Abstract
Membrane proteins comprise up to one-third of prokaryotic and eukaryotic genomes, but only a very small number of membrane protein structures are known. Membrane proteins are challenging targets for structural biology, primarily due to the difficulty in producing and purifying milligram quantities of these proteins. We are evaluating different methods to produce and purify large numbers of prokaryotic membrane proteins for subsequent structural and functional analysis. Here, we present the comparative expression data for 37 target proteins, all of them secondary transporters, from the mesophilic organism Salmonella typhimurium and the two hyperthermophilic organisms Aquifex aeolicus and Pyrococcus furiosus in three different Escherichia coli expression vectors. In addition, we study the use of Lactococcus lactis as a host for integral membrane protein expression. Overall, 78% of the targets were successfully produced under at least one set of conditions. Analysis of these results allows us to assess the role of different variables in increasing “expression space” coverage for our set of targets. This analysis implies that to maximize the number of nonhomologous targets that are expressed, orthologous targets should be chosen and tested in two vectors with different types of promoters, using C-terminal tags. In addition, E. coli is shown to be a robust host for the expression of prokaryotic transporters, and is superior to L. lactis. These results therefore suggest appropriate strategies for high-throughput heterologous overproduction of membrane proteins.
Keywords: membrane proteins, transporters, structural genomics, protein production, high-throughput screen
Biomembranes are essential components in the genesis of life and crucial structuring elements of cells. They preserve the functional integrity of cell organelles and regulate the exchange of solutes and signals between the different functional areas of the cell and between the cell and its environment. Membrane proteins make up 25%–30% of the genes in most organisms. These proteins are often very complex, function in many different ways and play key roles in processes including signal transduction, cell growth and differentiation, transport, and metabolism. In addition, their importance is enhanced by the fact that a large number of today's drugs are targeted at membrane proteins (Hopkins and Groom 2002; McKeegan et al. 2002). Despite the significance of these proteins, just a handful of membrane protein structures have been investigated in detail. Only ∼100 unique structures of membrane proteins are known (http://www.mpibp-frankfurt.mpg.de/michel/public/memprotstruct.html), compared with >13,000 nonredundant structures of soluble proteins (http://www.rcsb.org). It is clear that much more structural data is needed to form a detailed understanding of membrane protein function. However, because of the unique challenges involved in working with membrane proteins, our knowledge of membrane protein structure, function, and assembly is still limited.
One of the main problems surrounding studies of membrane proteins is the difficulty in producing enough material for biochemical and structural analyses. Currently, no ideal overexpression system exists for membrane proteins, as their overproduction may damage membrane integrity and is often lethal to the cell (Miroux and Walker 1996). In addition, matching the protein source organism as closely as possible with the host organism can be critical for proper membrane insertion and folding as well as posttranslational modifications. Producing prokaryotic membrane proteins in the simple, flexible, and inexpensive Escherichia coli system has already proven successful in many cases (Hockney 1994; Grisshammer and Tate 1995; Wang et al. 2003). One of the key advantages of this system is the large variety of different vectors accompanied with a diversity of promoters for expression control. In addition, the lack of posttranslational modifications and the likelihood that heterologous prokaryotic proteins will interface with the insertion machinery and be properly folded make production of prokaryotic membrane proteins in E. coli an attractive choice.
High-throughput screening has been successfully applied to the rapid identification of soluble proteins that are amenable to high-level production and crystallization (Christendat et al. 2000; Braun et al. 2002; Lesley et al. 2002; Yee et al. 2002; Heinemann et al. 2003). In the year 2005, structural genomics initiatives on soluble proteins accounted for only 20% of total protein structures, but 40%–50% of these structures were considered novel (Chandonia and Brenner 2006). Structural genomics efforts have also begun with membrane proteins, in most cases with proteins from prokaryotic organisms (Dobrovetsky et al. 2005; Eshaghi et al. 2005), but also with eukaryotic proteins (Luan et al. 2004; Busso et al. 2005; Andre et al. 2006). While high-throughput techniques enable more rapid screening of production conditions, there is as yet no consensus on which types of vectors or expression systems will provide the best results to increase the number of expressing membrane proteins and the overall amount of protein that is produced. Therefore, rational strategies that focus on the basic problem of producing sufficient amounts of protein for subsequent purification and crystallization trials have to be established for structural studies of membrane proteins.
We have undertaken the expression of ∼250 prokaryotic or archaeal proteins from 42 distinct transporter families from the source organisms Salmonella typhimurium LT2, Aquifex aeolicus VF5, and Pyrococcus furiosus DSM3638. The heterologous production of these proteins is being tested in E. coli to identify proteins produced in large enough amounts to enable structural studies. A subset of these proteins has been chosen for comparative analysis of a variety of production conditions, and this data is presented here. The production of 37 transport proteins from various families has been tested in E. coli using three different E. coli expression vectors with two sets of affinity tags to identify the most favorable system that would ensure the production of representative nonhomologous proteins from individual transporter families. In addition, production has been tested in the Gram-positive bacterium Lactococcus lactis (Kunji et al. 2003, 2005; Monne et al. 2005) to compare this relatively new expression system with the more traditional E. coli system. By analyzing the expression data obtained using these variables and subsequently constructing the expression space coverage, we are able to suggest efficient production strategies to pursue for heterologous membrane protein structural genomics.
Results
Target selection and expression strategy
The aim of this study is to evaluate the use of orthologous targets, various expression vectors and different expression hosts for the heterologous production of prokaryotic transporter proteins in order to identify appropriate conditions that allow for production of the maximum number of nonhomologous proteins (i.e., proteins from different transporter families) for structural studies. For this analysis, 14 transporter families predicted to have at least three transmembrane helices and to function as monomeric or homo-oligomeric inner membrane secondary transporters were selected (Ren et al. 2004). Within the 14 families, a total of 37 transporters were chosen from three organisms—S. typhimurium, A. aeolicus, and P. furiosus. Transporters from S. typhimurium are of interest due to the organism's pathogenicity. As membrane proteins from hyperthermophilic organisms may be more stable outside of the membrane as compared to those from mesophiles, proteins from the hyperthermophilic bacterium A. aeolicus and the hyperthermophilic archaeon P. furiosus were selected also.
In order to compare production in different host systems, both E. coli and L. lactis were chosen as hosts for protein production. E. coli is the most commonly used host for heterologous protein production of soluble proteins, and a variety of expression vectors exist for use with this organism (Hockney 1994; Grisshammer and Tate 1995; Wang et al. 2003). L. lactis has recently shown promise as a host for production of membrane proteins (Kunji et al. 2003, 2005; Monne et al. 2005), and its inclusion in this study allows for a direct comparison with production in E. coli.
For protein production in E. coli, three expression vectors with different promoters were chosen for comparison. The first is the pTTQ18 vector, which uses the moderately strong tac promoter for gene expression (Stark 1987). There is some evidence that more moderate promoters may be preferable for membrane protein gene expression, and this vector has been used previously for the successful production of membrane proteins with yields of up to 1–2 mg/L (Ward et al. 2001). The second is the pQE vector (Qiagen), in which the stronger T5 promoter drives expression. Additionally, this vector contains two lac operator binding sites, to prevent gene expression prior to induction. Finally, the pBAD vector (Invitrogen) was selected. This vector uses the araBAD promoter, which is very tightly repressed before induction with arabinose, and is therefore useful for the expression of potentially toxic genes (Siegele and Hu 1997). In addition, induction with arabinose instead of IPTG allows for a more precise linear control of protein production (Guzman et al. 1995). For protein production in L. lactis, the pNZ8048 vector system, in which the nisA promoter drives gene expression, was used (Kunji et al. 2003).
