Abstract
We have adapted and extended the decoy oligonucleotide technique for use in prokaryotes. To identify cis-acting regulatory elements within a promoter, we developed a DNase I/T7 exonuclease footprinting technique and applied it to actII-orf4 from Streptomyces coelicolor A3(2), which encodes the pathway-specific activator for production of the antibiotic actinorhodin. Our in vivo mapping data allowed us to create decoy oligonucleotides incorporating the identified regulatory elements and to test whether their introduction into S. coelicolor affected antibiotic production. We mapped the promoter region when in a transcriptionally inactive state before the onset of actinorhodin production with the aim of designing decoy oligonucleotides capable of interfering with potential repressor binding and so stimulate actinorhodin production. Mapping identified five candidates for decoy oligonucleotides, and these were tested in a plate-based assay to rapidly validate their activity. A transfection protocol was developed for liquid cultures that enabled efficient uptake of decoys, and quantitative real-time PCR demonstrated decoy persistence for >70 h. Measurement of the effects on growth, expression of actII-orf4, and antibiotic production demonstrated that one of the decoys, in concordance with the plate assay, was more efficacious than the others in increasing actinorhodin production. Two of the identified regulatory elements occurred upstream of gene SCO5812, deletion of which reduced actinorhodin production, confirming that experimental analysis of regulatory motifs can provide new insights into factors influencing antibiotic production in streptomycetes.
Keywords: actinorhodin, regulation, transcription factors, actII-orf4, secondary metabolism
Decoy oligonucleotides are designed to mimic the binding sites of transcription factors and prevent the latter from binding to their cognate genomic targets, with a consequent modification of gene expression. As such, they represent a simple and generic tool for manipulating the DNA–protein interactions that regulate specific genes and that consequently determine phenotypes. Their utility has been demonstrated mostly in eukaryotic systems, where a spur to their development was their potential to function as novel classes of therapeutic agents (1). To this end, decoy oligonucleotides have been used to demonstrate that transcription factor EF2 represses smooth muscle proliferation in rats (2), to block STAT3-mediated proliferation of carcinomas (3), and to show that targeting of the cAMP response element can control cancer proliferation in vivo (4). However, getting decoys to work in eukaryotes can be problematic; they can be rapidly degraded in serum and nuclear extracts (5), cellular uptake of the decoy and its transition across the nuclear membrane can be inefficient (6), and some treatments can trigger nonspecific or toxic effects. In principle, using decoys in prokaryotes should circumvent many of these problems and, as such, they might prove to be an effective tool for the rapid identification of cis-acting regulatory sequences, such as transcription factor binding sites controlling both specific genes and regulatory networks. A successful demonstration of the approach was the use of an AT-rich decoy to alter the expression of CO2-responsive genes in Cyanobacterium (7). In this example, the decoy was added directly to the medium from where it efficiently entered the cells. In this article, we combine the decoy approach with a simple in vivo footprinting protocol to rapidly identify candidate cis-acting regulatory motifs. Functional validation of these sequences was achieved by incorporating them into dumbbell decoy oligonucleotides whose circular format suppresses degradation by exo- and endonucleases (8), and by testing their effect on phenotype in vivo.
We used decoy oligonucleotides to study the regulation of the blue-pigmented antibiotic actinorhodin in Streptomyces coelicolor A3(2). This organism contains (for a prokaryote) a relatively large genome (8.7 Mb) with a complex and adaptive pattern of gene regulation, particularly with respect to the developmental and environmental cues that control antibiotic production (9). S. coelicolor is also the model organism for the actinomycetes, and increasing the level of understanding of the regulation of antibiotic production in this strain may inform new strategies for gaining access to the wide variety of secondary metabolites produced by these organisms. Many of these compounds have important applications in medicine (for example, as antibiotics) and in agriculture (10), and actinomycetes continue to be a profitable source of new drugs and enzymes (11).
Perhaps as a consequence of the complex regulation of antibiotic production, many pleiotropic mutants identified by genetic screens are conditional; for example, the antibiotic nonproducing phenotype of a relA null-mutant is highly medium-dependent (12). The occurrence of nutritionally conditional phenotypes implies that genetic screens may underestimate the number of regulatory factors influencing antibiotic production. Inactivation of a bona fide transcription factor may be missed if, under the conditions used, activation of target genes can be mediated by an alternative transcription factor or regulatory pathway. One of the advantages of decoys is that they identify and manipulate the sequence component of DNA–protein interactions controlling gene expression, potentially blocking the interaction of several transcription factors with a single promoter. In this article, we demonstrate the relative ease of targeting regulatory sequences and the ability to rapidly identify novel genes involved in antibiotic production in a manner that is complementary to conventional genetic screens.
