Abstract
Reducing body myopathy (RBM) is a rare disorder causing progressive muscular weakness characterized by aggresome-like inclusions in the myofibrils. Identification of genes responsible for RBM by traditional genetic approaches has been impossible due to the frequently sporadic occurrence in affected patients and small family sizes. As an alternative approach to gene identification, we used laser microdissection of intracytoplasmic inclusions identified in patient muscle biopsies, followed by nanoflow liquid chromatography–tandem mass spectrometry and proteomic analysis. The most prominent component of the inclusions was the Xq26.3-encoded four and a half LIM domain 1 (FHL1) protein, expressed predominantly in skeletal but also in cardiac muscle. Mutational analysis identified 4 FHL1 mutations in 2 sporadic unrelated females and in 2 families with severely affected boys and less-affected mothers. Transfection of kidney COS-7 and skeletal muscle C2C12 cells with mutant FHL1 induced the formation of aggresome-like inclusions that incorporated both mutant and wild-type FHL1 and trapped other proteins in a dominant-negative manner. Thus, a novel laser microdissection/proteomics approach has helped identify both inherited and de novo mutations in FHL1, thereby defining a new X-linked protein aggregation disorder of muscle.
Introduction
Reducing body myopathy (RBM) is a rare muscle disorder causing progressive muscular weakness and characteristic intracytoplasmic inclusions in myofibers, first described more than 30 years ago (1). Clinical presentations of RBM have ranged from early onset fatal to childhood onset to adult onset cases (1–12). It is noteworthy that the most severe cases frequently have included girls (13). In the rare familial cases on record, male-to-male transmission has never been documented (11, 13, 14). The typical histopathological findings in muscle biopsy specimens are intracytoplasmic inclusion bodies (Figure 1A) that reduce nitro-blue tetrazolium (NBT) and thus stain strongly with the menadione-NBT stain (Figure 1B and Supplemental Figure 2B; supplemental material available online with this article; doi:10.1172/JCI34450DS1). The inclusions are found throughout the cytoplasm but have been observed to be frequently positioned adjacent to a myonucleus (7, 13–16). Immunohistochemical analysis of muscle samples from RBM patients has suggested that the inclusions have features of aggresomes, structures formed following activation and overflow of the unfolded protein response and the ubiquitin-proteasome system, similar to inclusions seen in a number of neurodegenerative conditions (17–19). Notably, the inclusions labeled prominently for the intermediate filament desmin, ubiquitin, the luminal endoplasmic reticulum chaperone GRP78, as well as for γ-tubulin, all of which are consistent with aggresome formation (18). Although a large number of diverse proteins have also been reported to be contained within the inclusions in muscle, their relative role, if any, in the generation of the aggregates has remained unclear (2, 7, 13, 16, 18). Immunohistochemical analysis also does not allow for a determination of the relative contribution of each protein to suggest its potential importance in the pathogenesis of the inclusions. Small family sizes and the frequent occurrence of sporadic cases have made genetic linkage analysis not feasible, while candidate gene analysis has been unsuccessful (18).
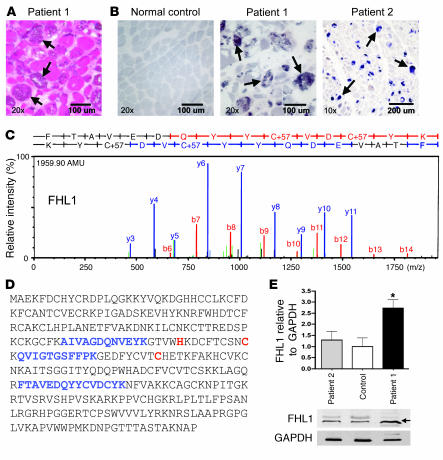
Figure 1. General histology, menadione-NBT stain and selected MS/MS spectra of FHL1.
(A and B) H&E stain of the muscle biopsy of patient 1 with evident intracytoplasmic inclusions (A, arrows) and menadione-NBT staining of normal muscle and of muscle biopsy sections from patients 1 and 2 (B) demonstrating positive staining of reducing bodies in both biopsies (arrows). (C) MS/MS spectra with at least 1 continuous y ion or b ion series of greater than 5 residues. (D) Amino acid sequence of human FHL1. FHL1 peptide fragments identified in reducing bodies using MS are shown in blue, localizations of the mutations in the patients in red. (E) Immunoblot analysis of FHL1 in muscle biopsy material from patients 1 and 2. There is a 2.7-fold higher protein content of FHL1 (arrow) in patient 1 and a slightly higher FHL1 protein content in patient 2. Densitometry normalized to GAPDH. *P < 0.001.
FHL1 is a 32-kDa protein containing an N-terminal zinc finger/half Lin-11, Isl-1, Mac-3 (LIM) domain, followed by 4 complete LIM domains. This arrangement of domains has led to the designation of this class of proteins as four and a half LIM domain (FHL) proteins. LIM domains are cysteine-rich, tandem zinc-finger protein interaction motifs, first recognized in the 3 homeodomain transcription factors Lin-11, Isl-1, and Mac-3 (20–24). Of the 5 known members constituting the FHL family (FHL 1–4 and CREM in testis [ACT]), FHL1–3 are expressed in striated and cardiac muscle (25–28). FHL proteins in general appear to be involved in cytoskeletal scaffolding as well as in regulation of transcription factors. Most information about cellular interactions and functions has been assembled for FHL2, for which interaction partners and subcellular localization with both cytoplasmic as well as nuclear functions have been well documented (26). FHL1 in skeletal muscle localizes to the sarcomere and also to the sarcolemma and is believed to participate in muscle growth and differentiation as well as in assembly of the sarcomere (27). Known interactions of FHL1 include the slow and cardiac isoforms of myosin binding protein C (MyBP-C) as well as interactions with ERK2, HPC2, RING1, and KBP1 (27, 29–32). There appear to be 3 basic isoforms of FHL1, generated by alternative splicing (25, 33). FHL1A, also known as skeletal muscle LIM protein 1 (SLIM1), is the full-length protein; FHL1B, or SLIMMER, contains LIM domains 1–3 plus nuclear localization and export signals and an RBP-J binding region; the shortest isoform, FHL1C or KyoT2, only contains LIM domains 1 and 2 plus an RBP-J binding region, but no nuclear localization signal (30). KyoT2 also interacts with PIAS1 (34).
Here we use laser microdissection followed by proteomic analysis to identify the X chromosomal FHL1 as the most prominent protein in the aggregates. Previously FHL1 had not been known to be part of the inclusions. We detected mutations in highly conserved zinc-coordinating histidine and cysteine residues in the second LIM domain of FHL1 in 2 female sporadic patients and two familial cases. We provide evidence that RBM-associated FHL1 mutations precipitate the formation of intracytoplasmic aggregations, trapping both mutant as well as wild-type FHL1 together with various other proteins involved in normal muscle function and cellular responses to misfolded proteins. Therefore, RBM can now be understood as an X-linked protein aggregation disorder of muscle caused by mutation in FHL1.
Results
Proteomic identification of FHL1 as a major component of the intracytoplasmic inclusions.
