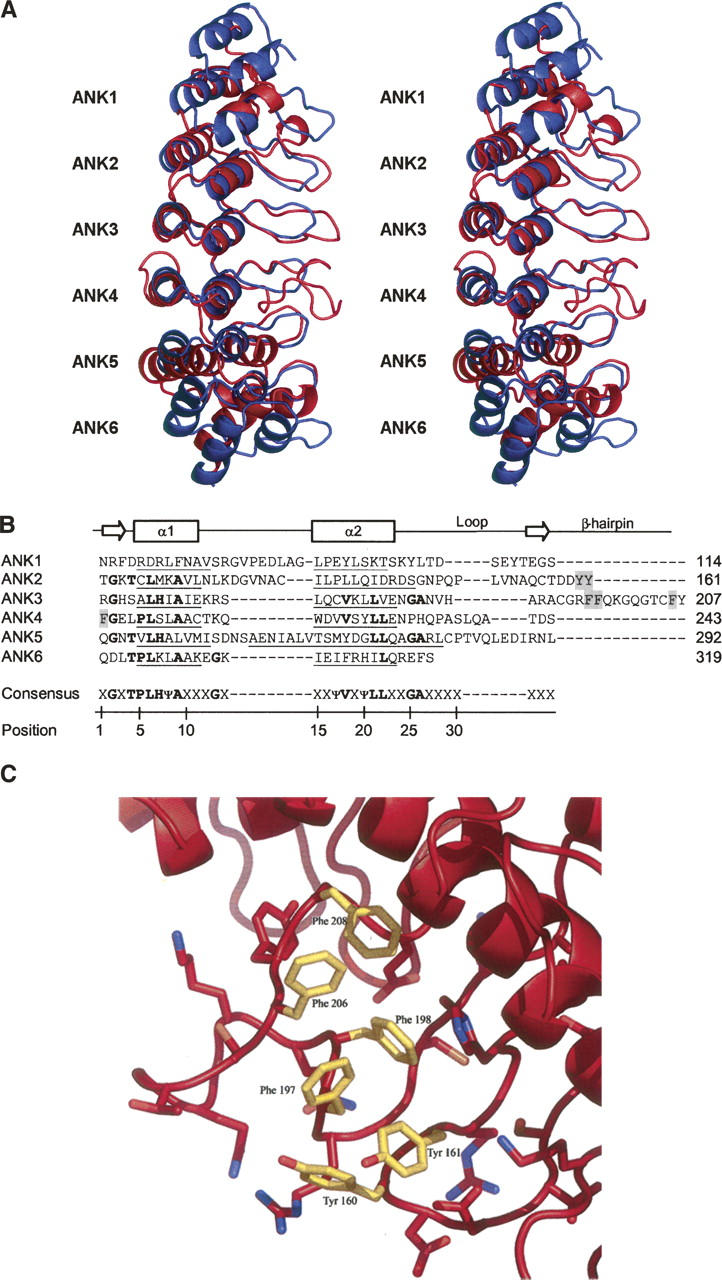
Ankyrin repeat domain (ARD) of human TRPV2. (A) Stereo ribbon representation of the human TRPV2 ARD superposed with a canonical (ankyrinR) ARD (Michaely et al. 2002) when viewed along the ankyrin groove of the first four repeats. TRPV2 is shown in red with each ankyrin repeat labeled ANK1 through ANK6, and ankyrinR is colored blue. Superposition is based on alignment of the helical Cα atoms for ANK1 through ANK4 of TRPV2 with the corresponding ankyrinR Cα atoms and results in a root mean square deviation of 2.6 Å. The two N- and C-terminal repeats of ankyrinR are not shown for clarity. (B) Structure-based sequence alignment of the TRPV2 ankyrin repeats with the ankyrin consensus sequence. α1 and α2 represent the two helices of each repeat with the exact limits of each helix underlined. The arrows show the beginning and end of the β-hairpin structure, and the residues of the hydrophobic patch (C) are shaded gray. In the consensus sequence, Ψ represents an aliphatic residue and X a nonconserved residue. (C) Hydrophobic patch in the β-hairpin regions of ANK2 and ANK3 with the exposed aromatic residues colored yellow.