Results and Discussion
Mapping Regulatory Elements Within the actII-orf4 Promoter.
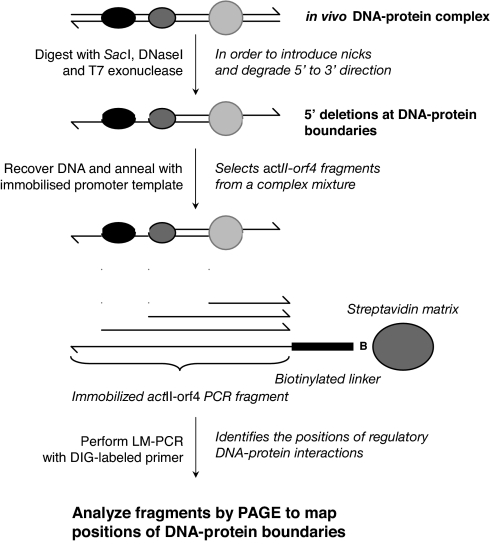
DNA–protein interactions controlling expression of actII-orf4 were studied by in vivo DNase I/T7 exonuclease mapping. This method (Fig. 1) was developed to identify the boundaries of cis-acting regulatory elements so that the deduced sequences could be used to design decoy oligonucleotides for functional studies. In vivo DNase I digestion of intact cells introduced nicks in the genomic DNA between the DNA–protein complexes; T7 exonuclease bound at these nicks and digested the DNA in the 5′ to 3′ direction until a boundary with a protein complex was reached. The position of the boundary was then mapped by PCR: Recovered fragments were hybridized to a biotinylated forward or reverse single strand from a PCR product incorporating the actII-orf4 promoter and amplified by using ligation-mediated (LM)-PCR with a digoxigenin (DIG)-labeled primer [see supporting information (SI) Text]. The labeled PCR fragments were size-fractionated by 12% polyacrylamide gel electrophoresis (PAGE), and their sizes were determined after chemiluminescent detection. The unprecedented combination of T7 exonuclease and DNase I in a footprinting protocol detects all of the boundaries of DNA–protein complexes within a promoter region and not just those adjacent to the restriction site (in this case, that for SacI) used to fragment the DNA, which is the case with standard mapping assays.
Fig. 1.
Schematic overview of the protocol for in vivo T7 exonuclease/DNase I mapping. To map the in vivo boundaries of DNA–protein complexes at specific locations within the S. coelicolor genome, DNase I and T7 exonuclease were added to freshly harvested cells. The DNase I introduced “nicks” into the DNA surrounding the complexes that then served as substrates for the 5′ to 3′ exonuclease activity of T7 exonuclease. DNA was recovered from the treated cells, and fragments from the targeted actII-orf4 promoter were captured by hybridization to an immobilized strand of a PCR fragment of the promoter that incorporated a biotinylated linker. Boundaries within the population of captured fragments were mapped by performing a PCR with a second DIG-labeled primer that hybridized to the complementary strand of the biotinylated linker. The sizes of the labeled products were determined by PAGE and chemoluminescent detection. Boundaries on the opposite strand were mapped in a similar manner, using a PCR product with the biotinylated linker at the opposite end.
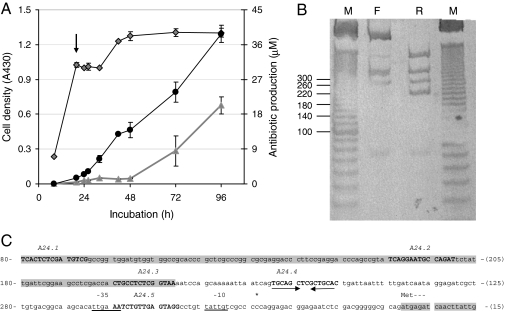
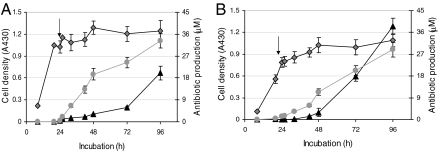
Because our aim was to discover regulatory elements involved in the repression of actII-orf4 expression, we mapped the transcriptionally down-regulated promoter in a rich liquid medium (R5) after 20 h of growth. Under these conditions, repressors may occupy the promoter and prevent expression. Growth curves were derived by measuring cell density (A430 of the culture), and production of the two pigmented antibiotics [the blue actinorhodin, and the red undecylprodigiosin (13)] and the transcriptional activity of actII-orf4 were determined. The latter is induced during later stages of growth, and thus samples for mapping were harvested before the visually detectable onset of actinorhodin production (indicated by the arrow in Fig. 2A). The amounts of the enzymes required for digestion were determined empirically, with the concentration of DNase I needing careful optimization. For example, an excess of DNase I resulted in loss of signal clarity, whereas too little enzyme was ineffectual for mapping. Two hundred fifty units of T7 exonuclease per reaction worked well in most cases; an excess of enzyme was required because it is not highly processive, nor were the buffer conditions optimal for activity. The resulting fragments were visualized by chemoluminescent detection (Fig. 2B), and the boundaries of the DNA–protein complexes in the transcriptionally silent actII-orf4 promoter region were deduced by comparison with a size ladder (Fig. 2C). Long and short runs were performed to accurately size the protected bands and thus to identify the center of binding sites to ±8 bp. We refer to the sequences defined by these regions as regulatory elements, and five were seen. The regulatory elements were labeled A24.1 to A24.5 (positioned 213 bp and 14 bp, respectively, upstream of the transcriptional start site), and these sequences were used to design the 15-bp decoy oligonucleotides (centered on the predicted binding sites) that were used in the subsequent functional studies.