We decided on an unbiased approach to determine the predominant component contained within the reducing bodies by isolating material directly from the inclusions contained in frozen sections of muscle biopsies from 2 severely affected sporadic female patients using laser capture microdissection (LCM). The captured material was subjected to trypsin digestion followed by nanoflow liquid chromatography–tandem mass spectrometry (liquid chromatography–MS/MS). The highest scoring peptides identified in the reducing bodies from both patients but not in the material from surrounding or normal muscle belonged to the FHL1 protein (also known as SLIM1 or KyoT), hitherto not known to be a component of reducing bodies (Figure 1, C and D) (18, 27). We also identified peptides belonging to proteins that had been previously identified as components of inclusions in RBM, including desmin, actin, and ubiquitin, thus validating our approach (Supplemental Table 1) (13, 18).
Localization of FHL1 immunoreactivity in patient muscle.
Immunoblotting of FHL1 protein in muscle from the 2 patients showed it to be present at levels greater than or equal to normal muscle (Figure 1E). Immunohistochemistry using antibodies directed against FHL1 confirmed that the intracytoplasmic inclusion bodies were highly enriched for FHL1 immunoreactivity (Figure 2B and Supplemental Figure 2D), whereas it was distributed without focal accumulations in normal muscle sections (Figure 2A and Supplemental Figure 2C). Under higher magnification, the FHL1-positive inclusions were seen throughout myofibers but were often also localized in close association with the myonuclei (Figure 2C), consistent with their appearance on electron microscopy (Figure 2D and Supplemental Figure 2A) and with previous histopathological observations in this condition (7, 13–16). We also confirmed that the intracytoplasmic inclusions labeled positively for other proteins identified in the mass spectrometry analysis, including desmin and ubiquitin (Figure 2, E and F) (2, 7, 13, 16, 18). Further analysis of the localization of FHL1 immunoreactivity within skeletal muscle from the RBM patients showed FHL1 to be localized to the I-band/Z disk in fibers without obvious inclusions, indistinguishable from the localization in normal muscle (Figure 2G). In contrast, in patient fibers containing prominent inclusions there was a severe reduction of FHL1 immunoreactivity at the contractile apparatus in the vicinity of the inclusions, suggesting that the inclusion bodies were depleting the fiber of normally localized FHL1 protein (Figure 2H). This was observed around multiple inclusions, suggesting that it was not solely the result of the frequently advanced pathological alteration of the affected fibers. FHL1 has been shown to bind to slow-type skeletal MyBP-C (27). We found MyBP-C to be a component of the inclusions (Supplemental Figure 2E). However, its localization to the contractile apparatus in RBM patient muscle compared with normal was unchanged (Supplemental Figure 2, F and G).
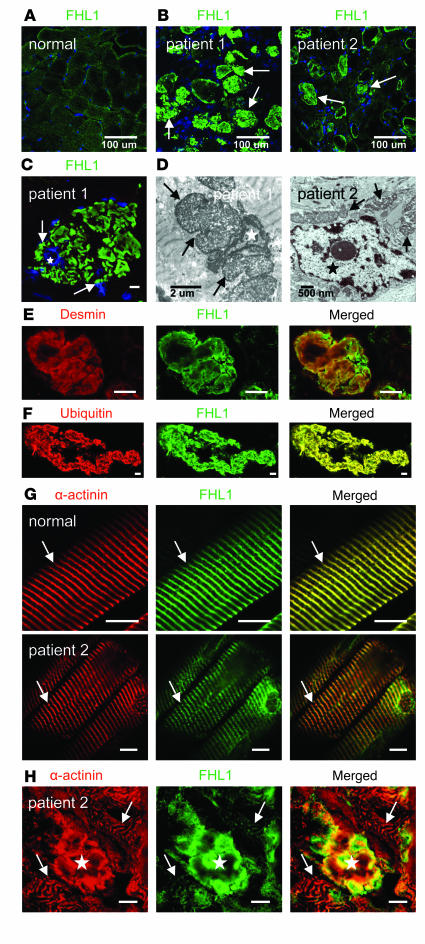
Figure 2. FHL1 localization in muscle from RBM patients.
(A) Distribution of FHL1 immunoreactivity in normal muscle (B) FHL1 immunoreactivity prominently decorates the intracytoplasmic inclusions, which are more numerous in patient 1 (arrows). (C) Higher magnification of FHL1-labeled inclusions (arrows) in the biopsy from patient 1 demonstrates their often close relationship with myonuclei (blue, star), confirmed on electron microscopy (D) where the inclusions (arrows) abut a myonucleus (patient 1) or are close by (patient 2; star indicates myonucleus). (E and F) Inclusions colabel for FHL1 and the intermediate filament desmin (E) as well as ubiquitin (F) (patient 1). (G) In fibers without prominent inclusions, FHL1 localizes properly to the contractile apparatus, mainly locating to the I-line and Z disk, which is colabeled with α-actinin (arrows). (H) In fibers with prominent inclusions, FHL1 immunoreactivity at the contractile apparatus in the vicinity of the inclusion is severely reduced. Star denotes inclusion, arrows point to the contractile apparatus with Z disks labeled with α-actinin (which is also associated with the inclusion). The fiber architecture is also severely distorted (patient 2). Scale bars: 10 μm.
Mutation identification in FHL1 in sporadic and familial patients.
Given that FHL1 is predominantly expressed in skeletal muscle and that it was the major component found in our proteomic analysis, we considered it to be an excellent candidate to be mutated in RBM, hypothesizing that an X-linked dominant-negative mechanism could give rise to the severe disease observed in the sporadic female patients. Direct sequencing of RT-PCR products from skeletal muscle from affected patients covering the entire FHL1 coding sequence revealed heterozygous mutations in both patients (c.367C→T resulting in p.H123Y in patient 1, c.395G→T resulting in p.C132F in patient 2) (Figure 3A and Supplemental Figure 1). Both mutations occur in the second LIM domain of FHL1, affecting invariant consensus residues (Figure 3B) involved in coordinating zinc binding (Figure 3, C and D). The mutations were confirmed by genomic sequencing of the third coding exon of the FHL1 gene and were not present in the parents or siblings of these patients and thus arose de novo in the patients.
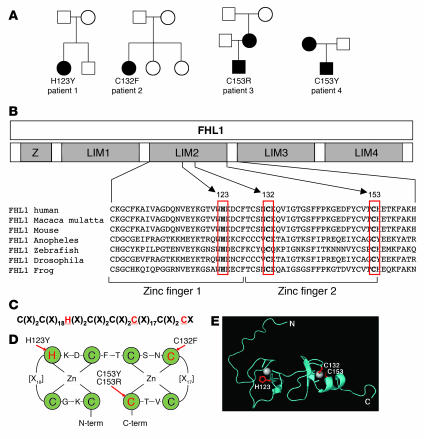
Figure 3. Mutations in FHL1 identified in patients with RBM affect conserved cysteine and histidine residues within the second LIM domain.