Each of the vectors was modified to include an identical multiple cloning site as well as one of two sets of tags. The first set, termed the “A” version, consists of a C-terminal TEV protease cleavage site followed by a poly-Histidine (poly-His) tag to facilitate purification. The second set, the “C” version, consists of an N-terminal poly-His tag and TEV protease cleavage site as well as a C-terminal Strep tag-II to allow for a second affinity purification step. Thus, for each target, eight different constructs were generated. However for some targets, cloning into certain vectors, in particular the pNZ8048 vector, was unsuccessful despite several attempts (see Table 1 and Materials and Methods). Therefore, the total number of constructs obtained was 235, and not the expected 296.
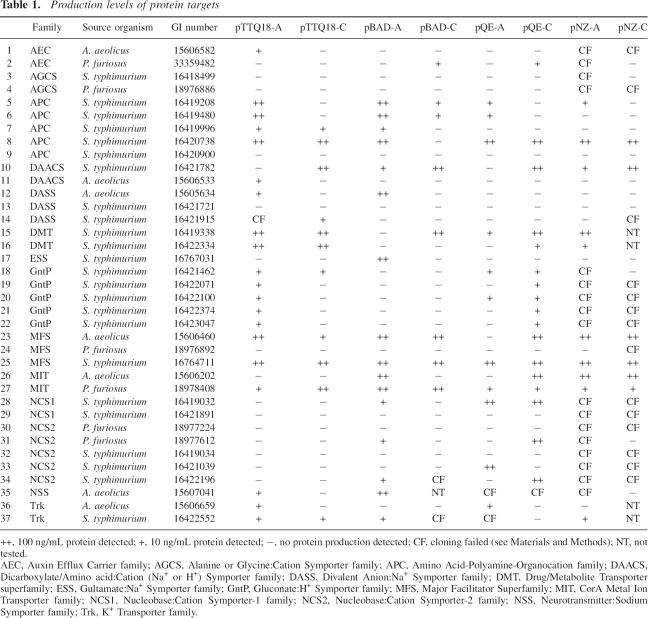
Table 1.
Production levels of protein targets
Detection of protein production and expression results
As membrane insertion of prokaryotic secondary transporters is linked to protein translation by the E. coli signal recognition particle (Luirink et al. 2005), it is likely that any successfully produced transporters will be localized at least in part, if not in total to the membrane. While there have been reports of prokaryotic integral membrane proteins that form insoluble aggregates when overproduced, most of these proteins have between zero and three predicted transmembrane helices, and therefore many cannot be considered to be true membrane integrated proteins, but rather membrane anchored proteins only (Korepanova et al. 2005; Columbus et al. 2006). In the study by Korepanova et al. (2005), proteins with four or more predicted transmembrane helices were always localized in total or in part to the membrane, and never to the insoluble fraction alone. In addition, there is other evidence that bacterial integral membrane proteins overproduced in E. coli are able to partition in total into the membrane (Drew et al. 2001). It is in fact unclear whether insoluble aggregates containing membrane proteins represent inclusion bodies, or rather a more dense protein-containing membrane fraction that sediments even during low-speed centrifugation, as has been reported previously (Arechaga et al. 2000).
As the aim of this study was to use the most efficient and high-throughput methods for evaluating production of integral membrane proteins, we chose to evaluate protein production levels based on analysis of whole cell lysates, as it is clear from the above evidence that the membrane transporters in this study, most of which have at least eight predicted transmembrane helices, and only three of which have three predicted helices, will most likely be partially or fully localized to the membrane. In addition, while production in the membrane is preferred, there are several studies in which in vitro refolding of eukaryotic integral membrane proteins has been successful (Kiefer et al. 1996; Rogl et al. 1998; Ma et al. 2002; for review, see Kiefer 2003), and thus it may be possible to purify protein from inclusion bodies as well.
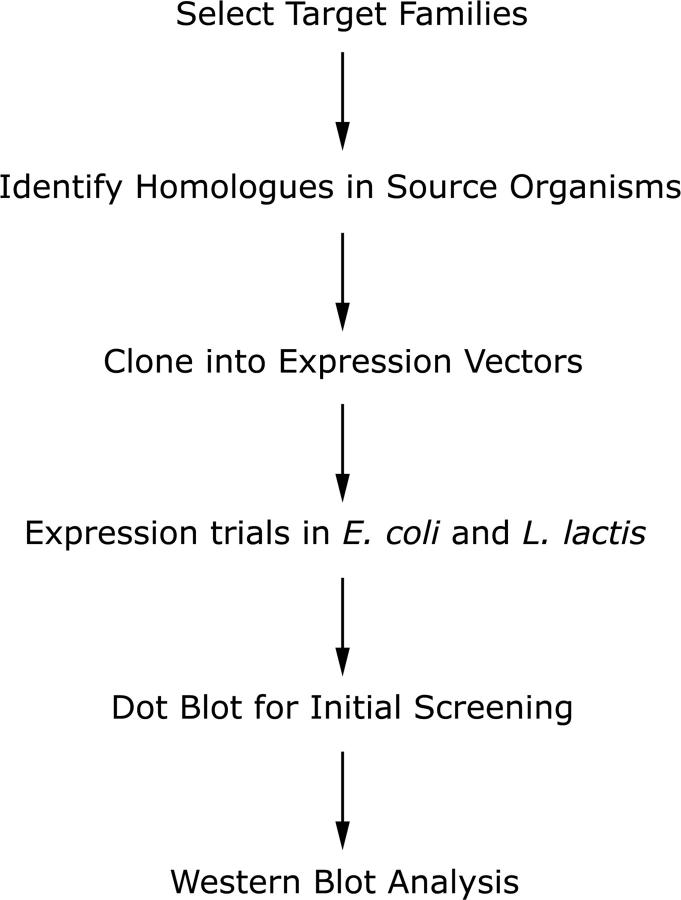
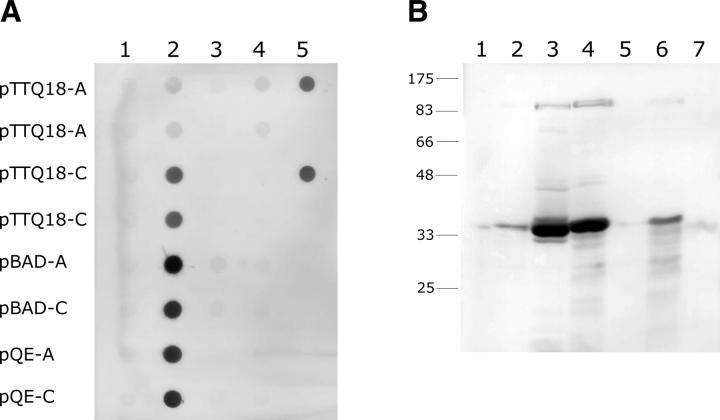
An outline of the expression strategy is shown in Figure 1. After selecting the protein targets, each gene was cloned into eight different vectors comprising A and C versions of the pTTQ18, pQE, pBAD, and pNZ8048 vectors, as discussed above. Each construct was then tested for expression in E. coli and L. lactis as described in the Materials and Methods. Briefly, two to four clones from each construct were grown in small-scale cultures, and after induction the cells were harvested and lysed to analyze the protein content. Target protein production was first detected via dot blot analysis of whole cell lysates using an antibody to the poly-His tags. Positive signals on the dot blot were then verified by SDS-PAGE and Western blotting. A representative dot blot showing production of several target proteins in different vectors is shown in Figure 2A. As seen in this figure, protein production levels vary significantly for different proteins. Additionally, different constructs containing the same protein also show varying production levels, as confirmed in Figure 2B, which shows the production of Protein 27 in several different vectors, as detected by Western blotting.
