Abstract
GOPC (Golgi-associated PDZ and coiled-coil motif-containing protein) represents a PDZ domain-containing protein associated with the Golgi apparatus, which plays important roles in vesicular trafficking in secretory and endocytic pathways. GOPC interacts with many other proteins, such as the Wnt receptors Frizzled 8 and neuroligin via its PDZ domain. Neuroligin is a neural cell-adhesion molecule of the post-synapse, which binds to the presynapse molecule neurexin to form a heterotypic intercellular junction. Here we report the solution structure of the GOPC PDZ domain by NMR. Our results show that it is a canonical class I PDZ domain, which contains two α-helices and six β-strands. Using chemical shift perturbation experiments, we further studied the binding properties of the GOPC PDZ domain with the C-terminal motif of neuroligin. The observations showed that the ensemble of the interaction belongs to fast exchange with low affinity. The 3D model of the GOPC PDZ domain/neuroligin C-terminal peptide complex was constructed with the aid of the molecular dynamics simulation method. Our discoveries provide insight into the specific interaction of the GOPC PDZ domain with the C-terminal peptide of Nlg and also provide a general insight about the possible binding mode of the interaction of Nlg with other PDZ domain-containing proteins.
Keywords: GOPC PDZ domain, NMR spectroscopy, C-terminal motif of Nlg, titration, dissociation constant, molecular dynamics simulation
GOPC (Golgi-associated PDZ and coiled-coil motif-containing protein) represents a PDZ domain containing protein associated with the Golgi apparatus (Charest et al. 2001; Yao et al. 2001). GOPC is expressed in all tissues analyzed in mice and in humans, suggesting that it plays a housekeeping role and takes part in multiple functions such as in vesicular trafficking in secretory and endocytic pathways (Charest et al. 2001; Cheng et al. 2002). A mouse GOPC is involved in acrosome formation by facilitating protein cargo transport from the Golgi apparatus. The GOPC-deficient mice showed complete lack of acrosomes due to the failure of vesicle transport from the Golgi apparatus (Yao et al. 2002). GOPC interacts specifically with golgin-160, a peripheral membrane protein belonging to the golgin family of Golgi-localized proteins, indicating that GOPC participates in golgin-160-dependent trafficking of cargo (Hicks and Machamer 2005). GOPC modulates intracellular trafficking via binding of its coiled-coil domain to the small GTPase Tc10, a member of the Rho-GTPase family (Neudauer et al. 2001). Activation of Tc10 leads to increased transport of GOPC and its associated cystic fibrosis transmembrane conductance regulator from the Golgi to the plasma membrane, showing that the Tc10/GOPC system determines the distribution of specific membrane proteins between the plasma membrane and intracellular compartments (Cheng et al. 2005).
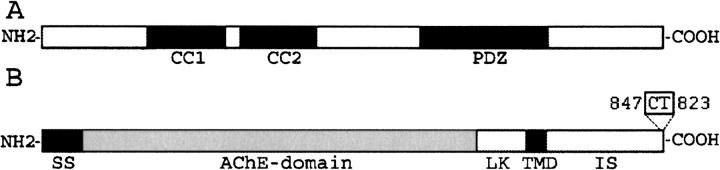
GOPC possesses two coiled-coil domains and a single PDZ domain (Fig. 1A). The coiled-coil domain could bind to plasma membrane protein syntaxin 6, which suggested a relationship between GOPC with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor-mediated membrane recognition and fusion (Charest et al. 2001). Previous work showed that the PDZ domain plays an important role in GOPC function (Cheng et al. 2002, 2004; Yue et al. 2002; Gentzsch et al. 2003; Ives et al. 2004; Wente et al. 2005). Many interactions of GOPC with other proteins, such as the Wnt receptors Frizzled 5 and 8 (Yao et al. 2001), δ2 glutamate receptor (Yue et al. 2002), and neuroligin (Nlg) (Meyer et al. 2004), occur in a PDZ domain-dependent manner. Here we focused on the interaction of GOPC with Nlg via its PDZ domain. At the same time, we also looked at its interaction with the C-terminal motif of Frizzled 8.
Figure 1.
Schematic representation of hGOPC and hNlg. (A) Structure of GOPC. CC, coiled-coil domain; PDZ, PDZ domain. (B) Structure of Nlg. SS, signal sequence; AchE-domain, acetylcholinesterase-homology domain; LK, linker domain; TMD, transmembrane domain; IS, intracellular sequence; CT, C terminus.
Nlg is a post-synaptic cell-adhesion molecule of synapses that contain five distinct domains: a cleaved N-terminal signal sequence, a large extracellular acetylcholinesterase homology domain (but enzymatically inactive), a linker domain resembling an O-glycosylation cassette, a highly conserved single transmembrane domain, and a short intracellular sequence with a highly conserved C terminus (Fig. 1B; Ushkaryov et al. 1992, 1994; Hata et al. 1996; Ichtchenko et al. 1996; Irie et al. 1997; Sugita et al. 2001). The extracellular domain of Nlg tightly binds to the extracellular domain of β-neurexins (also cell-surface proteins) to form a heterotypic intercellular junction (regulated by alternative splicing). The two intracellular sides of the Nlg/β-neurexin junction are assembled by similar but distinct PDZ domain-containing proteins (Sugita et al. 2001). The Nlg/β-neurexin interaction could increase the lifetime of the nascent synapse and induce further maturation of the synapse by signaling through their PDZ domain interactions (Scheiffele et al. 2000; Bolliger et al. 2001). Nlgs control the formation and functional balance of excitatory and inhibitory synapses and regulate the excitatory:inhibitory synaptic ratio through interaction with their post-synaptic binding partner, such as PSD-95 (Ichtchenko et al. 1995; Kurschner et al. 1998; Levinson et al. 2005; Nam and Chen 2005).
Recently, by using a yeast two-hybrid screen, GOPC was identified as a putative binding partner of Nlgs (Meyer et al. 2004). Although many biological functions have been elucidated for GOPC, the structural basis of GOPC and its interaction with Nlg remain unclear.
Understanding the function and specificity of GOPC requires detailed knowledge of its structure. Here we report the solution structure of the GOPC PDZ domain and describe the characterization of its interaction with the C-terminal motif (SHSTTRV) of hNlg-1 (Nlg (817–823)) using nuclear magnetic resonance (NMR). The data revealed that the GOPC PDZ domain is a typical class I PDZ domain. The interface of the GOPC PDZ domain interaction with the C terminus of Nlg was identified. The GOPC PDZ domain recognized the C-terminal sequence of Nlg with 10−4 M affinity. A model of complex of the GOPC PDZ domain with the C-terminal peptide of Nlg was established by molecular dynamics (MD) simulation from the solution structure of the GOPC PDZ domain. This model not only gives a better explanation for the NMR chemical shift perturbation experiment results but also provides a general picture of how the post-synaptic cell-adhesion molecule Nlg recognizes its partner PDZ domain-containing proteins.