(A) Pedigrees of the 4 families with affected members, indicating mutations in FHL1. Closed symbols designate affected individuals. (B) Schematic representation of the domain structure of FHL1 consisting of 4 LIM domains (LIM1–4) with an additional N-terminal half-LIM domain (Z). All 4 mutations are located in the second LIM domain and affect residues (boxed in red) that are absolutely conserved in orthologs from species down to mosquitoes. (C) Location of the mutations (highlighted in red) within the consensus sequence of the LIM domain and its 8 zinc coordinating residues. X denotes any amino acid. (D) Topology of zinc coordination. Green circles indicate zinc-binding residues. Arrows point to mutated residues. (E) Mutated residues (highlighted in red) superimposed on the NMR structure of the second LIM domain of FHL1 (RIKEN; http://www.genome.jp/dbget-bin/www_bget?pdb+1X63). Zinc atoms are indicated as gray spheres. Note that C153 falls into the α-helical domain at the C-terminal end of the LIM domain.
We then studied 2 additional families with biopsy-confirmed RBM in which the index patients were significantly affected boys, although less severely so when compared with the first 2 sporadic female patients in our analysis. Dilative cardiomyopathy was present in one of the boys. Sequencing identified the mutations c.457T→C resulting in p.C153R in patient 3 and c.458G→A resulting in a p.C153Y change in the same amino acid in patient 4 (Figure 3A, Supplemental Figure 1). These additional mutations affected an invariant conserved cysteine residue of the second zinc finger of the second LIM domain (Figure 3, B–D). Both mothers of these 2 male patients were comparatively less severely affected and were heterozygous for the respective mutation, while the available maternal grandparents of patient 3 showed no mutation, indicating that the mutation had arisen de novo in the mother of patient 3. The fact that the mothers of patients 3 and 4 were more mildly affected compared with their sons suggests that the presence of wild-type FHL1 can ameliorate the disease. We also considered the possibility that the milder disease in the mothers was due to skewed X-inactivation preferentially silencing the mutant X chromosome. However, X-inactivation studies on lymphocyte DNA did not detect significant skewing (Supplemental Figure 1E). In contrast, in the severely affected female patient 2, we found markedly skewed X-inactivation of 78% likely preferentially silencing the wild-type X chromosome, as in the muscle, most of the transcript detected on RT-PCR was from the mutant allele (Supplemental Figure 1E), thus perhaps contributing to the severity of the disease in this patient. No significant skewing was evident in patient 1.
Molecular modeling of FHL1 mutations in the LIM2 domain.
Analysis of NMR spectroscopy data on the second LIM domain of FHL1 (RIKEN, http://www.genome.jp/dbget-bin/www_bget?pdb+1X63) allowed mapping of the mutations in relation to the structure of the second LIM domain (Figure 3E). The mutations in the severely affected female patients 1 and 2 would be predicted to result in the complete disruption of the zinc binding sites and collapse of the LIM domain. In contrast, the mutations of C153 identified in patients 3 and 4, while still severely interfering with the structure of the second zinc finger motif of LIM domain 2, are predicted to be less severe due to their localization within the C-terminal α-helix, possibly loosening rather than collapsing the structure.
Transfection of FHL1 mutations H123Y and C123F into COS-7 cells.
To assess the ability of mutant FHL1 to precipitate the characteristic inclusions, we generated tagged expression constructs of the 2 severe mutations H123Y and C132F for transfection into 2 cell types, COS-7 cells and the muscle-derived cell line C2C12. First we used COS-7 cells, which have no detectable endogenous FHL1 expression (27), using GFP- and V5-tagged constructs of the mutations as well as of the wild-type sequence. After 24 hours in culture, wild-type GFP-FHL1–expressing cells developed GFP-positive inclusions only rarely. The rare inclusions detected in the transfection of the wild-type likely form as a result of overexpression of a non-native protein in the COS-7 cells. In contrast, GFP-FHL1 mutant–transfected cells exhibited significantly more frequently dense and large appearing inclusions, which labeled positive with antibodies against FHL1 as well as against GFP (Figure 4A and Supplemental Table 2). FHL1 labeling in most wild-type FHL1 transfected cells was inconspicuous in a lattice-like pattern (Figure 4B), corresponding to the absence of major inclusion in these cells by electron microscopy (Figure 4C). In contrast, electron microscopy of mutant FHL1-expressing COS-7 cells revealed electron-dense inclusions adjacent to the nucleus (Figure 4D), reminiscent of their position in patients (Figure 2D) and of the position of aggresomes (17). However, given the limited cytoplasmic space of COS-7 cells, this position could also be the result of constraint within the cell. To investigate whether aggregates of mutant FHL1 would also include wild-type FHL1, mutant GFP-FHL1 and wild-type V5-FHL1 constructs were cotransfected. Colabeling with anti-GFP and anti-V5 antibodies confirmed that both mutant as well as wild-type FHL1 protein aggregated in the same inclusions (Figure 4E). Thus, mutant FHL1 initiates aggregation and traps wild-type FHL1 in the inclusion bodies, consistent with a dominant-negative mode of action on the wild-type protein.
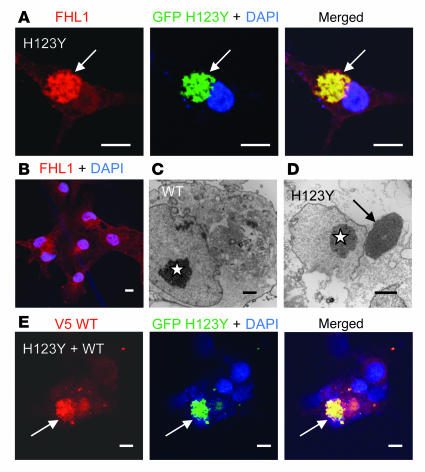
Figure 4. Transfection of mutant FHL1 H123Y and C132F in COS-7 cells.
(A) COS-7 cells transfected with GFP-tagged mutant constructs developed dense juxtanuclear inclusions (arrow), which colabeled with FHL1 antibody. (B) Most COS-7 cells transfected with wild-type FHL1 and labeled for FHL1 generated a lattice-like network within the cells but without clear aggregates in the majority of cells. (C and D) Electron microscopy images of COS-7 cell transfected with wild-type FHL1 without formed inclusions (C) in contrast to an H123Y FHL1 mutant transfected cell with a dense juxtanuclear inclusion (arrow) (D) Stars indicate nuclei. (E) COS-7 cells cotransfected with mutant GFP-tagged construct and wild-type V5-tagged FHL1 constructs. Detection with V5 antibody (red) and GFP antibody (green) shows that both mutant and wild-type constructs aggregated in the same inclusions, indicating that mutant FHL1 traps wild-type FHL1 in the aggregates (arrows). Scale bars: 10 μm (A, B, and E); 2 μm (C and D).
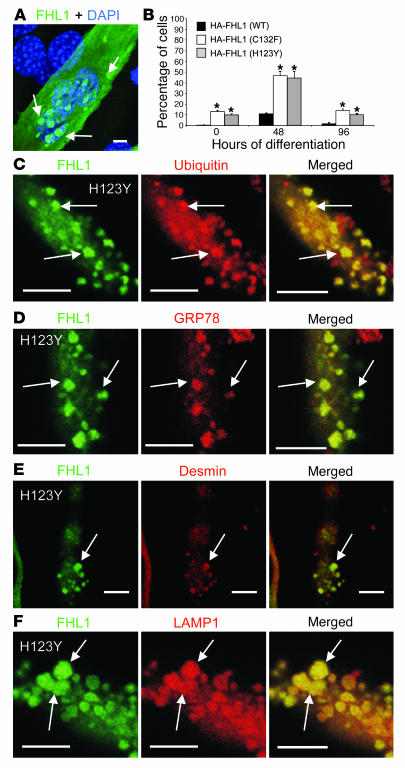
Transfection of FHL1 mutations H123Y and C123F into C2C12 cells.