Our crystal structure has two molecules in the asymmetric unit that associate in a head-to-tail orientation. It has been reported that TRPV ANK repeats are involved in channel subunit assembly (Erler et al. 2004; Arniges et al. 2006). However, the dimerization appears to be an artifact of crystallization as TRPV2 ARD behaves as a monomer in solution, as judged by size exclusion chromatography.
Unique features of the TRPV2 ARD
Insertions in the ANK repeats of reported ARD structures are not uncommon but are usually small, typically one to six residues, and confined to one or two repeats (Gorina and Pavletich 1996; Jacobs and Harrison 1998; Mandiyan et al. 1999; Michel et al. 2001; Padmanabhan et al. 2004). The TRPV2 ARD is unique, as every ANK repeat (with the exception of ANK6, which is not completely visible in our structure) contains insertions ranging from three to 16 residues (Fig. 1B).
The differences between the TRPV2 ARD and other reported ARD structures are clearly seen when superposing TRPV2 ARD with a canonical ARD, such as ANKR (Michaely et al. 2002), as shown in Figure 1A. Of note are the insertions between the helices of ANK1 and ANK2 and in the loop region following the helices, the extended β-hairpin structures (in particular that of ANK3), and the unusually long α2 helix of ANK5.
These insertions result in a novel kinked structure that has not yet been observed in other ARD structures. In canonical ARDs, the uniform arrangement of ANK repeats is maintained by a 2–3° rotation of each repeat with respect to the preceding repeat, regular spacing of adjacent helical pairs, and a continuous antiparallel β-sheet formed between adjacent β-hairpins. In TRPV2, main chain hydrogen bonding is maintained between the β-hairpin structures of ANK1, ANK2, and ANK3, but the large insertion in the β-hairpin region of ANK3 disrupts similar bonding between ANK3 and ANK4. Steric hindrance caused by the largely extended β-hairpin of ANK3 displaces the β-hairpin of ANK4 by ∼5 Å. This results in a distortion of the regular repeat stacking with ANK5 rotated by an additional ∼10° with respect to ANK4 (Fig. 1A).
Potential binding sites in the ARD
ARD–ligand complex structures demonstrate that ligand recognition almost exclusively occurs through a so-called ANK groove encompassing the entire length of the domain, comprising the β-hairpins, loop regions, and α1 helices (Mosavi et al. 2004). The ANK groove of TRPV2 with its large extended β-hairpins would provide a suitable surface for protein recognition. The kink between ANK4 and ANK5 and the large protruding insertion in the β-hairpin of ANK3 may function to create two separate binding sites in the ANK groove (Fig. 1A), a feature that is so far unique among reported ARD structures.
Another interesting feature of the ANK groove is an unusual hydrophobic patch, lined with aromatic residues Tyr160, Tyr161, Phe197, Phe198, Phe206, and Phe208, in the solvent-exposed β-hairpin regions of ANK2 and ANK3 (Fig. 1C).
TRPV2 ARD as a prototype for the TRPV channel family
Based on sequence alignment (see Supplemental Material) and topology predictions, all six ANK repeats are likely to be present in the six mammalian TRPV channels. The insertions observed in the TRPV2 ARD structure are conserved between TRPV1 through TRPV4, classifying them as a distinct subfamily of ARDs, whereas TRPV5 and TRPV6 form the shorter canonical repeats. In addition, the aromatic residues in the hydrophobic patch formed by the β-hairpin regions of ANK2 and ANK3 of TRPV2 are highly conserved only in TRPV1, TRPV3, and TRPV4 (see Supplemental Material). In TRPV1, phosphorylation of Tyr200 (equivalent to Tyr161 in TRPV2) in this hydrophobic patch has been shown to be critical for channel activation (Zhang et al. 2005). Interestingly, TRPV1 through TRPV4 display common functionalities such as temperature sensing, suggesting that the additional features observed in TRPV2 may be involved in a common mechanism for mediating such functions.