Results
NMR structure determination of the GOPC PDZ domain
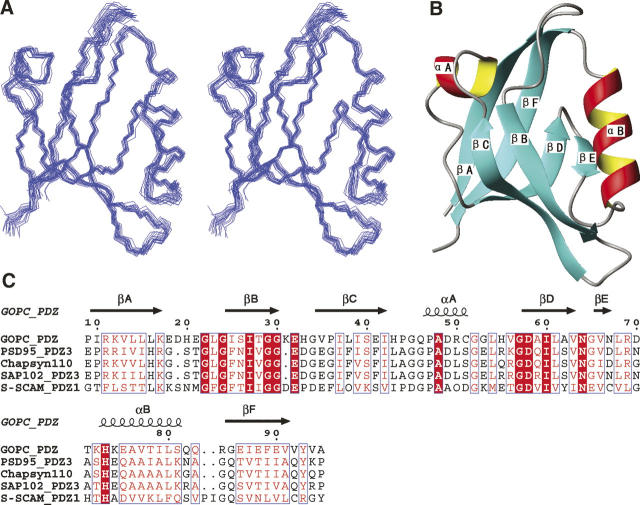
The complete backbone and >95% of the side-chain 1H resonances were assigned. A 15N–1H HSQC spectrum of 15N-labeled GOPC PDZ domain with corresponding residue assignments indicated is shown in Supplemental Figure S1. A total of 1645 experimentally derived distances and dihedral angle restraints was used in the structure calculations, and the 20 lowest-energy structures have an RMSD for backbone atoms of residues 9–95 of 0.46 Å (structural statistics summarized in Table 1). The N terminus (residues 1–8) and the C-terminal His tag (-LEHHHHHH) were not used in RMSD calculations. Figure 2A shows a stereo view of the superposition of a family of the 20 lowest-energy NMR structures, selected from 200 accepted structures by requiring no NOE violation >0.5 Å and no dihedral angle violations >5°.
Table 1.
Structural statistics for the family of 20 lowest-energy structures
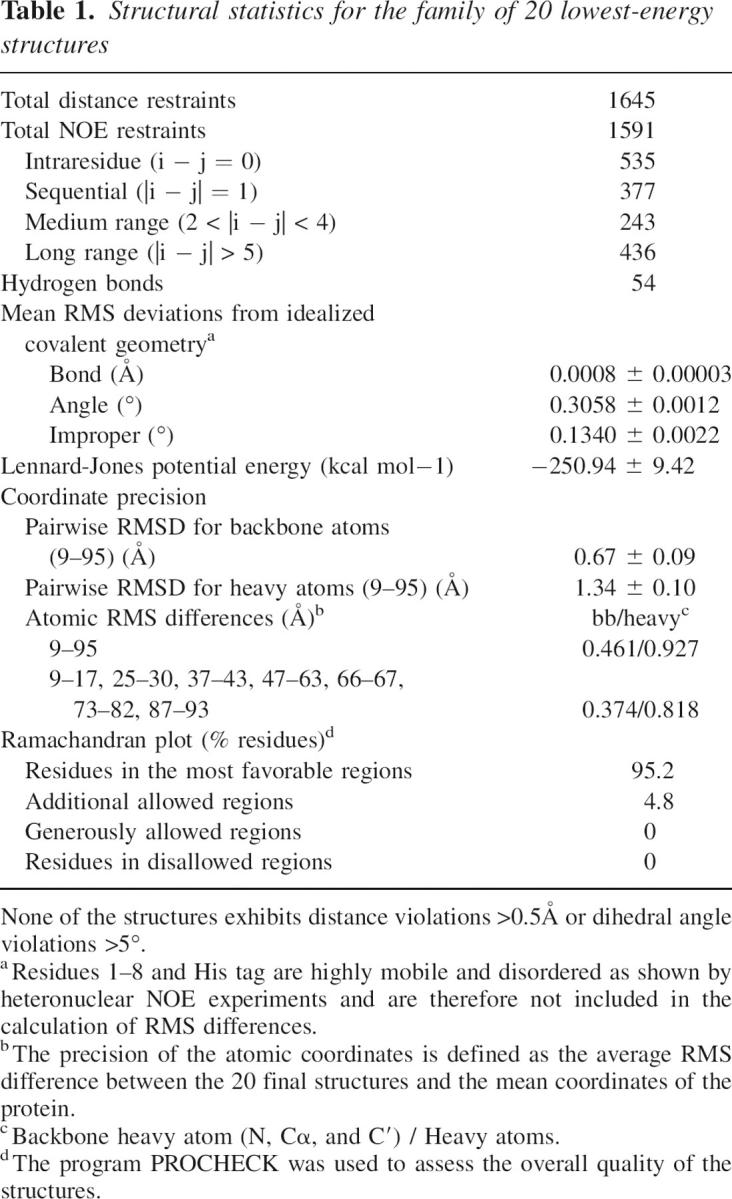
Figure 2.
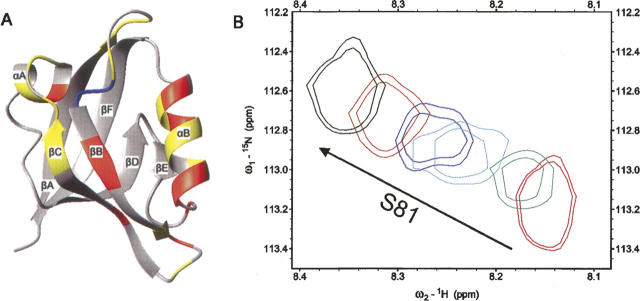
Structure of the GOPC PDZ domain determined by NMR spectroscopy. (A) Stereo view of the final 20 lowest-energy structures of the GOPC PDZ domain, superimposed using backbone atoms (N, Cα, and C′). (B) One representative structure of the GOPC PDZ domain with the secondary structure elements highlighted. The secondary structure elements are labeled following the scheme used in the crystal structure of PSD-95 PDZ3. Both figures were generated using MOLMOL (Koradi et al. 1996). (C) Amino acid sequence alignment of selected PDZ domains. The secondary structure of the GOPC PDZ domain determined is also included at the top of the sequence. Residues identical in all five sequences are shown in red columns, and conserved residues are shown in red letters. The sequence alignment was made using ClustalW.
The final ensemble of 20 refined structures has been deposited in the Protein Data Bank (PDB) under accession code 2DC2 and the chemical shifts have been deposited in the BMRB (accession code 7072).
Description of the structure
The NMR-derived tertiary structure of the GOPC PDZ domain is a canonical PDZ fold that contains two α-helices (αA and αB) and six β-strands (from βA to βF) (Fig. 2A,B). As part of the tertiary structure, the β-strands, βA/βF, βB/βC, and βD/βE, form antiparallel β-sheets, with βA, βB, βC, and αA forming one face of the shell-shaped structure and βD, βE, βF, and αB forming the other face. As in other PDZ domains, a deep binding groove for substrates is formed by strand βB, its N-terminal loop, and αB (see Figs. 2B, 3).
Figure 3.