To model the mutations in a more appropriate cell type, we generated fusion constructs of the same mutations with an in-frame N-terminal HA tag and transfected these constructs into C2C12 myoblasts, a mouse-derived myogenic cell line with endogenous expression of FHL1 (27). Transfected myoblasts were switched to differentiation medium for up to 96 hours to induce myotube formation and were labeled with a HA-specific antibody. Whereas only few inclusions were seen in cells expressing wild-type FHL1, the C132F and H123Y mutant constructs precipitated inclusions in a significant subset of transfected cells, frequently but not exclusively in a juxtanuclear position (Figure 5A and Supplemental Figure 3, A–E). Inclusion body–positive cells increased from 10% at the onset of differentiation to 50% at 48 hours, and then decreased again at 96 hours, suggesting that the formation of inclusions is a time-dependent process and also indicating that cells with inclusion may not survive to 96 hours, although the latter point will require further investigation (Figure 5B). The inclusions were also labeling positively for components characteristically found in aggresomes such as ubiquitin and GRP78 (Figure 5, C and D) as well as for other muscle proteins known to be contained in the inclusion bodies in RBM patients, including the intermediate filament desmin, the lysosome-associated protein LAMP1, and dystrophin (Figure 5, E and F, and Supplemental Figure 3F), confirming the human disease situation is faithfully reproduced in our in vitro model.
Figure 5. Transfection of mutant FHL1 H123Y and C132F in C2C12 cells.
(A) C2C12 myotubes transfected with HA-tagged FHL1 mutant H123Y. Arrows indicate inclusions clustered around a nucleus (blue). (B) Quantification of inclusion-carrying cells as percentage of transfected cells. n = 3; 100 cells counted each; error bars represent SEM. *P < 0.05. (C–F) Inclusions (arrows) in differentiated C2C12 cells induced by transfection of mutant FHL1 H123Y-HA or C132F-HA (data not shown) recapitulate findings seen in muscle biopsy sections of patients by including proteins typically associated with aggresomes, including ubiquitin (C) and the endoplasmic reticulum chaperone GRP78 (BiP) (D) as well as other endogenous muscle proteins such as the intermediate filament desmin (E) and the lysosome-associated protein LAMP1 (F). Scale bars: 10 μm.
Discussion
In this work we use a proteomic approach to establish the Xq26.3 gene FHL1 as the gene mutated in a progressive myopathy characterized by specific intracytoplasmic inclusions known as reducing bodies for their histochemical staining characteristics. The 4 mutations we identified in our patients are localized exclusively in the second LIM domain of the protein and are therefore expected to be included in all 3 isoforms of FHL1 (full-length FHL1A as well as the shorter isoforms FHL1B and FHL1C). The mutations affect absolutely conserved histidine and cysteine residues involved in zinc coordination within the zinc-finger motifs. The mutations H123Y and C132F found in the 2 severely affected girls are particularly likely to destabilize the LIM domain. The mutations of C153 (C153R and C153Y) found in the 2 familial cases may be milder given that the location of this residue already falls within the α-helical structure at the tail end of the domain. However, all mutations are likely to significantly affect the secondary structure and the folding of the protein. Mutations in LIM domain–containing proteins such as Cypher/ZASP or muscle LIM protein (MLP) have been associated with myopathy and cardiomyopathy, respectively, but have not been associated with the formation of the type of inclusions seen in RBM (35, 36).
Consistent with the X chromosomal location of the disease gene FHL1, male-to-male transmission has never been documented in familial occurrences of this condition. The fact that there are severely affected female patients suggests an X-linked dominant genetic mechanism for some of the mutations in the gene. In the 2 familial occurrences reported here, the mothers of the significantly affected boys were less affected, in particular the mother of patient 4, who just started to experience difficulties with walking in her 30s. Thus, it is possible that for milder mutations the pattern of inheritance may appear to be consistent with X-linked recessive inheritance with manifesting carriers. Furthermore, skewed X-inactivation may potentially play a role as a modifier of severity. We observed skewing toward the affected allele in one of our severe patients. However, it is also conceivable that females may have very favorable skewing of X-inactivation, thereby preventing manifestation of the disease. Thus, it is possible that clinically entirely unaffected transmitting mothers may be identified in the future. The characterization of additional mutations and phenotypes as well as the development of animal models will be necessary to understand the full spectrum of disease resulting from mutations in FHL1.
Our in vitro transfection data in COS-7 and C2C12 cells further support the notion that the mutant FHL1 protein is able to exert a strong dominant-negative effect by progressively inducing intracytoplasmic inclusions, including both mutant as well as wild-type FHL1, thereby likely interfering with normal function and localization of both. This notion is consistent with the findings in patient muscle showing that FHL1 is able to localize correctly to the contractile apparatus in muscle fibers that have not developed prominent inclusions (Figure 2G). However, in cells with large inclusions, FHL1 seems no longer be available for proper localization, as it appears to be progressively incorporated into the aggregations (Figure 2H). Consistent with the prominent accumulation of FHL1 in the inclusions, we find normal to elevated total FHL1 content in the biopsies. Myopathies that appear to have accumulations of a certain protein rather then a lack of it have also been referred to as protein-surplus myopathies (37).
As has been pointed out recently, the inclusions in RBM have many features of aggresomes (18). Aggresomes are subcellular structures that are formed in response to cellular overload of abnormally folded protein after the unfolded protein response and the ubiquitin proteasome pathway become overwhelmed and before autophagy is fully engaged. (17–19). The aggresomal feature noted in reducing bodies include their frequently (but not exclusively) juxtanuclear position (the location of the microtubular organizing center in some cell types), their incorporation of ubiquitin, the appearance of the ER luminal chaperone GRP78 as an early marker of aggresome formation as well as the incorporation into the inclusion of an intermediate filament (desmin in muscle), in addition to many other chaperones associated with the unfolded protein response and proteasomes (17, 38). Aggresomes are emerging as an important pathophysiological mechanism, most notably in many neurodegenerative conditions characterized by inclusions and in certain muscle disorders such as valosin-containing protein (p97/VCP) mutations (19, 39). The inclusions forming in our cellular transfection systems have many of the same characteristics, including the juxtanuclear localization in addition to their cytoplasmic localization, as well as the incorporation of ubiquitin, GRP78, and the intermediate filament desmin (Figure 5, C–E). In addition, the aggregates trap the interacting protein MyBP-C as well as other proteins that do not normally interact, such as dystrophin and LAMP1 (Figure 5F, Supplemental Figure 2E, and Supplemental Figure 3F). These proteins have previously been demonstrated to be part of the inclusions in patient biopsies by immunohistochemical analysis (2, 7, 13, 16, 18). Our transfection experiments in C2C12 cells demonstrate that the presence of the mutant FHL1 initiates this process, leading to the incorporation of these other bystander proteins into the inclusions. It has been postulated that the progressive formation of aggresomes in cells could thus have profound cytotoxic effects (17, 19). In RBM such effects may occur in addition to the depletion of FHL1 and relate to sequestering of such other muscle proteins as well as of crucial components of the cellular protein degradation machinery, thereby accounting for the progressive nature of this myopathy. Many details of these pathways remain to be worked out so that FHL1 mutations will likely serve as an important model system for further investigations into the nature of aggresome formation.