Figure 1.
Overview of expression strategy. The strategy for screening production of the selected membrane proteins is shown above. After selecting the target families, existing homologs in the source organisms A. aquifex, P. furiosus, and S. typhimurium were identified and cloned into expression vectors. Expression trials were performed in both E. coli and L. lactis, and the results analyzed by dot blot and Western blot.
Figure 2.
Dot blot and Western blot analysis of whole cell lysates. E. coli or L. lactis cells were lysed as described in the Materials and Methods. Lysate from 1 mL of cells (dot blot) or 100 μL of cells (Western) was loaded onto a PVDF membrane for dot blot analysis or SDS-PAGE gel for Western blot analysis. (A) Sample dot blot showing production levels for three transporters in each of the vectors indicated on the left. Protein was detected using an α-poly-His antibody. Lane 1, vector alone; lane 2, Protein 27; lane 3, Protein 17; lane 4, Protein 12; lane 5, 50 ng eGFP (upper dot) and 100 ng eGFP (lower dot). (B) Western blot of whole cell lysates of Protein 27 produced in various vectors. Protein was detected using an α-His antibody. Molecular weight markers are shown in kilodaltons. Lane 1, pTTQ18-A; lane 2, pTTQ18-C; lane 3, pBAD-A; lane 4, pBAD-C; lane 5, pQE-A; lane 6, pQE-C; lane 7, pNZ-A.
Each construct was scored as showing no expression (−), low expression (10 ng/mL; +), or high expression (≥100 ng/mL; ++) as compared to control samples containing a known amount of protein (Table 1). Among the 37 targets, eight proteins could not be produced at all while a total of 29 proteins were produced under one or more conditions, giving a 78% rate of success. Out of those 29 proteins, 17 were overproduced, which represents 46% of the targets.
At least one target from each of 13 families showed expression out of the 14 families attempted. The only family showing no expression under the conditions tested is the Alanine or Glycine:Cation Symporter (AGCS) family. Among the other families, there are clear differences in the number of proteins that are produced and in the level of production. For example, while all five proteins of the Gluconate:H+ Symporter (GntP) family were only expressed at low levels, four out of five targets of the Amino acid-Polyamine-Organocation (APC) family were overexpressed in more than one vector, in yields sufficient to attempt purification (Table 1). This suggests that certain transporter families have inherent characteristics that may facilitate their production.
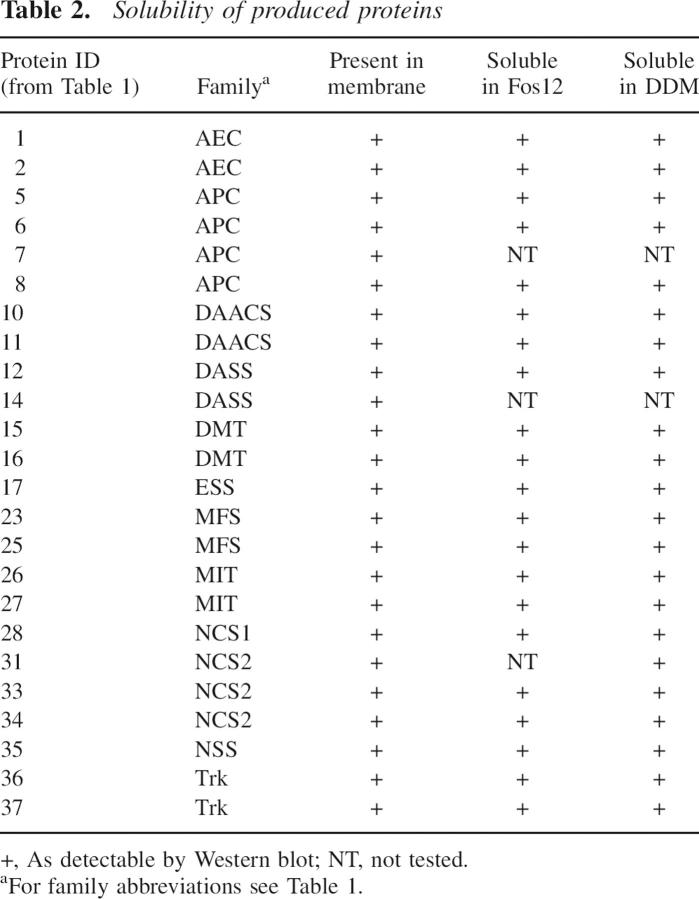
To verify that monitoring of protein production in whole cell lysates is indicative of protein that is inserted into the membrane, membrane fractions of at least one construct of 24 of the successfully produced proteins were prepared and analyzed for protein content. All of the tested proteins were present in the membrane fraction (Table 2). The solubility of these proteins in two commonly used detergents, n-dodecyl-β-D-maltoside (DDM) and Fos-choline 12 (Fos12), was also tested. In all of the cases tested, protein production detected in whole cell lysates corresponded to the production of membrane-inserted proteins that were easily solubilized by two different classes of detergents (Table 2).
Table 2.
Solubility of produced proteins
Comparison of expression by source organism
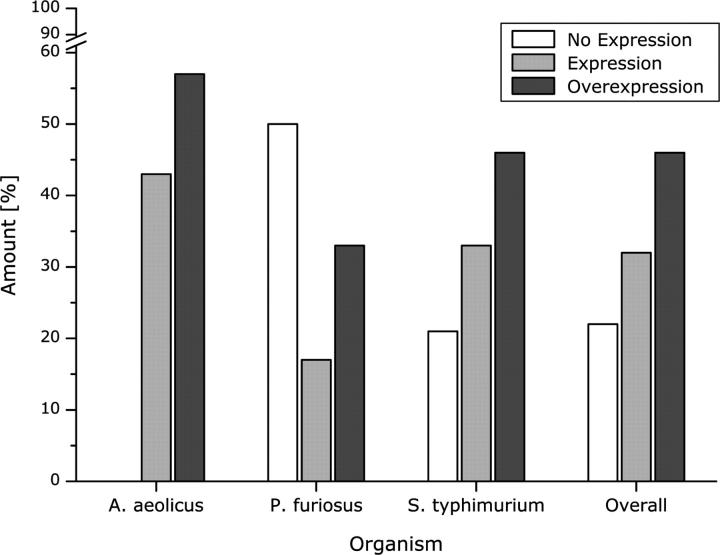
Out of the 29 genes successfully expressed in E. coli, seven genes are from A. aeolicus, three are from P. furiosus, and 19 are from S. typhimurium. As seen in Figure 3, 79% of the genes from S. typhimurium are expressed in total, with 33% expressed and 46% overexpressed. These high percentages may be due to the phylogenetic similarity between the source and host organisms (Wheeler et al. 2000). For the proteins from the hyperthermophilic organisms, 50% of P. furiosus genes are expressed in total, with 17% expressed and 33% overexpressed, while 100% of the genes from A. aeolicus are expressed in total, with 43% expressed and 57% overexpressed (Fig. 3). While this may indicate that A. aeolicus is a more appropriate source organism for the production of hyperthermophilic proteins, the targets cannot all be directly compared, as in approximately half of the cases the A. aeolicus and P. furiosus proteins are members of distinct transporter families (Table 1). Therefore, the lower percent expression of P. furiosus genes may also be due to the fact that genes from certain transporter families express more readily than others, as discussed above. Although a more extensive comparison is needed, these results suggest that despite the greater evolutionary divergence between the two organisms (Wheeler et al. 2000), many proteins from P. furiosus may also be readily produced in E. coli. From the data generated thus far, it appears that proteins from hyperthermophiles have no intrinsic properties that increase or hamper their production, as has also been seen for soluble proteins and for membrane proteins from the thermophile Thermotoga maritima (Savchenko et al. 2003; Dobrovetsky et al. 2005).