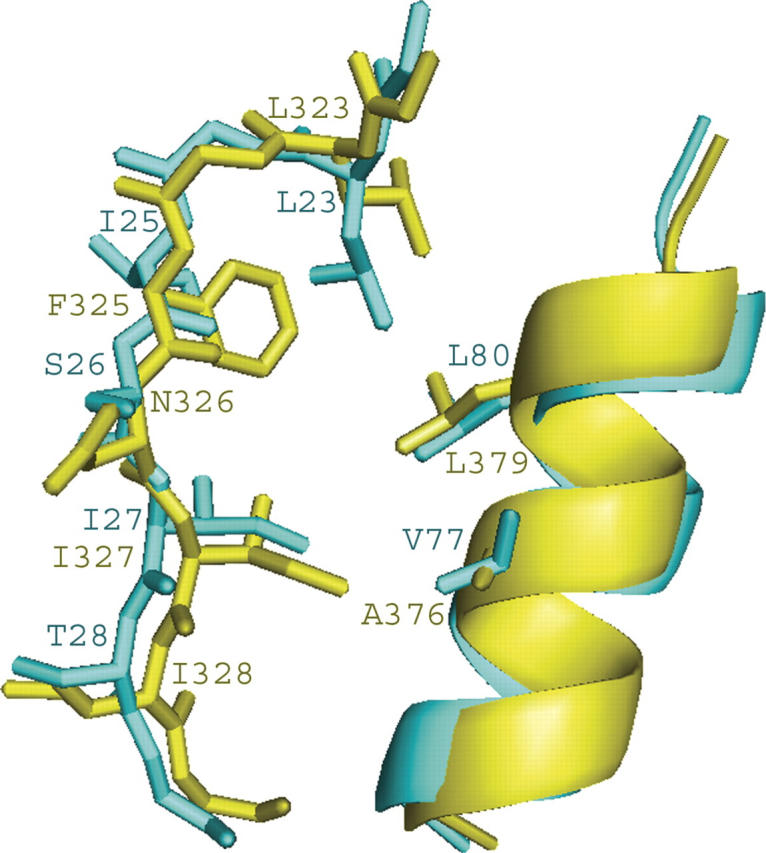
Comparison of the αB/βB groove conformation. GOPC PDZ domain (cyan), PSD-95 PDZ3 (yellow). The program MOLMOL (Koradi et al. 1996) was used to generate the figure.
The dynamic properties of GOPC PDZ domain were probed by measuring 15N relaxation parameters at 303 K. Longitudinal T1 and transversal T2 relaxation times as well as 1H–15N heteronuclear NOE values were obtained for a total of 76 backbone amide protons (88% of all possible) (see Supplemental Fig. S2). The average 15N–1H NOE value of 0.72 (±0.05) indicates that most regions of GOPC PDZ domain are relatively rigid, which is consistent with the narrow distribution of conformers in the calculated ensemble. Squared order parameter (S2), the effective correlation time for fast internal motions (τe), and the refined τm value were obtained by FAST-ModelFree program v1.0 (Cole and Loria 2003). A global τm of 7.77 nanoseconds was given while the order parameters S2 were obtained for the 63 remaining residues. The mean value of the order parameter for residues is 0.89 and most of the order parameters are >0.8 (S2 > 0.8) for residues located in α-helices and β-strands. However, there are several residues giving smaller order parameters, including R84, G85, and E86. The τe values in this region are higher than the average value, indicating that these residues have greater flexibility on the picosecond timescale. Coincidentally, they are located on loops or turns linking secondary structures.
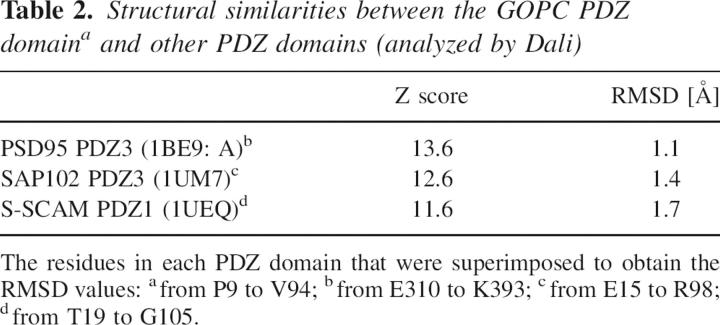
Dali analyses (Holm and Sander 1993) revealed that the structure of the GOPC PDZ domain is closest to that of PDZ3 of PSD95 with an RMSD value of 1.1 Å for the entire PDZ domain. The sequence identity and similarity between these two PDZ domains are 42% and 56%, respectively.
A previously reported study revealed that Nlg could interact with the GOPC PDZ domain and many other PDZ domain-containing proteins screened by the yeast two-hybrid system (Meyer et al. 2004). It is meaningful to see the structural similarities and differences among these proteins. Our GOPC PDZ domain structure (entry 2DC2) was superposed with other structures, including PSD95 PDZ3 (1BE9: A), SAP102 PDZ3 (1UM7), and S-SCAM PDZ1 (1UEQ), all of which are partners of Nlg. The superposition shows that those partners have similar tertiary structures (see Supplemental Fig. S3). Table 2 gives the Z score and RMSD between these structures.
Table 2.
Structural similarities between the GOPC PDZ domaina and other PDZ domains (analyzed by Dali)
The conformation of the binding cleft of GOPC PDZ domain and the other partners of Nlg were compared. Although the GOPC PDZ domain and the other partners of Nlg are similar, the difference was found in GLGF repeats (Fig. 2C), which are located at the βA/βB connecting loop containing the highly conserved sequence Gly–Leu–Gly–Phe in class I and class III PDZ domains. These residues, which occur at the top of the peptide-binding cleft before strand βB, turn out to play an important functional role in binding the C-terminal carboxylate group of the peptide. Therefore, the loop is referred to as the carboxylate-binding loop. In the GOPC PDZ domain, the carboxylate-binding loop is replaced by the sequence GLGI. By a comparison of the conformation of the αB/βB groove of the GOPC PDZ domain with that of PSD-95 PDZ3 (see Fig. 3), it is found that the side chain of the fourth amino acid in the GLGI motif is pointed toward the interior of the protein and is invariably hydrophobic despite the difference in sequence. Thus the structure of the binding site is not altered by this replacement. From Figure 3, it has also been found that the orientations of the side chains of amino acids that are located at the binding cleft besides the GLGF repeats are the same. These results imply that the C terminus of Nlg interaction with its partners may be using a similar pattern as with the GOPC PDZ domain.