Summary
We present the first structure from the TRPV family that can be used as a template for other TRPV channel ARDs. The structure reveals a basic framework of six ANK repeats with numerous insertions throughout that generate several structural features distinguishing it from other known ARD structures. Solution studies indicate that the TRPV2 ARD is monomeric, suggesting that the ARD alone is not sufficient to assemble the tetramers of the full-length channel. However, this does not exclude a potential role in forming intramolecular interactions within the tetrameric channel. The ARD provides a large surface for protein recognition and is most likely involved in binding regulatory proteins. The determination of this first structural component of the TRPV channel should facilitate further investigation into the role of the ARD in sensory transduction.
Materials and methods
Expression and purification
The cDNA sequence corresponding to residues 1–319 from human TRPV2 was amplified by PCR and cloned into plasmid pMH4 (Lesley et al. 2002) using restriction sites PmlI and PacI. The resulting construct encodes an expression and purification tag (MGSDKIHHHHHH) at the N terminus of the protein. Protein was overexpressed in a selenomethionine-containing minimal media using the Escherichia coli methionine auxotrophic strain DL41. The cells were induced at 37°C with 0.15% (w/v) arabinose for 3 h. Lysozyme was added to the culture at the end of induction to a final concentration of 250 μg/mL. Bacteria were lysed by sonication after a freeze/thaw procedure in Lysis Buffer (20 mM Tris at pH 7.9, 500 mM NaCl, 5 mM imidazole, 0.5 mM Tris[2-carboxyethyl]phosphine hydrochloride [TCEP]), and the cell debris was pelleted by centrifugation at 3400g for 60 min. The soluble fraction was applied to a nickel-chelating resin (Amersham Biosciences) pre-equilibrated with Lysis Buffer. The resin was washed with 20 mM Tris (pH 7.9), 500 mM NaCl, 50 mM imidazole, and 0.5 mM TCEP, and the protein was eluted with 20 mM Tris (pH 7.9), 300 mM imidazole, and 0.5 mM TCEP. The eluate was buffer-exchanged into Buffer Q (20 mM Tris at pH 7.9, 5% [v/v] glycerol, 0.5 mM TCEP) containing 50 mM NaCl and was applied to a RESOURCE Q column (Amersham Biosciences) pre-equilibrated with the same buffer. The protein was eluted by using a linear gradient of 50–500 mM NaCl in Buffer Q. The molecular weight and oligomeric state of the protein were determined by using a 1.0 × 30 cm Superdex 200 column (Amersham Biosciences) in combination with static light scattering (Wyatt Technology). The mobile phase consisted of 20 mM Tris (pH 7.9), 150 mM NaCl, and 0.02% (w/v) sodium azide.
Crystallization and data collection
Selenomethionine incorporated human TRPV2 (1–319) was buffer-exchanged into 20 mM Tris (pH 7.9), 150 mM NaCl, and 0.5 mM TCEP and concentrated to 10 mg/mL by centrifugal ultrafiltration (Millipore). Crystals were grown at 20°C by nano-drop vapor diffusion using 0.1 M HEPES (pH 7.5) and 1.5 M Li2SO4 as a precipitant. The asymmetric unit contains two molecules with a solvent content of 48%. Diffraction data were collected from crystals cooled to 100 K after addition of 20% (v/v) glycerol as a cryoprotectant. Single wavelength anomalous dispersion (SAD) data from a single selenomethionine-substituted crystal were collected at the selenium K-edge (peak) at Beamline 5.0.2 (Advanced Light Source). All data were processed with HKL2000 package (Otwinowski 1997).
Phasing, model building, and refinement
The scaled intensity SAD data were input into SOLVE (Terwilliger and Berendzen 1999), and the positions of six, out of an expected 10, selenium atoms in the asymmetric unit were found. Phases were calculated to 2.5 Å by SOLVE with a figure of merit of 0.31, and subsequent solvent flattening with RESOLVE (Terwilliger and Berendzen 1999) improved the figure of merit to 0.67. An atomic model (∼75% complete) was built with RESOLVE and completed manually by using COOT (Emsley and Cowtan 2004). A second higher resolution data set of a selenomethionine-substituted crystal was collected to 1.7 Å resolution, which was used to refine the model within the CCP4 program suite (CCP4 1994). All data collection and refinement statistics are shown in Table 1. The final model has two molecules in the asymmetric unit with residues 71–318 in molecule A (Gln202 is omitted) and residues 69–319 in molecule B. SDS-PAGE analysis of several dissolved crystals indicates that the first ∼50 amino acids are truncated during the crystallization process. A total of 90.2% of the main chain torsion angles of the nonglycine residues lie in the most favored regions of the Ramachandran plot with 9.4% in the additionally allowed regions as evaluated by PROCHECK (Laskowski et al. 1993). Glu210 in both molecules is in the disallowed region but fits well to the electron density. The structural coordinates have been deposited in the Protein Data Bank (accession code 2F37). All figures representing the TRPV2 model were prepared with PyMOL (DeLano 2002).