We have thus established FHL1 as the protein mutated in RBM, classifying this condition as a novel sporadic and familial X-linked progressive protein aggregation disorder of skeletal and cardiac muscle. To our knowledge our report also represents the first example in which a proteomic approach has directly led to the identification of a disease gene. This technique may become important in the molecular characterization of other rare or mostly sporadic disorders with cellular morphological alterations, such as inclusions, for which traditional genetic approaches have been unsuccessful.
Methods
Patients.
Four families were included in this study. Patients 1 and 2 were sporadic female patients with classic early onset severe RBM. Patient 1 is a female with onset of weakness in early childhood leading to loss of ambulation at 3 years of age, followed by respiratory insufficiency. She now at 8 years of age has a tracheostomy and G-tube and no antigravity strength. Patient 2 had onset of progressive weakness at 2 years of age, leading to loss of ambulation at 4.5 years of age, followed by loss of all antigravity strength and death in respiratory failure at 6.5 years of age. Patient 3 (a 21-year-old male) presented with progressive weakness at 5 years of age, losing ambulation at 8 years of age and requiring ventilatory support by 11 years of age. Dilated cardiomyopathy was detected at 18 years of age. He had reduced fractional shortening by echocardiography (ECHO) and was started on angiotensin-converting enzyme (ACE) inhibitor treatment. His mother started to experience weakness in her early 30s, and in her 40s now requires a wheelchair for longer distances. She had a normal ECHO 6 years ago. Patient 4 (an 18-year-old male) started to experience weakness and a rigid spine at 10 years of age, followed by progressive weakness and contractures, leading to loss of ambulation at age 16. His mother experienced proximal muscle weakness beginning in her mid 30s. Both the patient and his mother had normal ECHO exams. Thus, only patient 3 has a cardiomyopathy at this point; ECHOs in the other patients are still normal. All patients had inclusions on their biopsies staining positively with the menadione NBT stain (Figure 1B and Supplemental Figure 2B). Creatinine kinases ranged from normal to 4-fold elevated. The parents of patients 1, 2, and 3, the mother of patient 4, as well as a healthy brother of patient 1, 2 healthy sisters of patient 2, and the grandparents of patient 3 were included in this study (Supplemental Figure 1, A–D). All patients were of Caucasian descent. Informed consent from all subjects was obtained (approved by the Institutional Review Boards of the Children’s Hospital of Philadelphia, the University of Utah, and Imperial College London).
LCM.
Nine-micrometer frozen muscle samples of patients 1 and 2 as well as of a normal healthy subject were stained with H&E. LCM was performed using PALM Microlaser Systems (Laser Pressure Catapulting and Laser Microbeam Microdissection; Zeiss). The reducing bodies were captured, and control surrounding muscle tissue as well as muscle from a healthy subject were captured separately. Six thousand dots per sample were collected in H2O and stored at 4°C.
Mass spectroscopy.
For digestion of LCM samples, captured cellular material was diluted with 1 M ammonium bicarbonate (Sigma-Aldrich) to a final concentration of 50 mM, pH 8.0. Samples were reduced with 2.5 mM TCEP (Pierce Biotechnology) at 50°C for 30 minutes, followed by alkylation with 10 mM iodoacetamide (Sigma-Aldrich) for 30 minutes at room temperature. The total volume was adjusted to 100 μl with 50 mM ammonium bicarbonate and acetonitrile (Burdick and Jackson) (final concentration, 10%). Proteolysis was initiated by the addition of 100 ng of sequencing-grade Trypsin (Promega) and allowed to proceed overnight at 37°C. The reaction was terminated by adding of 4 μl of neat formic acid (Fluka).
For liquid chromatography–MS/MS analysis, peptide digests were loaded directly onto a C18 capillary column (75 mm × 100 mm; New Objective Proteoprep 2) isocratically in 2% acetonitrile/0.1% formic acid at a flow rate of 1 μl per minute using an Eksigent 2D Liquid Chromatography (LC) system. A linear gradient was then initiated at a flow rate of 300 nl per minute (3%–40% buffer B over 42 minutes, 40%–100% buffer B over 3 minutes, then 100% buffer B for 5 minutes). Buffer A was 0.1% formic acid, and buffer B was 80% acetonitrile/0.1% formic acid. Mass spectrometry was performed on a Thermo-Finnegan LTQ mass spectrometer in a data-dependent fashion as peptides were eluted off of the capillary column. A top 5 method was performed, in which one survey scan was followed by MS/MS analysis of the 5 most intense ions. MS thresholds were set to an ion count of 1,500, and the MSn was set to 500. A mass range of 800–2,500 m/z was implemented for all runs. A repeat count of 3 was selected such that after 3 MS/MS repeats this ion was placed onto an exclusion list for 0.5 minutes. An exclusion window was set to 0.5 daltons below the target m/z and 1.5 daltons above. MS/MS experiments were performed with isolation width of 2, collision energy of 35, activation Q = 0.25, and activation time of 30. Peptides that showed a neutral loss of 98, 49.5, or 33.2 daltons were subjected for a further round of sequencing (MS3) (40).
Analysis of MS/MS data and database searching.
Raw files were searched against the species-specific component of the Swissprot database (accession number for FHL1_human Q13642) using both Sequest (Thermo) and MASCOT (Matrix Science). Two missed cleavages were allowed. A fixed modification of carbamidimethylation for cysteine, and variable modification for methionine oxidation was used. A parent mass window of 1.2 and a fragment tolerance of 0.6 daltons were utilized for all ion trap–based searches. Peptide spectra were only accepted after SEQUEST analysis using the following criteria: Xcorr > 1.5 (z = 1), 2 (z = 2), 2.5 (z = 3); ΔCn > 0.1; and a continuous ion series of at least 5 y ions and/or b ions. Additionally, the 5 major peaks in the MS/MS spectrum need to be assigned. Only proteins with a MASCOT score above 60 were accepted. Following the initial search, data were analyzed in detail using the program Scaffold (Proteome Software). Only proteins whose peptide probabilities were above 95% and whose protein probabilities were above 80% were accepted. Any protein with a 1-peptide match was manually inspected and only accepted if a contiguous y and/or b ion series was greater than or equal to 5 residues.
Western blotting.