The binding interface of the C terminus of Nlg on the GOPC PDZ domain
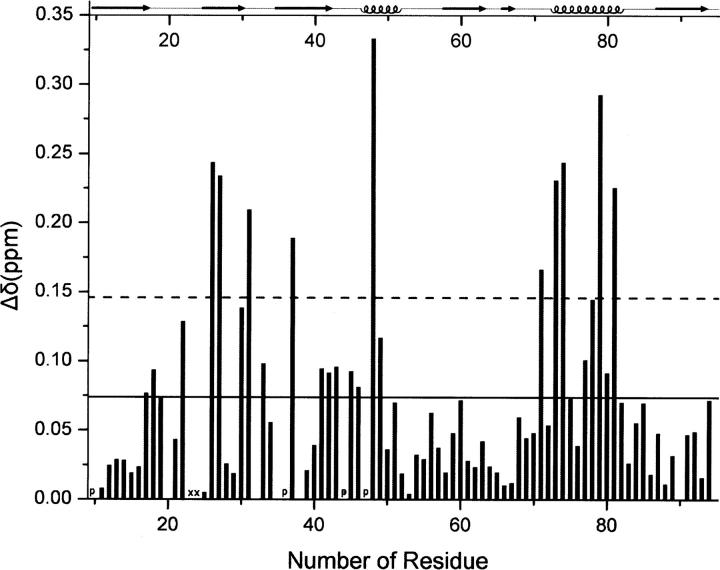
The residues that are involved in the interaction with the peptide were identified by NMR chemical shift mapping during titration of the protein with peptide. The chemical shift changes (Δδppm) of the backbone amide, between the free and the bound state, were plotted versus the GOPC PDZ domain sequence number for the final point in each titration (Fig. 4). There were 10 residues perturbed significantly whose chemical shift changes were greater than the sum of the mean and standard deviations during titration of the Nlg (817–823). Especially, resonances of Leu23 and Gly24 disappeared when the titration began and never re-emerged. Besides the residues with significant changes, the other residues whose chemical shift changes were larger than the mean value also were found, including Lys17, Glu18, Asp19, Gly22, Gly30, His33, Glu41, Ile42, His 43, Gly45, Gln46, Asp49, Val77, Thr78, and Leu80. We found that most perturbed residues clustered in the βB, βC, and αB regions, which is consistent with the canonical ligand-binding site of the PDZ domain. Taken together, the NMR results indicate that the peptide binds to the GOPC PDZ domain in the canonical fashion, with the peptidic ligand adopting an extended, β-strand-like conformation and occupying the cleft between αB and βB (with an antiparallel arrangement with the βB strand). Perturbation of Lys31 and His33 (both located in the βB/βC loop) indicates that the βB/βC loops of the PDZ domain might be involved in ligand interaction.
Figure 4.
Histogram showing differences in chemical shifts in the free GOPC PDZ domain and its complex with C-terminal peptide from Nlg. The mean value is shown as a continuous line; the mean value plus 1 standard deviation, as a broken line. P marks proline residues; X, missing resonances that are not visible in the HSQC spectra.
C terminus of Nlg binds to the GOPC PDZ domain with a low affinity
The spectral changes induced by the interaction of the peptide with the PDZ domain are typical for fast exchange on the NMR timescale (except for Leu23 and Gly24, they might belong to intermediate exchange). No slow exchange was observed. With increasing concentrations of Nlg (817–823), the amide proton and nitrogen resonances of affected residues shifted gradually from apo states to final binding states (Fig. 5B shows this change of chemical shift of Ser81, one of significant residues), indicating that the ensemble of the binary complexes was in fast exchange on the NMR timescale. The dissociation constant of the interaction between the GOPC PDZ domain and Nlg was obtained by monitoring the change in resonance chemical shifts of the residues that had significant chemical shift changes (except for A48, which is located out of the βB, βC, and αB regions and has poor “goodness of fit”) as a function of peptide concentration during titration with Nlg (see Supplemental Table S1). The chemical shift titration curves were fit to a two-state equilibrium model. The average value for KD is 260 (±86) μM calculated using Equations 2 and 3 in Materials and Methods. The error in the measured KD is quoted as the standard error of the fit. For accurate measurements, it is required that the KD be no more than 1 order of magnitude larger/smaller than the protein concentration, although dissociation constants 1 order of magnitude lower than the protein concentration can readily be distinguished with this method (Hu et al. 2004). In our chemical shift titration, the total concentration of the GOPC PDZ domain is 500 μM while the average value for KD is 260 (±86) μM. That is, both orders are the same, showing the reliability of our results.
Figure 5.
Results of the chemical shift perturbation. (A) Ribbon diagram of the chemical shift changes of the GOPC PDZ domain on titration with the C-terminal peptides of Nlg. The residues showing chemical shift changes larger than the mean value plus 1 standard deviation caused by addition of peptide are colored orange. The residues showing chemical shift changes larger than the mean value are colored yellow. Blue represents missing assignment; gray, otherwise. The figure was prepared using the program MOLMOL (Koradi et al. 1996). (B) 1H–15N HSQC spectra illustrating the significant chemical shift change of Ser81 on titration with the C-terminal peptides of Nlg. Red, green, cyan, blue, gold, and black are defined as molar ratio of the GOPC PDZ domain vs. Nlg peptide at 1:0, 1:0.3, 1:0.6, 1:0.9, 1:1.5, and 1:3.0, respectively.
Model of the GOPC PDZ domain/C-terminal peptide of Nlg complex
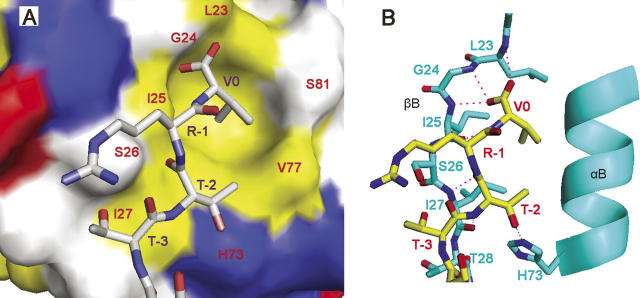
Due to the fast exchange between GOPC PDZ and the C-terminal peptide of Nlg, it is difficult to solve the structure of the complex directly by NMR. We employed the crystal structure of the complex of PSD95 PDZ3 with the peptide TKNYKQTSV (PDB ID 1BE9) as a template and got the 3D models of GOPC PDZ–Nlg complex by molecular dynamics (MD) simulation (Fig. 6A,B).
Figure 6.
Nlg-binding site on the surface of the GOPC PDZ domain. (A) Surface conformation of the GOPC PDZ domain interaction with C-terminal peptide of Nlg. The hydrophobic amino acid residues are drawn in yellow; the positively charged residues, in blue; the negatively charged residues, in red; and the uncharged polar residues, in white. (B) Intermolecular hydrogen bonds obtained from chemical shift perturbation and MD simulation are represented by a dotted pink line. The program PyMOL (http://www.delanoscientific.com) was used to generate the figure.
Inspection of the complex model reveals seven possible intermolecular hydrogen bonds between the GOPC PDZ domain and Nlg (Fig. 6B). Leu23, Gly24, and Ile25 backbone amides of the GOPC PDZ domain can form a hydrogen bond network with the C-terminal STTRV motif of Nlg. The Ile25 backbone carboxyl group pairs with V0 backbone amide. Ile27 carboxyl and amide backbone groups pair with the −2 position (T−2) backbone amide and carboxyl group, respectively. The GOPC PDZ domain side chains of Leu23, Gly24, and Ile25 can form a hydrophobic cluster with V0 of Nlg. His73 can, in principle, form a hydrogen bond with the side chain of T−2. V77 forms a polar interaction with the −2 position (T−2). This model can explain the results of chemical shift perturbation experiments.
In the canonical PDZ domain, the binding specificity is critically determined by the interaction of the first residue of αB helix with the side chain of the −2 residue of the ligand (Songyang et al. 1997). Class I PDZ interactions are characterized by hydroxyl groups (Ser or Thr) at the −2 position of the peptide ligand and His at the first position of αB helix of the PDZ domain. Amino acid sequence analysis reveals that a histidine (His73) takes the start position of αB in the GOPC PDZ domain (Fig. 2C), which indicates that the GOPC PDZ domain is a canonical class I PDZ domain. This is also confirmed by NMR titrations with peptidic ligand and is consistent with our model.