Table 1.
Data collection and refinement statistics
Electronic supplemental material
Supplemental material provides sequence alignment of the N terminus of human TRPV1 through TRPV6. ANK repeats are labeled ANK1 through ANK6, as seen in the crystal structure of TRPV2. The TRPV2 residues in each helical pair of the repeats are underlined with residues involved in the hydrophobic patch shaded gray. TRPV residues identical to the consensus ANK repeat sequence (as in Fig. 1B) are shown in bold. ClustalW was used to generate the sequence alignment.
Note added in proof
While this manuscript was in preparation, the structure of the rat TRPV2 ankyrin repeat domain has been published: Jin, X., Touhey, J., and Gaudet, R. 2006. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. J. Biol. Chem., published June 29, 2006 as doi:10.1074/jbc.C600153200.
Acknowledgments
We thank Heath Klock and Justin Haugen for help with protein expression and purification and the staff at the Advanced Light Source, BL502/503 (Berkeley, CA), for beamline assistance.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Andreas Kreusch, Genomics Institute of the Novartis Research Foundation, 10675 John Jay Hopkins Drive, San Diego, CA 92121; e-mail: kreusch@gnf.org; fax: (858) 812-1746.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062357206.
References
- Arniges M., Fernandez-Fernandez J.M., Albrecht N., Schaefer M., Valverde M.A. 2006. Human TRPV4 channel splice variants revealed a key role of ankyrin domains in multimerization and trafficking. J. Biol. Chem. 281: 1580–1586. [DOI] [PubMed] [Google Scholar]
- Bhave G., Zhu W., Wang H., Brasier D.J., Oxford G.S., Gereau R.W.T. 2002. cAMP-dependent protein kinase regulates desensitization of the capsaicin receptor (VR1) by direct phosphorylation. Neuron 35: 721–731. [DOI] [PubMed] [Google Scholar]
- Boels K., Glassmeier G., Herrmann D., Riedel I.B., Hampe W., Kojima I., Schwarz J.R., Schaller H.C. 2001. The neuropeptide head activator induces activation and translocation of the growth-factor-regulated Ca2+-permeable channel GRC. J. Cell Sci. 114: 3599–3606. [DOI] [PubMed] [Google Scholar]
- Caterina M.J. and Julius D. 2001. The vanilloid receptor: A molecular gateway to the pain pathway. Annu. Rev. Neurosci. 24: 487–517. [DOI] [PubMed] [Google Scholar]
- Caterina M.J., Rosen T.A., Tominaga M., Brake A.J., Julius D. 1999. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature 398: 436–441. [DOI] [PubMed] [Google Scholar]
- Clapham D.E. 2003. TRP channels as cellular sensors. Nature 426: 517–524. [DOI] [PubMed] [Google Scholar]
- . Collaborative Computational Project Number 4 (CCP4). 1994. The CCP4 Suite: Programs for crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. In The PyMOL Molecular Graphics System . 2002. Delano Scient ific, San Carlos, CA.