Figure 3.
Comparison of production levels by source organism. The number of transporter genes from each source organism that showed expression or overexpression was determined from the data in Table 1, and was divided by the total number of transporters tested from each organism to calculate the percent of successful production represented in the graph. The total number of targets from each organism is as follows: A. aeolicus, 7; P. furiosus, 6; S. typhimurium, 24.
Comparison of expression by host organism
Twenty-five of the 37 targets were tested for expression in both L. lactis and E. coli. Of these 25 targets, 84% showed expression in E. coli while only 40% showed expression in L. lactis (Table 1). Under the conditions tested here, the level of production in L. lactis was always equal to or less than that in E. coli (Table 1; Fig. 2). In addition, no protein that was not successfully produced in E. coli could be produced instead in L. lactis (Table 1). The much higher percent expression in E. coli may be due to the fact that only one type of L. lactis expression vector was tested here, versus three in E. coli. However, when comparing the performance of individual E. coli vectors to the L. lactis pNZ8048 vector, it is still clear that E. coli has an advantage. Using only the 25 proteins screened in L. lactis to calculate total expression levels for each vector gives 68% total expression for the pTTQ18 vector, 65% for the pBAD vector, and 56% for the pQE vector, which are all greater than the 40% total expression seen with the pNZ vector (Table 1).
In addition, these values are all higher than those calculated in Figure 5 (see below) for the E. coli vectors using data from all 37 target proteins. This may be because the 12 targets that were not tested in L. lactis represent targets from protein families (AEC, AGCS, GntP, NCS1, and NCS2) that often showed low or no expression, perhaps due to toxicity when overproduced (Table 1). For many of these targets, successful cloning into E. coli expression vectors required first cloning into the pDrive vector as well as screening of dozens of clones (data not shown), suggesting that the presence of these genes is somehow toxic to the cell. These results imply that for prokaryotic targets, E. coli is a much more robust production host, and may in fact be more suitable for producing toxic proteins than L. lactis. However, it should be noted that the E. coli strains used here also contained the pRARE plasmid, which codes for some tRNAs rarely used by E. coli to compensate for codon differences with heterologously expressed genes. As no such plasmid is yet available for L. lactis, codon differences between L. lactis and the heterologous genes may affect expression.
Figure 5.
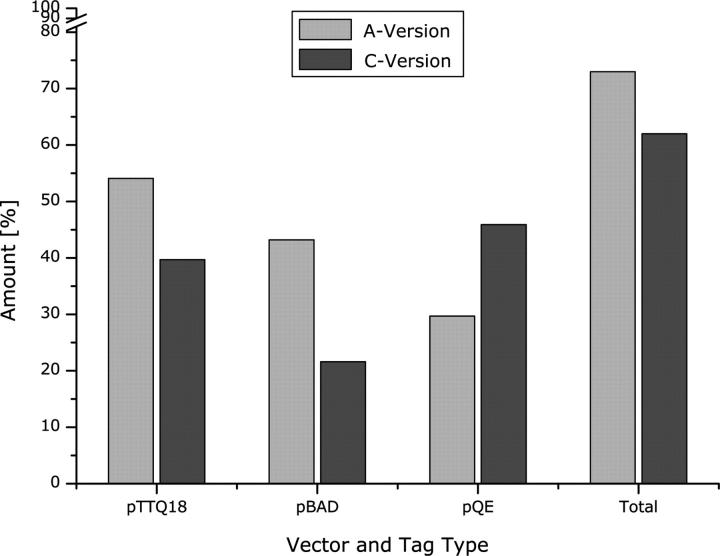
Comparison of production levels by tag position. The number of targets successfully expressed in either the A or C version of each vector was determined from the data in Table 1, and was divided by the total number of targets tested in that vector to calculate the percentages shown in the graph. Target genes that could not be successfully cloned into a particular vector were counted as negative.
Comparison of expression by vector
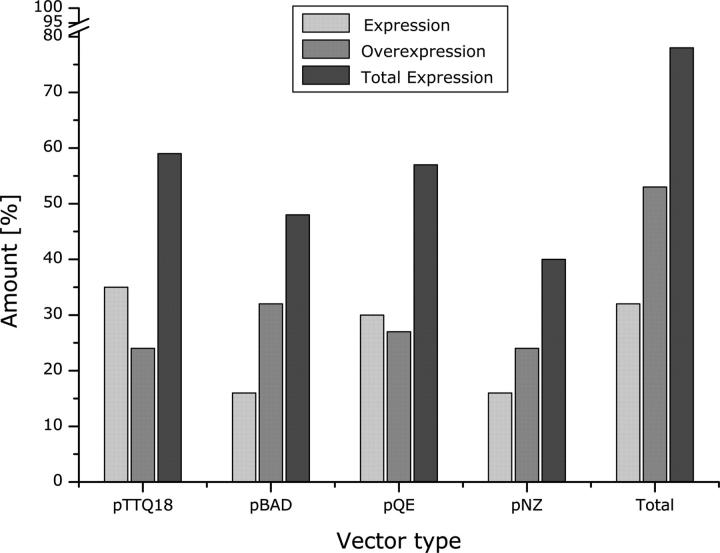
One important consideration for optimizing protein production is the choice of expression vector. In the present work, three E. coli expression vectors with different promoters were compared. When all vectors were tested, 78% of the targets were expressed in total, with 32% of targets expressed and 46% overexpressed (Fig. 4). Both numbers are significantly higher than those for the individual vectors alone. Therefore, testing two or more vectors clearly results in a higher percentage of protein production.
Figure 4.
Comparison of production levels by expression vector. The number of transporter genes that showed expression or overexpression in each vector was determined from the data in Table 1, and was divided by the total number of transporters tested in that vector to generate the percentages shown in this graph. All 37 targets were tested for expression in the pTTQ18 and pBAD vectors, 36 targets were tested in the pQE vector, and 25 targets were tested in the pNZ8048 vector.