The binding interface of the C terminus of Frizzled 8 on the GOPC PDZ domain
To understand more about the function of the GOPC PDZ domain, we also analyzed the PDZ domain interaction with the C-terminal peptide (KQMPLSQV) of hFrizzled 8 (687–694) using the same titration methods as in the above section. The frizzled gene is evolutionally conserved in a wide variety of organisms and the Frizzled is implicated in Drosophila development (Vinson et al. 1989; Strutt 2001). A previous study showed that the PDZ domain of GOPC could interact with the C-terminal motif of mouse Frizzled and that GOPC might play a role in vesicle transport of Frizzled from the Golgi apparatus to the plasma membrane (Yao et al. 2001). With our titration with the peptide, most resonances of interaction residues broadened markedly as the ligand concentration increased and became undetectable at around [ST]/[PT] = 2. Signals reappeared at high ligand concentration and may have shifted somewhat as the concentration was further increasing. Thus it belonged to intermediate exchange and we could not fit the dissociation constant accurately. Significant chemical shift changes were observed for residues Ser26, Thr28, Gly30, Lys31, Ile37, Leu38, Glu41, Ala48, Lys74, Val77, Ile79, and Ser81. Resonances of Leu23, Gly24, Ile27, and Gly29 disappeared with titration with the peptide. Other changes (larger than the mean value but smaller than significant changes) also were found, including residues Glu18, Gly22, Ile25, His33, Ile42, Leu68, His73, and Thr78. Thus the interaction interface between the PDZ domain and the C-terminal peptide of the Frizzled 8 is also located in the βB, βC, and αB regions (see Supplemental Figs. S4, S5).
Discussion
In this paper, we report the solution structure of the GOPC PDZ domain, which possesses six β-strands and two α-helices. Our results demonstrate that the GOPC PDZ domain is a canonical class I PDZ domain, which is consistent with the previous study (Piserchio et al. 2005). Structural comparison with other Nlg binding PDZ domains showed that the GOPC PDZ domain has similar tertiary structure with other canonical I PDZ domains. Especially, there are the same orientations of the side chains of the amino acids located at the binding groove (3), indicating that they perhaps have similar interaction mode with their partners.
We performed the relaxation experiments at 303 K (see Supplemental Fig. S2). Our relaxation experiments showed that the averages of T2 and T1 are 100 msec and 516 msec at 303 K, respectively. These results indicated that the GOPC PDZ domain might be not aggregated or that the aggregation is very little at 303 K. We also measured the relaxation times of the PDZ domain at 293 K (data not shown). Interestingly, the average value of T2 calculated is 67 msec, which is similar to that of Piserchio et al. (2005). The investigators performed these at 298 K (25°C) and suggested that the protein is partially aggregated according to the correlation time (10.5 nsec, calculated from their relaxation experiments). Taken together, it is possible that there is a balance between monomer and aggregation of the GOPC PDZ domain, and the shift of balance depends on the temperate.
The binding property of the GOPC PDZ domain with the C-terminal peptide of Nlg was characterized by the chemical shift perturbation experiments. The results indicate that the C-terminal peptide of Nlg extends in the cleft between the βB strand and the αB helix and is antiparallel with βB strand as an additional strand, as most PDZ-binding ligands do. The ensemble of the binary complexes belongs to fast exchange on the NMR timescale. The dissociation constants were calculated according to titrating dynamics. The results show that the GOPC PDZ domain binds to the C-terminal peptide of Nlg with 10−4 M affinity. Although, in most cases, interaction between a PDZ domain and its target is constitutive, with a binding affinity of 1–10 μM (Ponting et al. 1997), it has been reported that the affinities for other PDZ domain/peptide complexes are similar to that of the GOPC PDZ domain/Nlg peptide complex (Wiedemann et al. 2004). Due to GOPC taking part in multiple functions, the low affinity of the PDZ GOPC domain interaction with the C terminus of Nlg might be important for changing different binding partners. Nevertheless, the fast exchange makes it difficult to determine the structure of a complex using NMR experiments because too few intermolecular NOEs are observed. So MD simulation employing the solution structure of the complex of PSD95 PDZ3 with the peptide TKNYKQTSV (PDB ID 1BE9) as a template was also performed to verify the interaction between the GOPC PDZ domain and C-terminal peptide of Nlg, and the binding interfaces on the GOPC PDZ domain were identified. Dali analysis (Table 2) shows that the structure of the GOPC PDZ domain is most similar to that of PSD95 PDZ3. PSD95 is a prototypic PDZ domain-containing protein and interacts with Nlg via its PDZ3 (Cho et al. 1992; Irie et al. 1997). Our results will help to understand the function of GOPC and Nlg. Structural similarity between GOPC PDZ and PSD95 PDZ3 implies these two proteins may bind to Nlg with similar modes.
Both Nlg and neurexin are neural cell adhesion molecules located in the post-synapse and the presynapse, respectively. The extracellular domain of neuroligins tightly binds to the extracellular domain of neurexins and form a heterotypic intercellular junction. Nlg and β-neurexin play an important role during the development of synapses. The C terminus of Nlg lacks any signaling motifs and only contains a type I PDZ binding motif. GOPC, the protein related with vesicle transport from the Golgi apparatus, also binds the Frizzled 8 via its PDZ domain besides Nlg. Frizzled genes are related with the development of cell polarity in the Wnt pathway. The GOPC PDZ domain might function as an important link between Nlg/Neurexin and Wnt pathways.
Recently a novel protein complex linking the δ2 glutamate receptor and autophagy was discovered (Yue et al. 2002). GOPC played an important role in this complex. Its PDZ domain binds to δ2 glutamate receptor while its coiled-coil region binds to Beclin1, an important regulator of autophagy. GOPC and Beclin1 can synergize to induce autophagy. Evidence has accumulated that autophagy is an important player in neuronal cell death and neurodegenerative disease. The structure and ligand-binding interface of GOPC PDZ provide a structural basis for the further functional study of this important molecule.
Materials and methods
Expression and purification of the GOPC PDZ domain
The DNA fragment encoding amino acid residues 270–363, which constitutes the human GOPC PDZ domain (276–363) and six amino acid residues preceding it, was polymerase chain reaction–amplified from the human brain cDNA library with specific primers. The amplified DNA fragment was inserted into the plasmid pET22b (+) (Novagen). The recombinant vector harboring the target gene was transformed into Escherichia coli BL21 (DE3) host cells for large-scale protein production. Recombinant PDZ domain fragment was purified using HiTrap chelating column (Pharmacia) chromatography. The purified recombinant protein contained an N-terminal Met and a C-terminal His tag (-LEHHHHHH) carried over from the cloning vector. The concentrations of 15N-labeled and 13C, 15N-labeled GOPC PDZ domain were ∼0.8–1.0 mM, while the concentration of unlabeled GOPC PDZ domain was up to 2.0 mM. All samples for NMR contained 20 mM phosphate buffer (at pH 6.0, containing 1 mM EDTA and 4 mM DTT in 90% H2O, 10% D2O).