Fig. 2.
T7 exonuclease/DNase I mapping of candidate regulatory motifs within the promoter of actII-orf4. (A) S. coelicolor M145 mycelium was harvested from a culture grown in rich R5 medium at a time point (indicated by arrow) preceding visible actinorhodin production. Cell growth (diamonds) and production of actinorhodin (triangles) and undecylprodigiosin (circles) were monitored throughout. (B) Boundaries were mapped on both strands, as described in Fig. 1, and their positions were determined after size analysis by 12% nondenaturing PAGE, followed by chemoluminescent detection of DIG-labeled products. A 10- to 1,000-bp marker (lane M; sizes shown on the left of the gel) was used to determine the sizes of protected bands derived from the forward (F) and reverse (R) orientations. (C) Sequence of the actII-orf4 promoter showing the positions of the putative cis-regulatory elements (relative to the primers used in the mapping protocol). Boxed areas indicate the coding sequences of the upstream gene (actII-orf3) and of actII-orf4. Capitalized sequence marks the candidate regulatory elements with their names shown above. The underlined sequences indicate the −35 and −10 boxes for the actII-orf4 promoter, the asterisk shows the position of the transcriptional start site, and the convergent arrows indicate the inverted repeat present in A24.4. The sequence is numbered on the left side as distance downstream of the forward primer used in the PCR, and on the right, distance upstream of the reverse primer.
Rapid Screening for Decoy Function on Agar Plates.
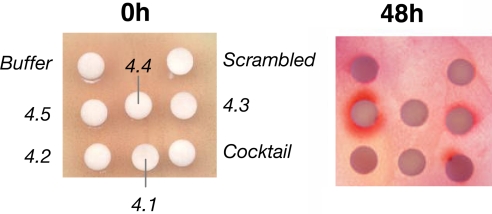
We developed a rapid agar-plate-based assay to determine whether the decoys had any effect on antibiotic production. R2YE agar was used because it promotes the production of both pigmented antibiotics at levels that are readily detectable by eye. Plates were inoculated with S. coelicolor M145 spores and incubated for 24 h at 30°C to produce confluent lawns of mycelial growth. At this stage, production of the red-pigmented undecylprodigiosin had begun but synthesis of actinorhodin (which is blue) had not. The plates were covered with a thin layer of soft nutrient agar (SNA) in 0.5% agarose, and before this set, small disks of Whatman paper (Antibiotic Assay disks) were laid onto the SNA and saturated with 15 μl of a 10 pmol/μl solution of decoy oligonucleotide or control solution (Fig. 3). Samples were prepared in a buffer containing 0.5% (vol/vol) of the two nonionic detergents Nonidet P-40 and Triton X-100 to improve transfection efficiency. Controls consisted of the buffer alone or a scrambled decoy oligonucleotide, where the sequence of the A24.5 decoy was randomized. The plate was incubated further at 30°C and inspected regularly to determine the effects on the amount or timing of antibiotic production. A purple/red halo was seen around the disk soaked with A24.5, and to a lesser extent A24.3 and the mixture of decoys containing an equimolar mixture of A24.1 to A24.5 (each at one-fifth of the concentration used for the individual samples) (Fig. 3 Right; note that the color of actinorhodin varies from blue to red depending on pH). To confirm that the purple/red halos reflected actinorhodin synthesis, the disks at 48 h were recovered and pigments were extracted and assessed spectrophotometrically; decoy A24.5 clearly activated early actinorhodin production, whereas A24.3 and the mixture of decoys did so to a lesser extent (data not shown). No precocious actinorhodin production was seen surrounding the control disks. By 96 h, the entire lawn of S. coelicolor M145 had produced both antibiotics (data not shown), and no zones of growth inhibition were apparent around any of the disks. The ability of some of the decoys to enhance production of actinorhodin at early time points, potentially by interfering with the binding of repressors to their identified sites within the actII-orf4 promoter region, stimulated us to perform further functional studies by introducing the decoys into cells grown in liquid culture.