When considering individual vectors, the pTTQ18 vector shows the greatest number of targets expressed, while the pBAD vector gives a higher percentage of targets that are overexpressed (Fig. 4). Including more than one vector in our expression screen greatly increased the percentage of overall expression. While 22 targets in total were successfully expressed using the pTTQ18 vector, adding one additional vector to the screen improves expression by five to six more targets, an increase of ∼25%. Thus, while there is no single vector that is the best in all cases, using multiple vectors can greatly increase the number of proteins produced, and may be worthwhile depending on the goal of the project. It should be noted that in some cases testing the different vectors also required the use of different E. coli host strains, which may also have effects on production levels, as seen previously (Eshaghi et al. 2005). Therefore, the results presented here should be considered applicable to certain vector and cell strain combinations (see Materials and Methods), and not the vectors alone.
In addition to considering the individual vectors, one can also analyze the effect of tag position on expression. We attempted expression using two sets of tags—the A version, with only a C-terminal tag, and the C version, with both N and C terminal tags. Overall, the use of the A vector resulted in a higher percentage of expression than the C vector, with 73% of targets expressed in the A vector, and 62% in the C vector (Fig. 5). For the individual vectors, only the pQE vector showed a different distribution. Using the A vector resulted in overall expression of 27 proteins, while adding the C vector enhanced production by only three targets, which represents an increase of 11%. Thus, from our limited sample size, it would seem that in most cases, screening different vectors is more advantageous than screening different tag positions.
The concept of expression space
The overall goal of our structural genomics project is to produce one or more proteins from each transporter family in order to have a large set of nonhomologous targets for structural studies. To evaluate the contribution of different variables in achieving this goal, we have incorporated the data in a simple Venn diagram to evaluate the effect of each variable on increasing “expression space coverage.” We define the term expression space as the number of protein families that is successfully expressed under certain conditions as compared to the total number attempted. By evaluating each variable for its effect on expression space coverage, one is able to easily assess the utility of varying different conditions at the level of protein production studies.
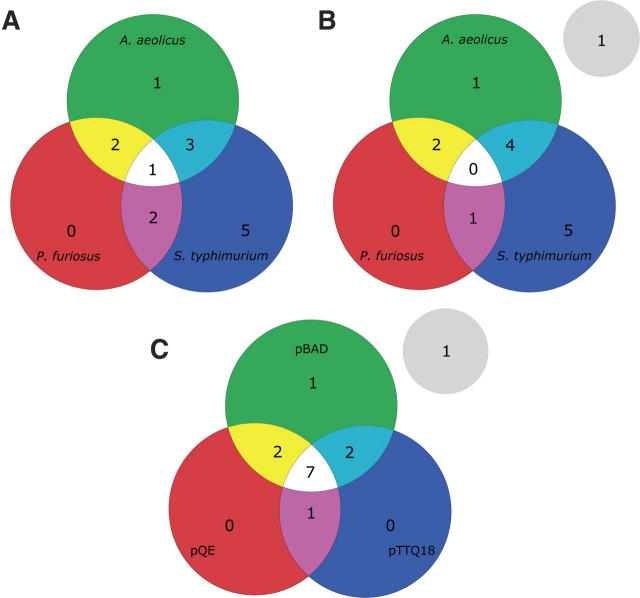
In Figure 6A,B, an expression space diagram was created from the data in Table 1 to determine whether including target proteins from different source organisms has increased expression space coverage. The diagram in Figure 6A shows the distribution of the 14 chosen transporter families in the three chosen source organisms, while the diagram in Figure 6B shows how many of these families were successfully produced by testing proteins from each source organism. Most of the families were successfully produced from each organism in which they are represented with two exceptions. The AGCS family, which is present in P. furiosus and S. typhimurium, was not expressed at all under this set of conditions and hence in Figure 6B this family has moved to the gray circle, which represents nonexpression space. In addition, from each source organism a transporter belonging to the MFS family was tested for expression. However, only the S. typhimurium and A. aeolicus homologs were expressed. Therefore, in Figure 6B the MFS family has changed location from the central white to the cyan colored region.
Figure 6.
Analysis of expression space coverage. (A) Venn diagram of the predicted expression space covered by using targets from three different source organisms, A. aeolicus, P. furiosus, and S. typhimurium. (B) Venn diagram of the experimentally determined expression space coverage showing the number of families that were successfully expressed by using targets from each organism. (C) Venn diagram of the expression space covered by using three different expression vectors. The diagram represents the number of transporter families whose targets were successfully expressed in each vector.
From Figure 6A,B, it is now clear that choosing more than one source organism has increased the expression space coverage of different transporter families. S. typhimurium, which occupies 77% of expression space, can be considered the most successful source organism for protein production. Including targets from A. aeolicus has increased the expression space coverage by 23% over S. typhimurium alone. The inclusion of targets from P. furiosus in addition to the other two organisms does not expand the expression space coverage. However, if only P. furiosus and S. typhimurium are considered, then the inclusion of P. furiosus also results in 15% more expression space coverage. Thus, even with the small sample size considered here, including orthologs from at least two different organisms can have a dramatic effect on the number of nonhomologous proteins produced.
Figure 6C shows a similar diagram used to evaluate whether testing expression in more than one E. coli vector has increased expression space coverage. The diagram shows how many transporter families were successfully expressed in each vector. As mentioned above, no member of the AGCS family was expressed under any condition, and therefore this family is represented in the gray circle, as in Figure 6B. From the distribution of the 13 other families, it is clear that all but one could be expressed using the pBAD vector. The addition of either the pQE or the pTTQ18 vector increases expression space coverage by 10%. Therefore, in our case, expression trials with a variety of vectors may not provide sufficient gains in the production of nonhomologous targets to justify the extra effort and resources needed for testing.
Discussion
Production of membrane proteins remains one of the key bottlenecks in membrane protein structure research. To achieve high-throughput production of membrane proteins in sufficient amounts, it is necessary to more accurately determine which classes of targets, expression systems, and vectors lead to increases in both the number of nonhomologous proteins produced and the total amount of protein produced. Here, we have evaluated the heterologous production of 37 targets from prokaryotic or archaeal organisms in several different ways, including the novel approach of expression space coverage. From these analyses it is clear that E. coli is able to express higher numbers of heterologous prokaryotic transporters at higher levels than L. lactis, and can therefore be considered a superior host organism for these types of proteins. Most important, ortholog screening is the most successful method for increasing the number of nonhomologous proteins that are produced, while comparing production in different expression vectors may also increase the total number of proteins produced. Our results show that by choosing an appropriate expression vector or vectors and a diverse group of nonhomologous targets, production rates comparable to those reported for soluble protein structural genomics could also be achieved for membrane proteins.
Several other recent studies have reported findings on structural genomics efforts for prokaryotic membrane proteins. One such study (Korepanova et al. 2005) has evaluated the production of 99 membrane proteins from Mycobacterium tuberculosis, expressed in E. coli with the pET vector system, which regulates expression using the T7 promoter. The authors report successful production of 64% of the proteins with three or more predicted transmembrane helices, a result similar to that described here. Another study has attempted the production of 45 small membrane proteins from T. maritima in E. coli with a single vector and single tag system (Columbus et al. 2006). Only 22% of these proteins were successfully produced. However, as most of the proteins included in this study contain relatively small numbers (0–4) of predicted transmembrane helices, this cannot be directly compared to the results presented here, in which the chosen targets all have at least three, and most have eight or more, predicted transmembrane helices, and therefore will most likely exhibit different expression behavior.