Frozen muscle tissues from patients 1 and 2 and from a normal healthy child were cut in a cryostat at 9 μm and weighed. After lysis (10% SDS, 10 mM EDTA, 10 mM EGTA, 50 mM DTT, 6% glycerol, 60 mM Tris-HCl) and sonication, 0.5 mg of muscle tissue in loading buffer (Laemmli Sample Buffer; Bio-Rad; 5% β-mercaptoethanol) were submitted to electrophoresis on a 10% SDS-PAGE gel and then transferred onto a PVDF membrane (Millipore). Nonspecific sites were blocked for 1 hour at room temperature in TBS with 1% bovine serum albumin. Incubation with primary antibody was performed overnight at 4°C for antibody against FHL1 (goat polyclonal to FHL1, Abcam 23957, 1:5,000) diluted in blocking buffer. After washing, the membrane was incubated 1 hour at room temperature with HRP-conjugated secondary antibody (donkey anti-goat HRP; Jackson ImmunoResearch Laboratories; 705-035-147, 1:15,000) diluted in blocking buffer. Detection was performed using the ECL system by Amersham.
After stripping the membrane (25 mM glycine-HCl at pH 2.0, 1% SDS) and washing in 1× PBS loading control was performed with same method using primary antibody against GAPDH (mouse monoclonal antibody; Chemicon; MAB374) and secondary antibody goat anti-mouse HRP (Santa Cruz Biotechnology Inc.; 1:3,000). Western blot band quantification was performed with ImageJ software version 1.38x (NIH).
Immunohistochemistry.
Nine-micrometer frozen muscle tissue sections were fixed in methanol at room temperature for 40 seconds and washed in PBS. Non specific sites were blocked in PBS with 5% FCS and 0.01% Triton X-100 for 1 hour and incubated overnight at 4°C, double-labeled with goat polyclonal FHL1 (Abcam 23957; 1:500) or rabbit polyclonal FHL1 (directed against a unique amino acid sequence located in the fourth LIM domain of the human FHL1; Department of Biochemistry and Molecular Biology, Monash University; 1:5) and monoclonal slow-type MyBP-C (0.1 μm/ml, ALD66; Developmental Hybridoma Bank; 1:5), α-actinin (Sigma-Aldrich; A7811, 1:100), desmin (Novocastra; NCL-DES-DERII, 1:200), dystrophin II (C terminus) (Novocastra; NCL-DYSII, 1:200), and ubiquitin (Streegen; SPA-200, 1:200) diluted in blocking buffer. After washing, the sections were incubated 1 hour at room temperature with the secondary antibodies (Molecular Probes) goat anti-mouse 568 (1:500), goat anti-rabbit 488 (1:500), or donkey anti-goat 594 (1:100) and donkey anti-rabbit 488 (1:100) diluted in blocking buffer. Sections were analyzed using laser scanning confocal microscopy with a ×63 objective (Leica DMRE). Images were acquired using Leica confocal software version 2.61 Build 1537 and merged using Adobe Photoshop version 7.0.
Menadione NBT staining.
Muscle sections were incubated at 37°C for 1 hour in menadione NBT solution (8 mg menadione; Sigma-Aldrich; M5625), 1.3 ml acetone, 20 mg NBT, 20 ml Gomori-Tris-HCl Buffer at pH 7.4) and followed by a series of washes in acetone (30%, 60%, 90%, 60%, 30%).
FHL1 sequencing.
RNA was isolated from frozen homogenized muscle tissue from the patients and a normal control (QIAGEN). One μg RNA was used for RT-PCR (Invitrogen). cDNA were used to design 3 overlapping primers covering the whole sequence. Primer sequences were as follows: primer set 1: FHL1 F1, 5′-TCCTATCTGCCACACATCCA-3′; FHL1 R1, 5′-AATGCTTGGCAAACTTGGTC-3′; primer set 2: FHL1 F2, 5′-GTGGCCAAGGACAACAAGAT-3′; FHL1 R2, 5′-CCTTCATAGGCCACCACACT-3′; primer set 3: FHL1 F3, 5′-CTGCGTGGATTGCTACAAGA-3′; FHL1 R3, 5′-TCACTGTGTGCCCTTATTTGA-3′. PCR was carried out with 200 ng cDNA. PCR reaction was performed using Taq DNA polymerase (Invitrogen) with the following cycling parameters: 3 minutes denaturation at 94°C, followed by 35 cycles of 94°C (30 seconds), 58°C (30 seconds), 72°C (1 minute), and a final 72°C extension for 7 minutes. Purified PCR products were analyzed in both strands by unidirectional sequencing with ABI PRISM BigDye Terminator cycle sequencing kit 3.1 and run on a 3730 DNA Analyzer (Applied Biosystems). Sequences were analyzed using Sequencing Analysis 5.2 software (Applied Biosystems). The sequences were blasted against the human FHL1 clone MGC:15297 (GI 33872192, BC010998). The mutation was confirmed by PCR of the third coding exon of the genomic DNA of the patients and their families following the protocol described above (primers 5′-GGGTTGGCACAAGCACAAGTAGAA-3′ and 5′-TGGGATAGTGTGTGTGGAAGTGCT-3′). The 4 mutations were not detected in 100 control chromosomes.
X chromosome inactivation studies.
X chromosome inactivation was determined using the human androgen receptor assay. We isolated genomic DNA from sort-purified T, B, and NKT cells from a normal female control and a female XLP carrier who was heterozygous for a G383C substitution in SH2D1A. We digested 200 ng of DNA with RsaI for 12 hours at 37°C. We then digested half of the DNA with the methylation-sensitive enzyme HpaII for 5 hours at 37°C. HpaII-digested and nondigested DNA samples served as templates to amplify PCR across a (CAG)n repeat within exon 1 of the human androgen receptor gene using fluorescent primers (sequences available upon request). PCR products were evaluated using an ABI 3100 DNA Fragment Analyzer (Applied Biosystems), and data were analyzed using Genescan and Genotyper software (Applied Biosystems). If randomly inactivated, a proportion of the methylated maternal and paternal X chromosomes will remain intact after HpaII digestion and can serve as templates for PCR amplification. The maternal and paternal alleles are then distinguished based on variations in the sizes of the amplified products, which reflect differences in the lengths of the maternal and paternal (CAG)n repeats. If not randomly inactivated, the allele on the active X chromosome will be completely digested by HpaII. Only the inactive allele will be amplified, resulting in a single PCR product (41).
Primers used to generate wild-type or mutant FHL1 constructs.
Primers used to generate wild-type FHL1 construct are as follows: FHL1-205F, 5′-caccatggcggagaagtttg-3′; FHL1-1048R, 5′-cagctttttggcacagtcg-3′. Primers used to generate mutant FHL1 construct are as follows: FHL1-156F, 5′-tcctatctgccacacatcca-3′; FHL1-1048R, 5′-cagctttttggcacagtcg-3′ (NM_001449, GI 34147646).
Generation of wild-type and mutant FHL1 constructs.
The full-length wild-type FHL1 cDNA was cloned into pcDNA3.1D/V5-His-TOPO vector in-frame with downstream V5 epitope. The full-length mutant FHL1 cDNAs (H123Y FHL1 or C132F FHL1) were first cloned into pCR2.1-TOPO, vector then subcloned into EcoRI site of pcDNA3.1/CT-GFP vector in-frame with C-terminal GFP. The HindIII to PstI fragment of the mutant FHL1 cDNAs encompassing the mutation site was exchanged with the HindIII to PstI fragment of wild-type FHL1 in the pCGN-FHL1 vector to generate the pCGN-FHL1 mutants.