Fig. 3.
Plate assays demonstrating that decoy oligonucleotides can influence antibiotic production in S. coelicolor. Filter discs were saturated with solutions of decoys or, as control, buffer and applied to a lawn of S. coelicolor M145 overlaid with SNA medium. As the bacteria continued to grow, enhanced antibiotic production was recognized by early accumulation of the pigmented antibiotics around the disks (evident 48 h after the addition of the decoys). Negative controls (buffer alone or a “scrambled” version of decoy A24.5) did not show precocious production.
Uptake and Stability of Decoys in Liquid Cultures.
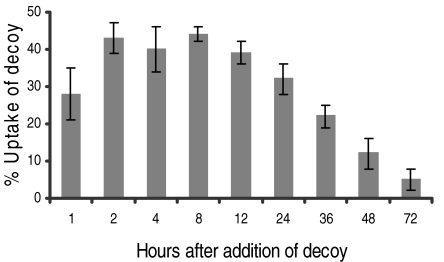
The next issues to address were whether a decoy could be efficiently introduced into mycelium in liquid culture, and if so, how long would it persist? Six cultures of S. coelicolor M145 were set up in solid minimal media (SMM) liquid medium and grown to mid-exponential phase. Cells were collected by gentle centrifugation and resuspended in one-tenth of the original volume of a permeabilizing buffer (containing the same concentrations of detergents used in the plate assays) before transfection with varying amounts of decoy A24.1 (0, 5, 10, 20, 50, and 100 mM). After a brief incubation, the cells were resuspended in the retained medium and incubation was continued. Uptake of the decoy oligonucleotide was measured by quantitative real-time PCR (qrt-PCR), using primers designed to amplify a small fragment (40 bp) spanning the join introduced in the formation of the dumbbell decoy. Reference to a standard curve was used to calculate the absolute number of copies of decoy present in the cells (after centrifugation and two washes), and this was corrected by reference to a genomic control. To assess stability, aliquots were withdrawn at various time points (0–72 h). The rate of uptake saturated above concentrations of 20 mM. Optimal uptake was achieved with a 20 nM transfection, 2 h after which 45% of the decoy had entered the cells (Fig. 4). The decoy could be detected intracellularly 72 h after addition, with 22% (corresponding to half that had entered the cells) persisting after 36 h. Because the cultures were transfected when nearing stationary phase, it was assumed that the decoys were being slowly degraded by endogenous nucleases as opposed to being diluted by continued growth. Thus, the decoy was able to enter the mycelium and persist for a prolonged period, suggesting that this approach could be used to alter gene expression in liquid cultures.
Fig. 4.
Uptake and stability of a decoy oligonucleotide. Actively growing cells were treated with a solution containing decoy and the uptake of the oligonucleotide and its stability estimated by qrt-PCR. After transfection with 20 nM A24.1, cells were harvested and washed before qrt-PCR was used to estimate copies of decoy remaining in the cell as a function of time. The data represent three independent determinations.
Use of Decoy Oligonucleotides to Control Antibiotic Production.