Two other studies have also attempted to compare the use of different vectors, tags, and expression strains in the production of prokaryotic integral membrane proteins. One study (Eshaghi et al. 2005) has analyzed the production of 49 E. coli proteins in E. coli using a series of three vectors with Gateway recombination sites, a T7 promoter and two lac operator binding sites for strong repression (Tobbell et al. 2002), which were modified with different N-terminal tags and a C-terminal poly-His tag. In addition, several different E. coli host strains were compared. In this study, the level of protein production was determined after solubilization of cell lysate with the detergent Fos-choline 12. Similar to our results, 71% of these proteins were successfully produced, although for any single vector tested the number of targets produced is ∼50% (Eshaghi et al. 2005).
A second study (Dobrovetsky et al. 2005) has characterized the expression of 203 targets from E. coli and 77 targets from the hyperthermophile T. maritima, using E. coli as an expression host. This study employed the pET expression vector, and all targets were tagged at the N terminus with a poly-His tag. The level of protein production for each target was measured after solubilization of the proteins from the cell pellet after lysis using the detergent Triton X-100. In contrast to the study described above, the overall expression rate for all of the targets in this study was 30%.
Fifteen E. coli proteins were part of both of the studies described above. When comparing expression data for the matching E. coli genes, the results are the same for only three of these 15 genes (Dobrovetsky et al. 2005; Eshaghi et al. 2005). Out of the remaining 12 proteins, nine were shown to express in the first study and not in the second, and for three of these proteins the results were vice versa. It is unlikely that these differences result from the different vectors used, as the two vectors drive gene expression from the same promoter. However, the additional lac operator site in the vector used in the first study may allow for tighter gene regulation, which may increase protein production levels. The lower level of overall production found in the second study may be a result of false negatives due to the fact that many membrane proteins are not efficiently solubilized by Triton X-100 (S. Surade, M. Klein, P. Stolt-Bergner, and C. Muenke, unpubl.; Eshaghi et al. 2005). In addition, the different tag positions utilized in the two studies may also have an effect on protein production.
In our study, we show an overall expression rate of 50%–60% for any individual vector (59% pTTQ18, 48% pBAD, 57% pQE), and 76% total expression for all vectors. As our results were determined from measuring protein levels in whole cell lysates, they do not depend on an additional membrane solubilization step. These percentages are in agreement with those established for individual vectors in the work from Eshaghi et al. (2005), and also with the rate of expression found by Korepanova et al. (2005), which supports the conclusion that high-throughput overproduction of integral membrane proteins can be achieved with an overall expression rate of at least 50%.
What strategy should be taken to achieve high-throughput production of membrane proteins? Obviously, for larger sets of targets screening multiple vectors and/or expression systems will become unmanageable. It is clear from our work and that of others that E. coli is a robust production host for both homologous and heterologous prokaryotic integral membrane proteins (Dobrovetsky et al. 2005; Eshaghi et al. 2005; Korepanova et al. 2005), and will be suitable for high-throughput production. However, it is difficult to describe a single E. coli expression vector as ideal for membrane protein production, as no vector evaluated thus far can be singled out as having overwhelming advantages. For our project, in which the goal is to express at least one protein from each transporter family, the expression space analysis from Figure 6C suggests that screening expression in the pBAD vector is a good approach. However, as should be clear from the above discussion, various protein production strategies with similar vectors can lead to different results, and it is unlikely that one particular vector will always give the best results. Thus, when feasible a choice of two or more different vectors may be more appropriate. Based on this study, the pTTQ18 and pQE vectors performed equally in the expression space analysis (Fig. 6C). While the pTTQ18 vector is more successful in expressing targets that the pBAD vector does not express, the pQE vector is better at overexpressing targets expressed at low levels from the pBAD vector (Table 1). Therefore, either one of these vectors would be complementary to the pBAD vector.
In addition, our results tentatively suggest that designing constructs with only C-terminal tags may lead to higher membrane protein production levels (Fig. 5), although studies including a larger number of targets are necessary to confirm this result. This also implies that screening more than one tag or tag position may not be as advantageous as screening different vectors, as in our study (Fig. 5) and in one described study (Eshaghi et al. 2005), adding one additional type of tag led to only an ∼10% increase in the number of proteins produced versus a minimum 25% increase seen in our study when adding one additional vector. However, it is noteworthy that an increase from 50% to 70% in the total number of proteins produced was also achieved in the described study by screening one vector with three types of tags in three E. coli host strains (Eshaghi et al. 2005).
Furthermore, as seen by our expression space analysis, screening of orthologs will prove useful in identifying target proteins from a particular membrane protein family that can be produced at high levels, as has been seen for soluble proteins (Savchenko et al. 2003). Even with the relatively small sample size used here, we were able to increase expression space by >20% by including targets from two different organisms. Thus, a good strategy for membrane protein target selection should be to choose the majority of genes from an organism that is compatible with the host organism, or whose proteins are of particular interest, and then add orthologs from other organisms.
We believe this is the first example of a membrane protein expression screen in which a variety of different variables were tested and analyzed for their effectiveness in this manner. By modifying the definition of expression space to fit the goal of each individual project, this type of straightforward analysis could prove helpful in evaluating the usefulness of variables tested in other structural genomics initiatives as well. While membrane proteins are much more difficult targets than soluble proteins, our results suggest strategies that will be valuable for the overproduction of membrane proteins, and that production of membrane proteins at sufficient levels for structural studies is certainly possible.
Materials and methods
Expression vectors
For E. coli expression, the pTTQ18 (a kind gift from P. Henderson, University of Leeds, United Kingdom; Stark 1987), pBAD (Invitrogen), and pQE (Qiagen) vectors were used and for L. lactis the pNZ8048 vector (a kind gift from L. Schmidt, Heinrich Heine University, Duesseldorf, Germany; Kunji et al. 2003) was used. The expression vectors were modified at the multiple cloning site to yield recombinant proteins with either a C-terminal poly-His tag preceded by a TEV cleavage site (A-variant), or an N-terminal poly-His tag combined with a TEV-cleavage site and a C-terminal Strep tag-II (IBA; C-variant).
Cloning of genes
Coding sequences for the proteins on the target list were obtained from the PEDANT, NCBI, or TIGR databases and used for primer design. Genes encoding the target proteins were amplified by PCR using the Phusion DNA polymerase (Finnzymes), genomic DNA from S. typhimurium (a kind gift from A. Kahnert and S.H.E. Kaufmann, Max Planck Institute for Infection Biology, Berlin, Germany), A. aeolicus, or P. furiosus (H. Huber, Regensburg University, Germany) and primers containing at least 15 gene-specific nucleotides (Invitrogen). Resulting PCR products were digested and ligated into the appropriate expression vector. Resulting constructs were used to transform chemically competent E. coli DH5α cells for identification of positive clones by colony PCR and preparation of plasmid DNA. In some cases, the PCR products were first cloned into the pDrive Vector (Qiagen) and then subcloned into the expression vectors.
In the case of the L. lactis expression system, the ligations were transformed by electroporation into electrocompetent L. lactis NZ9000 (a kind gift from L. Schmidt, Heinrich Heine University, Duesseldorf, Germany; Kunji et al. 2003), and were incubated for 2 h in M17 medium (Difco) supplemented with 0.5% glucose, 0.5 M sucrose, 20 mM MgCl2, and 2 mM CaCl2 at 30°C before plating on M17 agar containing 0.5% glucose, 0.5 M sucrose, and 5 μg/mL chloramphenicol. Plasmid DNA was isolated using the QIAprep Spin Miniprep Kit (Qiagen) according to the manufacturer's protocol modified with an additional lysis step by incubating the resuspended cell pellet with lysozyme to a final concentration of 10 μg/μL for 15 min at 55°C.