Peptide synthesis and purification
The C-terminal peptide (SHSTTRV) of hNlg-1 (Nlg (817–823)) was chemically synthesized using standard Fmoc (N-(9-fluorenyl) methoxycarbonyl) chemistry at Shanghai Sangon Biological Engineering & Technology and Service Co. Ltd. The synthetic peptide was purified by a reverse-phase high-pressure liquid chromatography (C18 column) eluted with an acetonitrile linear gradient of 15%–30%. The final product was lyophilized and verified by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry and NMR signal assignments.
NMR spectroscopy
The NMR experiments were performed on a Bruker DMX500 spectrometer with self-shielded Z-axis gradients. The following spectra were recorded at 303 K to obtain backbone and side-chain resonance assignments: 2D 1H–15N HSQC, 2D 1H–13C HSQC, 3D triple-resonance spectra HNCO, HN(CA)CO, CBCA(CO)NH, CBCANH, HBHA(CBCACO)NH, C(CO)NH-TOCSY, H(CCO)NH-TOCSY, 15N-TOCSY, HCCH-TOCSY, HCCH-COSY (Bax et al. 1990a,b,c; Clubb et al. 1992; Grzesiek and Bax 1992; Muhandiram and Kay 1994). The 15N-labeled sample was lyophilized and dissolved in 99.96% D2O, followed immediately by HSQC experiments to monitor the disappearance of NH signals at 293 K. NMR data processing was carried out using NMRPipe and NMRDraw software (Delaglio et al. 1995), and the data were analyzed using Sparky 3. All software was run on a Linux system.
NMR distance restraints were collected from three different NOESY spectra: 3D 15N-separated NOESY in water for amide protons, 3D 13C-separated NOESY in water for aliphatic protons (mixing time 130 msec), and 2D 1H-NOESY in D2O (mixing time 100 msec) for aromatic protons. The TALOS (torsion angle likelihood obtained from shift and sequence similarity) (Cornilescu et al. 1999) was calculated for five types of nuclear: 13Cα, 13Cβ, 13CO, 1Hα, and 15N. Only TALOS “good” predictions with nine or 10 matches in agreement were used and converted into restraints on ϕ and ψ angles. The chemical shift index (CSI) (Wishart and Sykes 1994), which was calculated for four types of nuclear 13Cα, 13Cβ, 13CO, and 1Hα, was applied as supplement. Hydrogen bond restraints were obtained by identifying the slow-exchange amide protons after overnight incubation following solvent exchange. For each hydrogen bond, two distance restraints (NH–O and N–O) were used.
Structures were calculated using the program CNS v1.1, employing a simulated annealing protocol for torsion angle dynamics. The calculated structures were analyzed by the programs PROCHECK and MOLMOL (Koradi et al. 1996; Laskowski et al. 1996).
15N relaxation experiments were carried out at 303 K on a Bruker DMX500 NMR spectrometer. 15N relaxation measurements were carried out using the published methods (Farrow et al. 1994). 15N T1 relaxation rates were measured with eight relaxation delays: 111, 61.3, 141.5, 241.8, 362.2, 522.7, 753.4, and 1144.5 msec. 15N T2 relaxation rates were measured with six relaxation delays: 17.6, 35.2, 52.8, 70.4, 105.6, and 140.8 msec. A recycle delay of 1 sec was used for measurement of T1 and T2 relaxation rates. The spectra measuring 1H–15N NOE were acquired with a 2-sec relaxation delay followed by a 3-sec period of proton saturation. The spectra recorded in the absence of proton saturation employed a relaxation delay of 5 sec. The exponential curve fitting and extract of T1s and T2s are processed by Sparky 3.
NMR titration
NMR titration of the GOPC PDZ domain with Nlg was performed using 500 μM 15N-labeled protein. Peptidic ligand stock solution of 50 mM was titrated such that the 500-μL NMR sample was diluted by no more than 10%. The 1H and 15N resonance variations were followed by HSQC experiments, and all NMR 15N-HSQC spectra were acquired at 303 K. Combined chemical shift perturbation was calculated using the following equation:
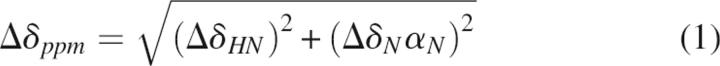 |
in which ΔδHN and ΔδN are the chemical shift variations in the proton and nitrogen dimensions, respectively, and αN is the scaling factor used to normalize the 1H and 15N chemical shifts, which equaled 0.17 (Tochio et al. 1999; Zhou et al. 2005).
Measurement of equilibrium dissociation constants
Dissociation constants of the interaction of the GOPC PDZ domain with the C terminus of Nlg were obtained by monitoring the chemical shift changes between the GOPC PDZ domain in the apo and bound conformation during titration experiments. When the exchange rate is greater than the chemical shift difference between the free and the bound states (as for the peptidic ligand in this study), the observed chemical shift at each titration point, δay, is a weighted average between the chemical shifts of the free and bound states (Lian et al. 1994; Liu et al. 1996; Hu et al. 2004) obtained by
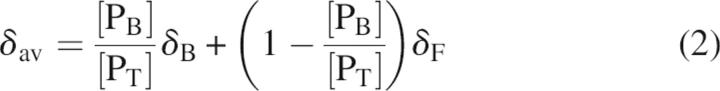 |
where δF is the chemical shift of the protein domain in the absence of peptidic ligand, δB is the chemical shift of the protein domain bound to peptidic ligand, [PB] is the concentration of ligand-bound protein domain, and [PT] is the total concentration of protein domain. The mole fraction used to fit the titration curve was calculated for the total substrate concentration at each point in the titration and from the fitted dissociation constant using Equations 2 and 3. In Equation 3, KD is the dissociation constant and [ST] is the total peptidic ligand concentration at each titration point.
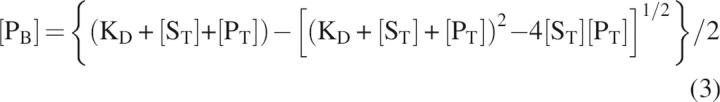 |
Molecular modeling of the GOPC PDZ domain with C-terminal penta-peptide of Nlg
Modeling of the complex of the GOPC PDZ domain with C-terminal peptide of Nlg was done using molecular replacement. We employed the solution structure of the complex of PSD95 PDZ3 with the peptide TKNYKQTSV (PDB ID 1BE9) as a template. Structure alignment was performed using the CE (Shindyalov and Bourne 1998) algorithm followed by molecular replacement. Molecular dynamics (MD) simulation were performed to optimize the models. First, 500 steps of the steepest-descent energy minimization followed by 300 psec MD simulation at a temperature gradually increased from 50 K to 300 K were carried out, with all heavy atoms of the protein and the peptidic ligand backbone restrained to their respective starting positions by a harmonic potential, with a restraint force constant gradually decreased from 1.0E4 to 5.0E3 KJ/(mol × nm2). Then another 500-psec MD simulation was performed with position restraints applied to the heavy atoms of the residues out of the binding region including residues 17–33, 37–43, and 71–82. In the next step, the entire system was optimized by 500 psec MD simulation without any restraints. Finally, the resulting configuration was minimized through stepwise simulated annealing: In a 500-psec simulation the temperature of the system was gradually reduced from 300 K to 100 K. The GROMOS96 package (van Gunsteren et al. 1996) with the GROMOS96 force field combined with an explicit solvent model (SPC) was used to perform the simulation. The resulting models were checked for quality using the program PROCHECK.