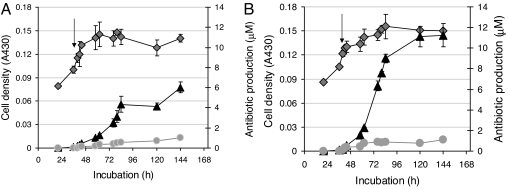
The five decoys, corresponding to the identified regulatory elements, were used to transfect cultures of S. coelicolor M145 grown in R5 or SMM liquid media. Transfection was timed to precede the expression of actII-orf4, providing the decoys with the opportunity to interact with their cognate transcription factors and potentially influence actinorhodin production. Control experiments were similar to those used in the plate assays and consisted of either a mock transfection procedure or the introduction of three scrambled decoys (based on the sequences of decoys A24.1, A24.3, and A24.5). In each experiment, the growth of the culture, the production of both pigmented antibiotics, and the expression of actII-orf4 were determined at fixed intervals after transfection. Treatment with the decoys led to a slight reduction in the extent of growth compared with the control sample (e.g., Fig. 5 with A24.5); interestingly, however, addition of some of the decoys enhanced actinorhodin production and, in some cases, also that of undecylprodigiosin. In R5 medium, where antibiotic production is relatively high, decoy A24.5 up-regulated actinorhodin production, increasing the yield by 95% at the 96-h time point (Fig. 5). Decoys A24.1 and A24.3 had milder effects, causing up-regulation of both pigmented antibiotics. In all of the treatments where an increase in actinorhodin production occurred, there was a corresponding increase in the absolute level of actII-orf4 expression (determined by qrt-PCR; data not shown). Hence, the results were largely consistent with those seen on agar plates. The decoys were also tested in SMM medium, a minimal medium that supports less antibiotic production than R5. Transfection with decoy A24.5 led to a doubling in actinorhodin production with no effect on the extent of growth (Fig. 6). In this medium, increases in actinorhodin production were also seen for decoys A24.1 and A24.3 (data not shown). Note that the delay between the addition of the decoy oligonucleotides and the stimulation of actinorhodin production (Figs. 5 and 6) is consistent with the delay observed between the activation of act gene transcription and actinorhodin biosynthesis observed in earlier studies (12), the reason for which is unknown. Thus, the decoys had served as convenient tools to validate the identification of cis-acting regulatory elements within the actII-orf4 promoter, allowing us to influence the onset of antibiotic production. Possible reasons for the concomitant increase in undecylprodigiosin production elicited by some of the decoys are discussed below.
Fig. 5.
Decoy-mediated increase in actinorhodin production in cultures grown in R5 liquid medium. S. coelicolor M145 was grown for 20 h before transfection (indicated by arrows) with a scrambled version of decoy A24.5 (A) or the A24.5 decoy (B). Cell growth (diamonds) and the amount of undecylprodigiosin produced (circles) were similar in the two cultures, and the only variation seen was in the accumulation of actinorhodin (triangles), which was stimulated after treatment with decoy A24.5. The data represent the average of three independent determinations, and the bars show the standard error.
Fig. 6.
Decoy-mediated increase in actinorhodin production in cultures grown in SMM liquid medium. Comparison of the data obtained after treating M145 cultures with a scrambled version of decoy A24.5 (A) and decoy A24.5 (B) revealed that the decoy oligonucleotide caused a pronounced increase in actinorhodin production. The data are presented as in Fig. 5 and are the average of three independent determinations; bars show the standard error.
Discovery of a Modulator of Actinorhodin Production.
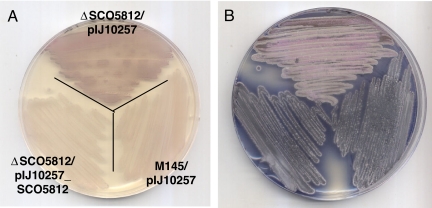
The combination of a previously unexplored mapping technique and decoy oligonucleotides identified three regulatory elements influencing actII-orf4.expression. We next asked whether these regulatory motifs occurred in the promoter regions of other genes; such genes might also be involved in the regulation of antibiotic production. BLAST searches with all five decoy sequences identified one such gene. SCO5812 contained strong matches to A24.1 (a run of 10 of 14 bp) and A24.3 (a run of 11 of 18 bp), and is a potential homologue of ribonuclease HII. As such, it may have a similar function to a previously identified gene that pleiotropically influences antibiotic production, absB [SCO5572 (13)]. AbsB is a homologue of RNase III and thought to be involved in transcript processing (14). To establish the role, if any, of SCO5812 in antibiotic production, we deleted the gene from M145 and compared production with the parental strain. Whereas deletion of SCO5812 dramatically reduced actinorhodin production on R5 agar (Fig. 7B), it enhanced undecylprodigiosin synthesis on SMMS (Fig. 7A). Reintroduction of SCO5812 into the deletion mutant after cloning in the conjugative, integrative pIJ10257 restored the M145 phenotype. It is of interest to note that cells transfected with either A24.1 or A24.3 showed a small but reproducible up-regulation of undecylprodigiosin production, suggesting that the transcription factor(s) that binds to these sites may influence the expression of both of the antibiotic biosynthetic gene clusters. One of the decoys tested, A24.4, which contains a 6-bp inverted repeat (Fig. 2), was found to have a match (8 of 12) in the promoter region of redD, the pathway-specific activator gene of the undecylprodigiosin biosynthetic cluster (15). This motif is a predicted binding site for the nucleoid protein IHF (based on sequence homology to previously identified sites), raising the possibility that the site has an architectural role in the nucleoprotein complex instead of directly affecting transcription.