Due to the large number of clones necessary for this study, if positive clones were not obtained after two attempts, efforts at cloning those constructs were abandoned.
Protein overproduction
The resulting expression constructs were used to transform E. coli C43(DE3) (Avidis; Miroux and Walker 1996; pTTQ18 and pQE vectors), NM554 (Stratagene; pTTQ18 and pQE vectors) and TOP10 or LMG194 (Invitrogen; pBAD vector) strains containing the pRARE plasmid (Novagen). Transformants were selected on LB agar plates containing 50 μg/mL carbenicillin and 34 μg/mL chloramphenicol.
For protein production, E. coli cells from overnight cultures from single colonies were transferred into 2 mL fresh LB in 24-well plates at 37°C using a 1:50 to 1:20 dilution and were grown until the cultures reached an OD600 of 0.6. The cultures then were induced with 0.5 mM isopropyl-β-1-thiogalactoside (IPTG; pTTQ18, pQE) or 0.02% arabinose (pBAD) and incubated for 5 h or cooled down to 20°C and induced overnight. The cells were harvested by centrifugation and finally stored at −20°C. Expression at either 37°C, 20°C, or both was recorded as positive expression.
The L. lactis cells were grown in a similar manner in 24-well plates at 30°C using M17-medium supplemented with 0.5%–1.0% glucose and 5 μg/mL chloramphenicol until the cultures reached the OD660 of 0.8. The cultures were then induced with ∼10 μg of nisin from culture supernatant produced by the L. lactis strain NZ9700 (a kind gift from L. Schmidt, Heinrich Heine University, Duesseldorf, Germany; Kunji et al. 2003), incubated for 3 h, cooled down to 4°C and incubated overnight. Cells from 1 mL fractions were harvested by centrifugation and finally stored at −20°C.
Cell lysis and sample analysis
For dot blot analysis the E. coli cell pellets obtained from 1 mL of culture were resuspended in 100 μL of lysis buffer containing 1 U/mL benzonase, 1 mM MgCl2, 200 μg/mL lysozyme, 50 mM Tris/HCl (pH 8), 1 mM PMSF, and 1 mM EDTA and incubated for 15 min at 20°C. Then, 300 μL of 8 M guanidinium hydrochloride solution were added followed by another 15 min incubation. The lysate was then centrifuged to remove the cell debris. For dot blot analysis, 150 μL of the supernatant were applied to the PVDF membrane (Millipore). For Western blot analysis the frozen E. coli cell pellets were resuspended in 50 μL of H2O and cell lysis and protein extraction were performed by adding 150 μL of a 20% SDS solution for 15 min at room temperature. The suspension was centrifuged and the supernatant was analyzed by SDS-PAGE and Western blot.
Membrane preparation and solubilization trials
E. coli cell pellets obtained from 50 mL of culture were resuspended in 1 mL of lysis buffer containing 1 U/mL benzonase, 1 mM MgCl2, 200 μg/mL lysozyme, 50 mM HEPES (pH 8.0), 1 mM PMSF, and 1 mM EDTA. One milliliter of glass beads (0.2–0.5 mm diameter) was added and the sample was vortexed for 10 min. After removal of the glass beads by filtration, the lysate was then centrifuged at 10,000g to remove the cell debris. The membranes were harvested by 1 h centrifugation at 100,000g and resuspended in a buffer containing 50 mM HEPES (pH 8) and 300 mM NaCl to a final concentration of 10 mg/mL. For the solubilization trials, 50 μL of the same buffer additionally containing 4% of DDM (Glycon Biochemicals) or Fos-12 (SynphaBase AG), respectively were added to 50 μL of the membrane solution. The extraction was carried out at 4°C for 1 h followed by another ultracentrifugation step. Fifteen microliters of the supernatant were analyzed by Western blot.
Dot blot and Western blot analysis
The poly-His tagged proteins were detected using a monoclonal α-poly-histidine-alkaline phosphatase conjugated antibody (Sigma) according to the manufacturer's protocols. The signals were detected using the colorimetric BCIP-NBT detection system. For quantification of the signal, a concentration curve of 0, 10, and 100 ng tagged eGFP was used as a standard to estimate the protein levels on the dot blot, and 10 and 100 ng were used on the Western blot.
Acknowledgments
We thank Dr. Terukazu Nogi and Dr. Jiang-bi Xie for assistance during the initial stages of the project. We also thank Hannelore Mueller for excellent technical assistance. In addition, we thank Dr. Edmund Kunji (MRC, Cambridge, England) and the laboratory of Dr. Lutz Schmidt (Heinrich Heine University, Duesseldorf, Germany) for helpful discussions on the use of Lactococcus lactis for protein production. This project was supported by funding from the Bundesministerium fuer Bildung und Forschung (ProAMP: Proteome-wide Analysis of Membrane Proteins), the Max-Planck-Gesellschaft, the Deutsche Forschungsgemeinschaft (SFB 628: Functional Membrane Proteomics), the Fonds der Chemischen Industrie, and the European Membrane Protein Consortium (E-MeP). P.C.S. was supported by an EMBO long-term fellowship for part of this work.
Footnotes
Reprint requests to: Dr. Hartmut Michel, Department of Molecular Membrane Biology, Max Planck Institute for Biophysics, Max-von-Laue Str. 3, D-60438 Frankfurt/M., Germany; e-mail: hartmut.michel@mpibp-frankfurt.mpg.de; fax: +49-69-6303-1002.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062312706.