- Emsley P. and Cowtan K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Erler I., Hirnet D., Wissenbach U., Flockerzi V., Niemeyer B.A. 2004. Ca2+-selective transient receptor potential V channel architecture and function require a specific ankyrin repeat. J. Biol. Chem. 279: 34456–34463. [DOI] [PubMed] [Google Scholar]
- Gorina S. and Pavletich N.P. 1996. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science 274: 1001–1005. [DOI] [PubMed] [Google Scholar]
- Hoenderop J.G., Voets T., Hoefs S., Weidema F., Prenen J., Nilius B., Bindels R.J. 2003. Homo- and heterotetrameric architecture of the epithelial Ca2+ channels TRPV5 and TRPV6. EMBO J. 22: 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M.D. and Harrison S.C. 1998. Structure of an IκBα/NF-κB complex. Cell 95: 749–758. [DOI] [PubMed] [Google Scholar]
- Kanzaki M., Zhang Y.Q., Mashima H., Li L., Shibata H., Kojima I. 1999. Translocation of a calcium-permeable cation channel induced by insulin-like growth factor-I. Nat. Cell Biol. 1: 165–170. [DOI] [PubMed] [Google Scholar]
- Kedei N., Szabo T., Lile J.D., Treanor J.J., Olah Z., Iadarola M.J., Blumberg P.M. 2001. Analysis of the native quaternary structure of vanilloid receptor 1. J. Biol. Chem. 276: 28613–28619. [DOI] [PubMed] [Google Scholar]
- Kim S., Kang C., Shin C.Y., Hwang S.W., Yang Y.D., Shim W.S., Park M.Y., Kim E., Kim M., Kim B.M. et al. 2006. TRPV1 recapitulates native capsaicin receptor in sensory neurons in association with Fas-associated factor 1. J. Neurosci. 26: 2403–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.J., MacArthur N.W., Moss D.S., Thornton J.M. 1993. PROCHECK: A program to check stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–290. [Google Scholar]
- Lesley S.A., Kuhn P., Godzik A., Deacon A.M., Mathews I., Kreusch A., Spraggon G., Klock H.E., McMullan D., Shin T. et al. 2002. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. 99: 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandiyan V., Andreev J., Schlessinger J., Hubbard S.R. 1999. Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein β. EMBO J. 18: 6890–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaely P., Tomchick D.R., Machius M., Anderson R.G. 2002. Crystal structure of a 12 ANK repeat stack from human ankyrinR. EMBO J. 21: 6387–6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Soler-Lopez M., Petosa C., Cramer P., Siebenlist U., Muller C.W. 2001. Crystal structure of the ankyrin repeat domain of Bcl-3: A unique member of the IκB protein family. EMBO J. 20: 6180–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morenilla-Palao C., Planells-Cases R., Garcia-Sanz N., Ferrer-Montiel A. 2004. Regulated exocytosis contributes to protein kinase C potentiation of vanilloid receptor activity. J. Biol. Chem. 279: 25665–25672. [DOI] [PubMed] [Google Scholar]
- Mosavi L.K., Cammett T.J., Desrosiers D.C., Peng Z.Y. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13: 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki K., Iwata Y., Katanosaka Y., Ito T., Ohya S., Shigekawa M., Imaizumi Y. 2003. TRPV2 is a component of osmotically sensitive cation channels in murine aortic myocytes. Circ. Res. 93: 829–838. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z.M.W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Padmanabhan B., Adachi N., Kataoka K., Horikoshi M. 2004. Crystal structure of the homolog of the oncoprotein gankyrin, an interactor of Rb and CDK4/6. J. Biol. Chem. 279: 1546–1552. [DOI] [PubMed] [Google Scholar]
- Patapoutian A., Peier A.M., Story G.M., Viswanath V. 2003. ThermoTRP channels and beyond: Mechanisms of temperature sensation. Nat. Rev. Neurosci. 4: 529–539. [DOI] [PubMed] [Google Scholar]
- Stokes A.J., Shimoda L.M., Koblan-Huberson M., Adra C.N., Turner H. 2004. A TRPV2-PKA signaling module for transduction of physical stimuli in mast cells. J. Exp. Med. 200: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55: 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voets T., Janssens A., Droogmans G., Nilius B. 2004. Outer pore architecture of a Ca2+-selective TRP channel. J. Biol. Chem. 279: 15223–15230. [DOI] [PubMed] [Google Scholar]
- Zhang X., Huang J., McNaughton P.A. 2005. NGF rapidly increases membrane expression of TRPV1 heat-gated ion channels. EMBO J. 24: 4211–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]