Transfection of wild-type or mutant FHL1 construct into COS-7 cells.
COS-7 cell line was grown in DMEM supplemented with 2 mM l-glutamine and 10% FBS. One day before transfection, 1 × 105 cells were plated in each well of a 24-well plate (with collagen I–coated aclar coverslip), so that cells were about 90% confluent at the time of transfection. The wild-type or mutant FHL1 constructs were transfected separately or cotransfected into above cells using Lipofectamine 2000 (Invitrogen).
Immunofluorescence labeling of COS-7 cells.
Seven days after being transiently transfected, COS-7 cells on aclar coverslips were fixed with Methanol at room temperature for 5 minutes, air dried, and blocked with vehicle (10% FBS with 0.1% Triton X-100 in 1× PBS) at room temperature for 30 minutes. Mutant FHL1 proteins that were fused with C-terminal GFP were detected using rabbit polyclonal anti-GFP (Invitrogen; A11122, 1:400), wild-type FHL1 protein that was fused with downstream V5 epitope was detected using rabbit polyclonal anti-V5 (Chemicon; AB3792, 1:400), followed by donkey anti rabbit Alexa Fluor 488–conjugated secondary antibody (1:200). The above FHL1 fusion protein was also colabeled with goat polyclonal anti-FHL1 (Abcam 23957, 1:100), followed by donkey anti-goat Alexa Fluor 594–conjugated secondary antibody (1:200). Sections were analyzed using laser scanning confocal microscopy with a ×63 objective (Leica DMRE). Images were acquired using Leica confocal software version 2.61 Build 1537 and merged using Adobe Photoshop version 7.0.
Electron microscopy of transfected COS-7 cells.
Transfected COS-7 cells (in 60-mm dish) were rinsed in 1× PBS, scraped off the plate, and spun down in 1× PBS. Supernatant was discarded, and the cell pellet was fixed in 2.5% glutaraldehyde (prepared from 70% glutaraldehyde [Sigma-Aldrich; G-7776]). Sample was stored at 4°C. Sections were osmicated and cut at 80 nm.
Electron microscopy was done on a Philips EM 208S microscope and images were acquired with AMT Advanced Microscope Technology Cooperation software, version 5.42.
Transfection of wild-type or mutant FHL1 construct into C2C12 cells.
C2C12 skeletal myoblasts (5 × 104 for myoblasts, 1 × 105 for differentiated cells) were seeded onto fibronectin–coated (5 μg/ml) coverslips in a 6-well dish in growth media (DMEM containing 20% FCS and 2 mM l-glutamine). Twenty-four hours after plating, cells were transiently transfected with 4 μg HA-FHL1 (C132F, H123Y, and wild-type) DNA using 4 μl Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, cells either remained undifferentiated (myoblasts) or differentiated for up to 96 hours by switching to low-serum differentiation media (DMEM containing 2% horse serum and 2 mM l-glutamine). Cells were fixed and permeabilized for 10 minutes using PBS containing 4% formaldehyde and 0.2% Triton X-100, washed thoroughly with PBS, then blocked for 10 minutes using PBS containing 1% bovine serum albumin. Cells were colabeled with mouse monoclonal HA antibody (Covance; 1:1,000) and either rabbit polyclonal desmin antibody (Sigma-Aldrich; 1:20), rabbit polyclonal dystrophin antibody (Abcam; 1:50), LAMP1 (Sigma-Aldrich; 1:100), Grp78 (Sigma-Aldrich; 1:1,000), or ubiquitin (Abcam; 1:500) for 1 hour. Cells were washed thoroughly and incubated with an anti-mouse Alexa Fluor 488–conjugated secondary antibody (Molecular Probes; 1:600) and an anti-rabbit Alexa Fluor 594–conjugated secondary antibody (Molecular Probes; 1:600) for 1 hour. Samples were viewed using laser scanning confocal microscopy (Leica TCS NT; performed at Monash Microimaging Facility, Monash University). Images were processed with Corel Photopaint software.
For the menadione NBT staining, cells were washed thoroughly with PBS, rinsed briefly with H2O, and stained with menadione NBT for 20 minutes at room temperature. Cells were washed briefly with H2O, destained by brief washes in acetone (30%, 60%, 90%, 60%, 30%), and rinsed again in H2O. Samples were viewed using an Olympus AX70 microscope fitted with a F-view II FW camera. Images were taken using the analySIS FIVE program (Olympus).
Statistics.
For statistical analysis, the significance of differences in the means was assessed by 2-tailed Student’s t test. For all tests, P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank the affected individuals and their families for their participation in this study, Daniel Martinez (Children’s Hospital of Philadelphia) for LCM support, and James Whisstock (Monash University) for pointing us to the molecular model of FHL1 and helpful discussion. Ian Krantz (Children’s Hospital of Philadelphia) provided the 50 control genomic DNA samples. Mark Paulosky (Children’s Hospital of Philadelphia) processed the electron microscopy samples, and Jennifer A. Richards (University of Pennsylvania) helped perform the X-inactivation experiments. We thank Tejvir S. Khurana, Elizabeth Barton, and Richard S. Finkel (University of Pennsylvania) for helpful discussions, Tom Crawford (Johns Hopkins Hospital) and Mena Scavina (Alfred I. duPont Hospital for Children) for clinical information. The monoclonal antibody slow-type MyBP-C (ALD66) developed by Donald A. Fischman (Cornell University Medical College) was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa. C.G. Bönnemann is supported by grants from MDA USA and NIH/NIAMS.