Fig. 7.
Deletion of SCO5812 leads to reduced production of actinorhodin on R5 agar and overproduction of undecylprodigiosin on SMMS agar. M145 ΔSCO5812/pIJ10257 (top), M145/pIJ10257 (right), and the complemented mutant M145 ΔSCO5812/pIJ10257+SCO5812 (left) were streaked on SMMS agar medium (A) and R5 agar medium (B) and incubated for 72 and 96 h, respectively.
Concluding Remarks.
We have used previously unexplored techniques to identify and validate a binding site for a putative repressor of antibiotic production in S. coelicolor. Validation was performed with decoy oligonucleotides: Transfection of copies of these regulatory elements led to derepression of the targeted gene and increased production of actinorhodin. The intention of our work was to demonstrate that decoy oligonucleotides can be a valuable tool in prokaryotes to rapidly delineate genetic networks. Has it been successful? In two senses, it has been. Three of the five decoys tested showed the expected activity. The decoy with the strongest effect, A24.5, encompasses the binding site for a TetR-like transcriptional regulator, AtrA, which stimulates promoter activity (16), and consequently the decoy might be expected to reduce, not enhance, actinorhodin production. We hypothesize that when the promoter is inactive, a repressor binds to the same site, preventing binding of AtrA or other TetR-like transcription factors. Addition of the decoy could assist in dislodging this repressor, allowing access to AtrA. Consistent with this, our own work, using affinity purification to identify repressors of actII-orf4, has similarly identified TetR-like transcription factors bound to this region (M.M. and M.J.B., unpublished work); it is our assumption that as decoy A24.5 is added before transcription of actII-orf4, it enhances expression by diminishing the binding of these putative repressors.
Many genes are controlled at multiple levels through the interaction of regulatory factors with cis-regulatory sites. The nucleoprotein complexes formed may do so in a tissue-specific, developmental-stage-specific, or stimulus-dependent manner. In short, in vivo DNA–protein interactions are physiologically determined and represent the interaction between two dynamic components: the DNA binding proteins that constitute the trans-acting environment, and the cis-regulatory sequences. Although our knowledge of the patterns of gene expression has grown substantially in recent years, this growth has not been paralleled by a comparable increase in our knowledge of regulatory factors that control specific genes affecting specific cellular processes. We anticipate that application of these techniques is likely to yield fresh insights into the complexities of prokaryotic gene regulation and establish them as a complementary approach to conventional genetic analysis.
Materials and Methods
Strains, Growth Conditions, and Strain Manipulation.
Spores of S. coelicolor A3(2) strain M145 were germinated synchronously by heat treatment (17) and grown in SMM or R5 medium (17) at 30°C with shaking. Growth of the culture and actinorhodin production were measured as described in ref. 17. R2YE (17), SMMS (17), and SNA (17) were used for agar plate assays of decoy-induced antibiotic production. Deletion of SCO5812 was accomplished by PCR targeting (18) and confirmed by PCR analysis; the entire SCO5812 coding region was replaced by a six base scar sequence yielding 5′-atg gct agc tga-3, where atg and tga are the SCO5812 translation start and stop codons, respectively. Complementation of the mutation was achieved by cloning SCO5812 as a NdeI–HindIII PCR fragment into the conjugative, integrative expression vector pIJ10257 (19) that had been cleaved with NdeI and HindIII. Table 1 lists the oligonucleotide primers used for deletion construction, confirmation, and complementation.
Table 1.
Oligonucleotide primers used in this study
| Name | Sequence |
|---|---|
| Decoy oligonucleotides | |
| A24.1 | 5′-P-atatcactctcgatgtcggcgttttcgccgacatcgagagtgatatagttttct-3′ |
| A24.2 | 5′-P-atatcaggaatgccagatgcgttttcgcatctggcattcctgatatagttttct-3′ |
| A24.3 | 5′-P-atactgcctctcggtaagcgttttcgcttaccgagaggcagtatagttttct- 3′ |
| A24.4 | 5′-P-atatgcagctcgctgcacgcgttttcgcgtgcagcgagctgcatatagttttct-3′ |
| A24.5 | 5′-P-ataaatctgttgagtagggcgttttcgccctactcaacagatttatagttttct-3′ |
| A24.5 scrambled | 5′-P-atagcaatttagatggtggcgttttcgccaccatctaaattgctatagttttct-3′ |
| For quantitative PCR for decoy copy number | |
| A24.1 copy f | 5′-tcgccgacatcgagagtgatat-3′ |
| A24.1 copy r | 5′-aaacgccgacatcgagagtga-3′ |
| For PCR deletion of SCO5812 | |
| 5812KOf | 5′-cgcccggcgatcgcggcggcgtacgctagggacgccatggctacgtgtaggctggagctgcttc-3′ |
| 5812KOr | 5′-tcggcgggtggcacatacctgcgagcgcgagcgggaccgtcagctagcattccggggatccgtcgacc-3′ |
| 5812Testf | 5′-tcccgcttttgggaccggggtgt-3′ |
| 5812Testr | 5′-atcaaacgttggtgcggctcgcg-3′ |
| For PCR cloning of SCO5812 for complementation | |
| 5812exf | 5′-aaagatcgaacatatgccgtacgaaccacctacgcac-3′ |
| 5812exr | 5′-ttacgatcaagctttcagaaatcgaatccgagctgaccc-3′ |
The cis-regulatory sequences in each of the decoy oligonucleotides are boldface.