Data bank accession codes
The coordinates of the 20 refined structures of the GOPC PDZ domain have been deposited in the PDB with the accession code 2DC2, and the chemical shifts have been deposited in the BMRB with accession code 7072.
Electronic supplemental material
Supplemental material includes dissociation constants of interaction of the GOPC PDZ domain with the C terminus of Nlg (Table S1); the ensemble of the GOPC PDZ domain (Fig. S1); NMR relaxation data, and S2, τe, and Rex results of analysis by FAST-ModelFree (Fig. S2); superposition of the PDZ domains (Fig. S3); and the GOPC PDZ domain interaction with C-terminal motif of the Frizzled 8 (Figs. S4, S5).
Acknowledgments
We thank Dr. F. Delaglio and Prof. A. Bax for providing the software NMRPipe, Prof. T.D. Goddard and Prof. D.G. Kneller for providing Sparky, Prof. A.T. Brünger for providing the program CNS, Dr. R. Koradi and Prof. K. Wüthrich for providing MOLMOL, M. Carson for providing Ribbons, Dr. W.L. DeLano for providing PyMOL, and Prof. W.F. van Gunsteren for providing GROMOS96 package. This work was supported by the Chinese National Fundamental Research Project (grants 2002CB713806 and 2004CB520500), the Chinese National Natural Science Foundation (grants 30270293, 30121001 and 30570361), and the Pilot Project of the Knowledge Innovation Program of the Chinese Academy of Science (grant KSCX1-SW-17).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Yunyu Shi or Jihui Wu, School of Life Science, University of Science and Technology of China, Hefei, Anhui 230026, People's Republic of China; e-mail: yyshi@ustc.edu.cn or wujihui@ustc.edu.cn; fax: +86-551-3601443.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062087506.
Abbreviations: GOPC, Golgi-associated PDZ and coiled-coil motif-containing protein; Nlg, neuroligin; MD, molecular dynamics.
References
- Bax A., Clore G.M., Driscoll P.C., Gronenborn A.M., Ikura M., Kay L.E. 1990a. Practical aspects of proton-carboncarbon-proton three-dimensional correlation spectroscopy of 13C labeled proteins. J. Magn. Reson. 87: 620–627. [Google Scholar]
- Bax A., Clore G.M., Gronenborn A.M. 1990b. 1H–1H correlation via isotropic mixing of 13C magnetization, a new three dimensional approach for assigning 1H and 13C spectra of 13C enriched proteins. J. Magn. Reson. 88: 425–431. [Google Scholar]
- Bax A., Ikura M., Kay L.E., Torchia D.A., Tschudin R. 1990c. Comparison of different modes of two-dimensional reverse correlation NMR for the study of proteins. J. Magn. Reson. 86: 304–318. [Google Scholar]
- Bolliger M.F., Frei K., Winterhalter K.H., Gloor S.M. 2001. Identification of a novel neuroligin in humans which binds to PSD-95 and has a widespread expression. Biochem. J. 356: 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest A., Lane K., McMahon K., Housman D.E. 2001. Association of a novel PDZ domain-containing peripheral Golgi protein with the Q-SNARE (Q-soluble N-ethylmaleimide-sensitive fusion protein (NSF) attachment protein receptor) protein syntaxin 6. J. Biol. Chem. 276: 29456–29465. [DOI] [PubMed] [Google Scholar]
- Cheng J., Moyer B.D., Milewski M., Loffing J., Ikeda M., Mickle J.E., Cutting G.R., Li M., Stanton B.A., Guggino W.B. 2002. A Golgi-associated PDZ domain protein modulates cystic fibrosis transmembrane regulator plasma membrane expression. J. Biol. Chem. 277: 3520–3529. [DOI] [PubMed] [Google Scholar]
- Cheng J., Wang H., Guggino W.B. 2004. Modulation of mature cystic fibrosis transmembrane regulator protein by the PDZ domain protein CAL. J. Biol. Chem. 279: 1892–1898. [DOI] [PubMed] [Google Scholar]
- 2005. Regulation of cystic fibrosis transmembrane regulator trafficking and protein expression by a Rho family small GTPase TC10. J. Biol. Chem. 280: 3731–3739 ———. [DOI] [PubMed] [Google Scholar]
- Cho K.O., Hunt C.A., Kennedy M.B. 1992. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 9: 929–942. [DOI] [PubMed] [Google Scholar]
- Clubb R.T., Thanabal V., Wagner G. 1992. A constant-time 3-dimensional triple-resonance pulse scheme to correlate intraresidue H-1(N), N-15, and C-13 chemical shifts in N-15-C-13-labeled proteins. J. Magn. Reson. 97: 213–217. [Google Scholar]
- Cole R. and Loria J.P. 2003. FAST-Modelfree: A program for rapid automated analysis of solution NMR spin-relaxation data. J. Biomol. NMR 26: 203–213. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio F., Bax A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Farrow N.A., Muhandiram R., Singer A.U., Pascal S.M., Kay C.M., Gish G., Shoelson S.E., Pawson T., Forman-Kay J.D., Kay L.E. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003. [DOI] [PubMed] [Google Scholar]
- Gentzsch M., Cui L., Mengos A., Chang X.B., Chen J.H., Riordan J.R. 2003. The PDZ-binding chloride channel ClC-3B localizes to the Golgi and associates with cystic fibrosis transmembrane conductance regulator-interacting PDZ proteins. J. Biol. Chem. 278: 6440–6449. [DOI] [PubMed] [Google Scholar]
- Grzesiek S. and Bax A. 1992. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J. Magn. Reson. 99: 201–207. [Google Scholar]
- Hata Y., Butz S., Sudhof T.C. 1996. CASK: A novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J. Neurosci. 16: 2488–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks S.W. and Machamer C.E. 2005. Isoform-specific interaction of golgin-160 with the Golgi-associated protein PIST. J. Biol. Chem. 280: 28944–28951. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138. [DOI] [PubMed] [Google Scholar]
- Hu H.Y., Horton J.K., Gryk M.R., Prasad R., Naron J.M., Sun D.A., Hecht S.M., Wilson S.H., Mullen G.P. 2004. Identification of small molecule synthetic inhibitors of DNA polymerase β by NMR chemical shift mapping. J. Biol. Chem. 279: 39736–39744. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K., Hata Y., Nguyen T., Ullrich B., Missler M., Moomaw C., Sudhof T.C. 1995. Neuroligin 1: A splice site-specific ligand for β-neurexins. Cell 81: 435–443. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K., Nguyen T., Sudhof T.C. 1996. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J. Biol. Chem. 271: 2676–2682. [DOI] [PubMed] [Google Scholar]
- Irie M., Hata Y., Takeuchi M., Ichtchenko K., Toyoda A., Hirao K., Takai Y., Rosahl T.W., Sudhof T.C. 1997. Binding of neuroligins to PSD-95. Science 277: 1511–1515. [DOI] [PubMed] [Google Scholar]