Footnotes
Nonstandard abbreviations used: FHL1, four and a half LIM domain 1; LCM, laser capture microdissection; LIM, Lin-11, Isl-1, Mec-3; MS/MS, tandem mass spectrometry; MyBP-C, myosin binding protein C; NBT, nitro-blue tetrazolium; RBM, reducing body myopathy.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:904–912 (2008). doi:10.1172/JCI34450
References
- 1.Brooke M.H., Neville H.E. Reducing body myopathy. Neurology. 1972;22:829–840. doi: 10.1212/wnl.22.8.829. [DOI] [PubMed] [Google Scholar]
- 2.Figarella-Branger D., et al. Adult onset reducing body myopathy. Neuromuscul. Disord. 1999;9:580–586. doi: 10.1016/s0960-8966(99)00058-9. [DOI] [PubMed] [Google Scholar]
- 3.Bertini E., et al. Reducing body myopathy and desmin storage in skeletal muscle: morphological and biochemical findings. Acta Neuropathol. (Berl.). 1994;87:106–112. doi: 10.1007/BF00386261. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter S., Karpati G., Holland P. New observations in reducing body myopathy. Neurology. 1985;35:818–827. doi: 10.1212/wnl.35.6.818. [DOI] [PubMed] [Google Scholar]
- 5.Dubowitz, V. 1978. Muscle disorders in childhood. W.B. Saunders. London, United Kingdom. 219–221. [Google Scholar]
- 6.Hubner G., Pongratz D. Granular body myopathy (so-called reducing body myopathy) [In German]. Pathologe. 1982;3:111–113. [PubMed] [Google Scholar]
- 7.Kiyomoto B.H., et al. Fatal reducing body myopathy. Ultrastructural and immunohistochemical observations. J. Neurol. Sci. 1995;128:58–65. doi: 10.1016/0022-510x(94)00204-2. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi Y., Nihei K., Kuwajima K., Nonaka I. Reducing body myopathy — a case report [In Japanese]. Rinsho Shinkeigaku. 1992;32:62–67. [PubMed] [Google Scholar]
- 9.Nomizu S., Person D.A., Saito C., Lockett L.J. A unique case of reducing body myopathy. Muscle Nerve. 1992;15:463–466. doi: 10.1002/mus.880150408. [DOI] [PubMed] [Google Scholar]
- 10.Oh S.J., Meyers G.J., Wilson E.R., Jr., Alexander C.B. A benign form of reducing body myopathy. Muscle Nerve. 1983;6:278–282. doi: 10.1002/mus.880060406. [DOI] [PubMed] [Google Scholar]
- 11.Reichmann H., Goebel H.H., Schneider C., Toyka K.V. Familial mixed congenital myopathy with rigid spine phenotype. Muscle Nerve. 1997;20:411–417. doi: 10.1002/(sici)1097-4598(199704)20:4<411::aid-mus2>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 12.Tome F.M., Fardeau M. Congenital myopathy with “reducing bodies” in muscle fibres. Acta Neuropathol. (Berl.). 1975;31:207–217. doi: 10.1007/BF00684560. [DOI] [PubMed] [Google Scholar]
- 13.Goebel H.H., et al. Reducing body myopathy with cytoplasmic bodies and rigid spine syndrome: a mixed congenital myopathy. Neuropediatrics. 2001;32:196–205. doi: 10.1055/s-2001-17374. [DOI] [PubMed] [Google Scholar]
- 14.Ohsawa M., et al. Familial reducing body myopathy. Brain Dev. 2006;29:112–116. doi: 10.1016/j.braindev.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Ikezoe K., Nakagawa M., Osoegawa M., Kira J., Nonaka I. Ultrastructural detection of DNA fragmentation in myonuclei of fatal reducing body myopathy. Acta Neuropathol. (Berl.). 2004;107:439–442. doi: 10.1007/s00401-004-0831-5. [DOI] [PubMed] [Google Scholar]
- 16.Shinde A., et al. Nucleolar characteristics of reducing bodies in reducing body myopathy. Acta Neuropathol. (Berl.). 2004;107:265–271. doi: 10.1007/s00401-003-0806-y. [DOI] [PubMed] [Google Scholar]
- 17.Kopito R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 18.Liewluck T., et al. Unfolded protein response and aggresome formation in hereditary reducing-body myopathy. Muscle Nerve. 2007;35:322–326. doi: 10.1002/mus.20691. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Mata R., Gao Y.S., Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 20.Kadrmas J.L., Beckerle M.C. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 21.Freyd G., Kim S.K., Horvitz H.R. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- 22.Curtiss J., Heilig J.S. DeLIMiting development. Bioessays. 1998;20:58–69. doi: 10.1002/(SICI)1521-1878(199801)20:1<58::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 23.Dawid I.B., Toyama R., Taira M. LIM domain proteins. C. R. Acad. Sci. III. 1995;318:295–306. [PubMed] [Google Scholar]
- 24.Jurata L.W., Gill G.N. Structure and function of LIM domains. Curr. Top. Microbiol. Immunol. 1998;228:75–113. doi: 10.1007/978-3-642-80481-6_4. [DOI] [PubMed] [Google Scholar]
- 25.Brown S., et al. Characterization of two isoforms of the skeletal muscle LIM protein 1, SLIM1. Localization of SLIM1 at focal adhesions and the isoform slimmer in the nucleus of myoblasts and cytoplasm of myotubes suggests distinct roles in the cytoskeleton and in nuclear-cytoplasmic communication. J. Biol. Chem. 1999;274:27083–27091. doi: 10.1074/jbc.274.38.27083. [DOI] [PubMed] [Google Scholar]
- 26.Johannessen M., Moller S., Hansen T., Moens U., Van Ghelue M. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell. Mol. Life Sci. 2006;63:268–284. doi: 10.1007/s00018-005-5438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGrath M.J., et al. Four and a half LIM protein 1 binds myosin-binding protein C and regulates myosin filament formation and sarcomere assembly. J. Biol. Chem. 2006;281:7666–7683. doi: 10.1074/jbc.M512552200. [DOI] [PubMed] [Google Scholar]
- 28.Morgan M.J., Madgwick A.J. The LIM proteins FHL1 and FHL3 are expressed differently in skeletal muscle. Biochem. Biophys. Res. Commun. 1999;255:245–250. doi: 10.1006/bbrc.1999.0179. [DOI] [PubMed] [Google Scholar]
- 29.Purcell N.H., et al. Extracellular signal-regulated kinase 2 interacts with and is negatively regulated by the LIM-only protein FHL2 in cardiomyocytes. Mol. Cell. Biol. 2004;24:1081–1095. doi: 10.1128/MCB.24.3.1081-1095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi Y., Furukawa T., Tun T., Han H., Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell. Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin H., et al. RING1 inhibits transactivation of RBP-J by Notch through interaction with LIM protein KyoT2. Nucleic Acids Res. 2004;32:1492–1501. doi: 10.1093/nar/gkh295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin H., et al. The PcG protein HPC2 inhibits RBP-J-mediated transcription by interacting with LIM protein KyoT2. FEBS Lett. 2005;579:1220–1226. doi: 10.1016/j.febslet.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 33.Ng E.K., et al. Characterization of tissue-specific LIM domain protein (FHL1C) which is an alternatively spliced isoform of a human LIM-only protein (FHL1). J. Cell. Biochem. 2001;82:1–10. doi: 10.1002/jcb.1110. [DOI] [PubMed] [Google Scholar]
- 34.Wang J., et al. The transcriptional repression activity of KyoT2 on the Notch/RBP-J pathway is regulated by PIAS1-catalyzed SUMOylation. J. Mol. Biol. 2007;370:27–38. doi: 10.1016/j.jmb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Selcen D., Engel A.G. Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann. Neurol. 2005;57:269–276. doi: 10.1002/ana.20376. [DOI] [PubMed] [Google Scholar]
- 36.Knoll R., et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 37.Goebel H.H., Warlo I.A. Surplus protein myopathies. Neuromuscul. Disord. 2001;11:3–6. doi: 10.1016/s0960-8966(00)00165-6. [DOI] [PubMed] [Google Scholar]
- 38.Waelter S., et al. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubbers C.U., et al. Pathological consequences of VCP mutations on human striated muscle. Brain. 2007;130:381–393. doi: 10.1093/brain/awl238. [DOI] [PubMed] [Google Scholar]
- 40.Mann M., Hendrickson R.C., Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu. Rev. Biochem. 2001;70:437–473. doi: 10.1146/annurev.biochem.70.1.437. [DOI] [PubMed] [Google Scholar]
- 41.Nichols K.E., et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.