In Vivo DNase I/T7 Exonuclease Mapping.
Before treatment with the footprinting reagent, cultures of S. coelicolor mycelium were supplemented with 0.5 mM CaCl2 and 50 units of DNase I (to remove extracellular DNA) and incubated for a further 15 min at 30°C. The mycelium was harvested by low-speed centrifugation and washed extensively in TES buffer [25 mM Tris[hydroxymethyl]methyl-2-amino-ethanesulphonic acid (pH 7.2) supplemented with 7.5 mM EDTA]. For DNase I/T7 footprinting, the cells were washed in TES supplemented with 0.5% (vol/vol) Nonidet P-40 and 0.5% (vol/vol) Triton X-100, and incubated at 30°C for 15 min. The cells were then washed in DNase I digestion buffer (DDB) preheated to 30°C and digested initially with 250 units/ml SacI for 30 min and then with a combination of typically 2 units/ml DNase I and 250 units/ml T7 exonuclease for a further 5 min. Reactions were stopped by addition of an equal volume of STOP buffer [50 mM Tris·HCl, 5 mM EDTA, 0.2% (wt/vol) SDS, 10 mg/ml proteinase K (pH 8)] and incubated overnight at 55°C. Nucleic acids were ethanol-precipitated after two rounds of phenol-chloroform extraction. The samples were digested with RNase A before reprecipitation and quantification. Specific genomic fragments were captured by hybridization to an immobilized DNA fragment containing the actII-orf4 promoter, and the positions of the boundaries detected by T7 digestion were determined by LM-PCR and analysis of fragment sizes by 12% PAGE. A detailed protocol is available in SI Text.
Decoy Oligonucleotide Studies.
Sequences of all oligonucleotides used are shown in Table 1. Decoy oligonucleotides were synthesized (Invitrogen) and ligated with CircLigase (EPICENTRE) to create dumbbells (8). One hundred picomoles of each decoy oligonucleotide was mixed in a 20-μl reaction volume of 1× CircLigase buffer supplemented with 50 μM ATP/2.5 mM MnCl2/500 units of CircLigase and incubated for 1 h at 60°C. The mixture was treated with an excess of exonuclease I to remove linear DNA, and the remaining covalent circles were precipitated. Each preparation was analyzed by 12% nondenaturing gel electrophoresis to check that the majority of products were monovalent covalent circles. The sample was resuspended at 200 pmol/μl in TE buffer [10 mM Tris·HCl (pH 8.0), 1 mM EDTA]. Ten microliters of each decoy was spotted onto 3-mm antibiotic assay disks that were then pressed gently into a thin layer of SNA agar that had been poured over a 24-h-old confluent lawn of S. coelicolor M145 grown on an R2YE agar plate. Induction of antibiotic production would result in a localized increase in pigmentation surrounding the disk. Alternatively, the dumbbell decoy oligonucleotides were used to transfect mycelium from exponentially growing cultures of S. coelicolor M145. Transfection involved washing the cells in equal volumes of TES buffer supplemented with 0.5% (vol/vol) Nonidet P-40 and 0.5% (vol/vol) Triton X-100, and then resuspending them into one-tenth of their original volume in TES plus detergents and supplemented with decoy oligonucleotide. The cells were incubated at 30°C for 15 min with gentle mixing and diluted in the retained culture medium, and incubation was continued. Samples were taken thereafter to assess the levels of production of the two pigmented antibiotics (17). Samples were simultaneously withdrawn for RNA analysis.
Gene Expression and Decoy Copy Number Analysis.