References
- Andre N., Cherouati N., Prual C., Steffan T., Zeder-Lutz G., Magnin T., Pattus F., Michel H., Wagner R., Reinhart C. 2006. Enhancing the functional production of G protein-coupled receptors in Pichia pastoris to levels required for structural studies via a single expression screen. Protein Sci. 15: 1115–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arechaga I., Miroux B., Karasch S., Huijbregts R., de Kruijff B., Runswick M.J., Walker J.E. 2000. Characterization of new intracellular membranes in Escherichia coli accompanying large scale over production of the B subunit of F(1)F(0) ATP synthase. FEBS Lett. 482: 215–219. [DOI] [PubMed] [Google Scholar]
- Braun P., Hu Y., Shen B., Halleck A., Koundinya M., Harlow E., LaBaer J. 2002. Proteome-scale purification of human proteins from bacteria. Proc. Natl. Acad. Sci. 99: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busso D., Poussin-Courmontagne P., Rose D., Ripp R., Litt A., Thierry J.-C., Moras D. 2005. Structural genomics of eukaryotic targets at a laboratory scale. J. Struct. Funct. Genomics 6: 81–88. [DOI] [PubMed] [Google Scholar]
- Chandonia J.-M. and Brenner S.E. 2006. The impact of structural genomics: Expectations and outcomes. Science 311: 347–351. [DOI] [PubMed] [Google Scholar]
- Christendat D., Yee A., Dharamsi A., Kluger Y., Savchenko A., Cort J.R., Booth V., Mackereth C.D., Saridakis V., Ekiel I. et al. 2000. Structural proteomics of an archaeon. Nat. Struct. Biol. 7: 903–909. [DOI] [PubMed] [Google Scholar]
- Columbus L., Lipfert J., Klock H., Millet I., Doniach S., Lesley S.A. 2006. Expression, purification, and characterization of Thermotoga maritima membrane proteins for structure determination. Protein Sci. 15: 961–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovetsky E., Lu M.L., Andorn-Broza R., Khutoreskaya G., Bray J.E., Savchenko A., Arrowsmith C.H., Edwards A.M., Koth C.M. 2005. High-throughput production of prokaryotic membrane proteins. J. Struct. Funct. Genomics 6: 33–50. [DOI] [PubMed] [Google Scholar]
- Drew D.E., von Heijne G., Nordlund P., de Gier J.-L. 2001. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 507: 220–224. [DOI] [PubMed] [Google Scholar]
- Eshaghi S., Hedren M., Nasser M.I.A., Hammarberg T., Thornell A., Nordlund P. 2005. An efficient strategy for high-throughput expression screening of recombinant integral membrane proteins. Protein Sci. 14: 676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisshammer R. and Tate C.G. 1995. Overexpression of integral membrane proteins for structural studies. Q. Rev. Biophys. 28: 315–422. [DOI] [PubMed] [Google Scholar]
- Guzman L., Belin D., Carson M., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Bussow K., Mueller U., Umbach P. 2003. Facilities and methods for the high-throughput crystal structural analysis of human proteins. Acc. Chem. Res. 36: 157–163. [DOI] [PubMed] [Google Scholar]
- Hockney R.C. 1994. Recent developments in heterologous protein production in Escherichia coli. Trends Biotechnol. 12: 456–463. [DOI] [PubMed] [Google Scholar]
- Hopkins A.L. and Groom C.R. 2002. The druggable genome. Nat. Rev. Drug Discov. 1: 727–730. [DOI] [PubMed] [Google Scholar]
- Kiefer H. 2003. In vitro folding of α-helical membrane proteins. Biochim. Biophys. Acta 1610: 57–62. [DOI] [PubMed] [Google Scholar]
- Kiefer H., Krieger J., Olszewski J.D., von Heijne G., Prestwich G.D., Breer H. 1996. Expression of an olfactory receptor in Escherichia coli: Purification, reconstitution, and ligand binding. Biochemistry 35: 16077–16084. [DOI] [PubMed] [Google Scholar]
- Korepanova A., Gao F.P., Hua Y., Qin H., Nakamoto R.K., Cross T.A. 2005. Cloning and expression of multiple integral membrane proteins from Mycobacterium tuberculosis in Escherichia coli. Protein Sci. 14: 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunji E.R.S., Slotboom D.-J., Poolman B. 2003. Lactococcus lactis as host for overproduction of functional membrane proteins. Biochim. Biophys. Acta 1610: 97–108. [DOI] [PubMed] [Google Scholar]
- Kunji E.R., Chan K.W., Slotboom D.J., Floyd S., O'Connor R., Monne M. 2005. Eukaryotic membrane protein overproduction in Lactococcus lactis. Curr. Opin. Biotechnol. 16: 546–551. [DOI] [PubMed] [Google Scholar]
- Lesley S.A., Kuhn P., Godzik A., Deacon A.M., Mathews I., Kreusch A., Spraggon G., Klock H.E., McMullan D., Shin T. et al. 2002. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. 99: 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan C.-H., Qiu S., Finley J.B., Carson M., Gray R.J., Huang W., Johnson D., Tsao J., Reboul J., Vaglio P. et al. 2004. High-throughput expression of C. elegans proteins. Genome Res. 14: 2102–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J., von Heijne G., Houben E., de Gier J.-W. 2005. Biogenesis of inner membrane proteins in Escherichia coli. Annu. Rev. Microbiol. 59: 329–355. [DOI] [PubMed] [Google Scholar]
- Ma C., Marassi F.M., Jones D.H., Straus S.K., Bour S., Strebel K., Schubert U., Oblatt-Montal M., Montal M., Opella S.J. 2002. Expression, purification, and activities of full-length and truncated versions of the integral membrane protein Vpu from HIV-1. Protein Sci. 11: 546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan K.S., Borges-Walmsley M.I., Walmsley A.R. 2002. Microbial and viral drug resistance mechanisms. Trends Microbiol. 10: s8–s14. [DOI] [PubMed] [Google Scholar]
- Miroux B. and Walker J.E. 1996. Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. Escherichia coli: 260: 289–298. [DOI] [PubMed] [Google Scholar]
- Monne M., Chan K.W., Slotboom D.-J., Kunji E.R.S. 2005. Functional expression of eukaryotic membrane proteins in Lactococcus lactis. Protein Sci. 14: 3048–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q., Kang K.H., Paulsen I.T. 2004. TransportDB: A relational database of cellular membrane transport systems. Nucleic Acids Res. 32: D284–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogl H., Kosemund K., Kuhlbrandt W., Collinson I. 1998. Refolding of Escherichia coli produced membrane protein inclusion bodies immobilized by nickel chelating chromatography. FEBS Lett. 432: 21–26. [DOI] [PubMed] [Google Scholar]
- Savchenko A., Yee A., Khachatryan A., Skarina T., Evdokimova E., Pavlova M., Semesi A., Northey J., Beasley S., Lan N. et al. 2003. Strategies for structural proteomics of prokaryotes: Quantifying the advantages of studying orthologous proteins and of using both NMR and X-ray crystallography approaches. Proteins 50: 392–399. [DOI] [PubMed] [Google Scholar]
- Siegele D.A. and Hu J.C. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. 94: 8168–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M.J.R. 1987. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene 51: 255–267. [DOI] [PubMed] [Google Scholar]
- Tobbell D.A., Middleton B.J., Raines S., Needham M.R.C., Taylor I.W.F., Beveridge J.Y., Abbott W.M. 2002. Identification of in vitro folding conditions for procathepsin S and cathepsin S using fractional factorial screens. Protein Expr. Purif. 24: 242–254. [DOI] [PubMed] [Google Scholar]
- Wang D.-N., Safferling M., Lemieux M.J., Griffith H., Chen Y., Li X.-D. 2003. Practical aspects of overexpressing bacterial secondary membrane transporters for structural studies. Biochim. Biophys. Acta 1610: 23–36. [DOI] [PubMed] [Google Scholar]
- Ward A., Hoyle C., Palmer S., Morrison S., Langton K., O'Reilly J., Rutherford N., Griffith J., Pos M., Poolman B. et al. 2001. Prokaryotic multidrug efflux proteins of the major facilitator superfamily: Amplified expression, purification, and characterization. In Microbial Multidrug Efflux (eds. Paulsen I.T. and Lewis K.) . pp. 123–141. Horizon Scientific Press, Wymondham, UK. [PubMed]
- Wheeler D.L., Chappey C., Lash A.E., Leipe D.D., Madden T.L., Schuler G.D., Tatusova T.A., Rapp B.A. 2000. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 28: 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee A., Chang X., Pineda-Lucena A., Wu B., Semesi A., Le B., Ramelot T., Lee G.M., Bhattacharyya S., Gutierrez P. et al. 2002. An NMR approach to structural proteomics. Proc. Natl. Acad. Sci. 99: 1825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]