- Ives J.H., Fung S., Tiwari P., Payne H.L., Thompson C.L. 2004. Microtubule-associated protein light chain 2 is a stargazin-AMPA receptor complex-interacting protein in vivo. J. Biol. Chem. 279: 31002–31009. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Wuthrich K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 29–32. [DOI] [PubMed] [Google Scholar]
- Kurschner C., Mermelstein P.G., Holden W.T., Surmeier D.J. 1998. CIPP, a novel multivalent PDZ domain protein, selectively interacts with Kir4.0 family members, NMDA receptor subunits, neurexins, and neuroligins. Mol. Cell. Neurosci. 11: 161–172. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann J.A., MacArthur M.W., Kaptein R., Thornton J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8: 477–486. [DOI] [PubMed] [Google Scholar]
- Levinson J.N., Chery N., Huang K., Wong T.P., Gerrow K., Kang R., Prange O., Wang Y.T., El-Husseini A. 2005. Neuroligins mediate excitatory and inhibitory synapse formation: Involvement of PSD-95 and neurexin-1β in neuroligin-induced synaptic specificity. J. Biol. Chem. 280: 17312–17319. [DOI] [PubMed] [Google Scholar]
- Lian L.Y., Barsukov I.L., Sutcliffe M.J., Sze K.H., Roberts G.C. 1994. Protein-ligand interactions: Exchange processes and determination of ligand conformation and protein-ligand contacts. Methods Enzymol. 239: 657–700. [DOI] [PubMed] [Google Scholar]
- Liu D., Prasad R., Wilson S.H., DeRose E.F., Mullen G.P. 1996. Three-dimensional solution structure of the N-terminal domain of DNA polymerase β and mapping of the ssDNA interaction interface. Biochemistry 35: 6188–6200. [DOI] [PubMed] [Google Scholar]
- Meyer G., Varoqueaux F., Neeb A., Oschlies M., Brose N. 2004. The complexity of PDZ domain-mediated interactions at glutamatergic synapses: A case study on neuroligin. Neuropharmacology 47: 724–733. [DOI] [PubMed] [Google Scholar]
- Muhandiram D.R. and Kay L.E. 1994. Gradient-enhanced triple resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B. 103: 203–216. [Google Scholar]
- Nam C.I. and Chen L. 2005. Postsynaptic assembly induced by neurexin–neuroligin interaction and neurotransmitter. Proc. Natl. Acad. Sci. 102: 6137–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudauer C.L., Joberty G., Macara I.G. 2001. PIST: A novel PDZ/coiled-coil domain binding partner for the rho-family GTPase TC10. Biochem. Biophys. Res. Commun. 280: 541–547. [DOI] [PubMed] [Google Scholar]
- Piserchio A., Fellows A., Madden D.R., Mierke D.F. 2005. Association of the cystic fibrosis transmembrane regulator with CAL: Structural features and molecular dynamics. Biochemistry 44: 16158–16166. [DOI] [PubMed] [Google Scholar]
- Ponting C.P., Phillips C., Davies K.E., Blake D.J. 1997. PDZ domains: Targeting signalling molecules to sub-membranous sites. Bioessays 19: 469–479. [DOI] [PubMed] [Google Scholar]
- Scheiffele P., Fan J., Choih J., Fetter R., Serafini T. 2000. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101: 657–669. [DOI] [PubMed] [Google Scholar]
- Shindyalov I.N. and Bourne P.E. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11: 739–747. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Fanning A.S., Fu C., Xu J., Marfatia S.M., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., Cantley L.C. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275: 73–77. [DOI] [PubMed] [Google Scholar]
- Strutt D.I. 2001. Asymmetric localization of frizzled and the establishment of cell polarity in the Drosophila wing. Mol. Cell 7: 367–375. [DOI] [PubMed] [Google Scholar]
- Sugita S., Saito F., Tang J., Satz J., Campbell K., Sudhof T.C. 2001. A stoichiometric complex of neurexins and dystroglycan in brain. J. Cell Biol. 154: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochio H., Zhang Q., Mandal P., Li M., Zhang M. 1999. Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide. Nat. Struct. Biol. 6: 417–421. [DOI] [PubMed] [Google Scholar]
- Ushkaryov Y.A., Petrenko A.G., Geppert M., Sudhof T.C. 1992. Neurexins: Synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science 257: 50–56. [DOI] [PubMed] [Google Scholar]
- Ushkaryov Y.A., Hata Y., Ichtchenko K., Moomaw C., Afendis S., Slaughter C.A., Sudhof T.C. 1994. Conserved domain structure of β-neurexins. Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J. Biol. Chem. 269: 11987–11992. [PubMed] [Google Scholar]
- van Gunsteren W.F., Billeter S.R., Eising A.A., Hunenberger P.H., Kruger P., Mark A.E., Scott W.R., Tironi I.G. In Biomolecular simulation: The GROMOS96 manual and user guide . 1996. Vdf Hochschulverlag, Zurich.
- Vinson C.R., Conover S., Adler P.N. 1989. A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature 338: 263–264. [DOI] [PubMed] [Google Scholar]
- Wente W., Stroh T., Beaudet A., Richter D., Kreienkamp H.J. 2005. Interactions with PDZ domain proteins PIST/GOPC and PDZK1 regulate intracellular sorting of the somatostatin receptor subtype 5. J. Biol. Chem. 280: 32419–32425. [DOI] [PubMed] [Google Scholar]
- Wiedemann U., Boisguerin P., Leben R., Leitner D., Krause G., Moelling K., Volkmer-Engert R., Oschkinat H. 2004. Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super-binding peptides. J. Mol. Biol. 343: 703–718. [DOI] [PubMed] [Google Scholar]
- Wishart D.S. and Sykes B.D. 1994. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4: 171–180. [DOI] [PubMed] [Google Scholar]
- Yao R., Maeda T., Takada S., Noda T. 2001. Identification of a PDZ domain containing Golgi protein, GOPC, as an interaction partner of frizzled. Biochem. Biophys. Res. Commun. 286: 771–778. [DOI] [PubMed] [Google Scholar]
- Yao R., Ito C., Natsume Y., Sugitani Y., Yamanaka H., Kuretake S., Yanagida K., Sato A., Toshimori K., Noda T. 2002. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc. Natl. Acad. Sci. 99: 11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Horton A., Bravin M., DeJager P.L., Selimi F., Heintz N. 2002. A novel protein complex linking the δ2 glutamate receptor and autophagy: Implications for neurodegeneration in lurcher mice. Neuron 35: 921–933. [DOI] [PubMed] [Google Scholar]
- Zhou H., Xu Y., Yang Y., Huang A., Wu J., Shi Y. 2005. Solution structure of AF-6 PDZ domain and its interaction with the C-terminal peptides from Neurexin and Bcr. J. Biol. Chem. 280: 13841–13847. [DOI] [PubMed] [Google Scholar]