Expression analysis was performed by qrt-PCR. Preparation of cDNA was performed as described in ref. 13. Briefly, 1 μg of RNA was thermally denatured (incubated at 70°C for 10 min and then placed on ice) and mixed with 0.5 units of AMV reverse transcriptase (Amersham), 1 mM dNTPs, 25 pmol of custom primer in 20 μl of the supplier's recommended buffer. The reaction was incubated at three successive temperatures, 45°C, 50°C, and 55°C, each for 30 min. Four microliters of this reaction was used as a template in a 20-μl qrt-PCR prepared in SYBR GreenER reaction mix (Invitrogen) supplemented with 10% (vol/vol) DMSO and 25 pmol of the custom forward and reverse primers for the target gene (actII-orf4) and an internal reference [SCO4742, a conserved hypothetical protein that shows little variation in expression after microarray analysis (A. Hesketh and M.J.B., unpublished results)] (Table 1). Amplification and analysis were performed on a Bio-Rad Chromo4 machine. Determination of the copy number of decoys was similarly performed by using SYBR Green detection. Primers were designed (Table 1) to detect circularized decoy oligonucleotides, and their copy number was determined after sonication of mycelium and comparison to known amounts of exonuclease-treated decoy; these values were corrected for the number of genomes in the sample by reference to the number of copies of SCO4742 present.
Supplementary Material
ACKNOWLEDGMENTS.
We were supported by a grant to the John Innes Centre from the Biotechnology and Biological Sciences Research Council.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710724105/DC1.
References
- 1.Mann MJ, Dzau VJ. Therapeutic applications of transcription factor decoy oligonucleotides. J Clin Invest. 2000;106:1071–1075. doi: 10.1172/JCI11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morishita R, et al. A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc Natl Acad Sci USA. 1995;92:5855–5859. doi: 10.1073/pnas.92.13.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leong PL, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci USA. 2003;100:4138–4143. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park YG, Nesterova M, Agawal S, Cho-Chung YS. Dual blockade of cyclic AMP response element- (CRE) and AP-1-directed transcription by CRE-transcription factor decoy oligonucleotide: gene-specific inhibition of tumor growth. J Biol Chem. 1999;274:1573–1580. doi: 10.1074/jbc.274.3.1573. [DOI] [PubMed] [Google Scholar]
- 5.Chu BCF, Orgel LE. The stability of different forms of double-stranded decoy DNA in serum and nuclear extracts. Nucleic Acids Res. 1992;20:5857–5858. doi: 10.1093/nar/20.21.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griesenbach U, et al. Cytoplasmic deposition of NFκB decoy oligonucleotides is insufficient to inhibit bleomycin-induced pulmonary inflammation. Gene Ther. 2002;9:1109–1115. doi: 10.1038/sj.gt.3301776. [DOI] [PubMed] [Google Scholar]
- 7.Onizuka T, et al. CO2 response for expression of ribulose-1,5-bisphosphate carboxylase/oxygenase genes is inhibited by AT-rich decoy in the cyanobacterium. FEBS Lett. 2003;542:42–46. doi: 10.1016/s0014-5793(03)00336-3. [DOI] [PubMed] [Google Scholar]
- 8.Ahn JD, et al. E2F decoy oligodeoxynucleotides effectively inhibit growth of human tumor cells. Biochem Biophys Res Commun. 2003;310:1048–1053. doi: 10.1016/j.bbrc.2003.09.124. [DOI] [PubMed] [Google Scholar]
- 9.Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Berdy J. Bioactive microbial molecules. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 11.Ward AC, Bora N. Diversity and biogeography of marine actinobacteria. Curr Opin Microbiol. 2006;9:279–286. doi: 10.1016/j.mib.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Chakraburtty R, Bibb MJ. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryding NJ, Anderson TB, Champness WC. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J Bacteriol. 2002;184:794–805. doi: 10.1128/JB.184.3.794-805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adamidis T, Champness W. Genetic analysis of absB, a Streptomyces coelicolor locus involved in global antibiotic regulation. J Bacteriol. 1992;174:4622–4628. doi: 10.1128/jb.174.14.4622-4628.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takano E, et al. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol Microbiol. 1992;6:2797–2804. doi: 10.1111/j.1365-2958.1992.tb01459.x. [DOI] [PubMed] [Google Scholar]
- 16.Uguru GC, et al. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol Microbiol. 2005;58:131–150. doi: 10.1111/j.1365-2958.2005.04817.x. [DOI] [PubMed] [Google Scholar]
- 17.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich: Innes Foundation; 2000. [Google Scholar]
- 18.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong H-J, Hutchings MI, Hill LM, Buttner MJ. The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J Biol Chem. 2005;280:13055–13061. doi: 10.1074/jbc.M413801200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.