Abstract
Alkylresorcinols and alkylpyrones, which have a polar aromatic ring and a hydrophobic alkyl chain, are phenolic lipids found in plants, fungi, and bacteria. In the Gram-negative bacterium Azotobacter vinelandii, phenolic lipids in the membrane of dormant cysts are essential for encystment. The aromatic moieties of the phenolic lipids in A. vinelandii are synthesized by two type III polyketide synthases (PKSs), ArsB and ArsC, which are encoded by the ars operon. However, details of the synthesis of hydrophobic acyl chains, which might serve as starter substrates for the type III polyketide synthases (PKSs), were unknown. Here, we show that two type I fatty acid synthases (FASs), ArsA and ArsD, which are members of the ars operon, are responsible for the biosynthesis of C22–C26 fatty acids from malonyl-CoA. In vivo and in vitro reconstitution of phenolic lipid synthesis systems with the Ars enzymes suggested that the C22–C26 fatty acids produced by ArsA and ArsD remained attached to the ACP domain of ArsA and were transferred hand-to-hand to the active-site cysteine residues of ArsB and ArsC. The type III PKSs then used the fatty acids as starter substrates and carried out two or three extensions with malonyl-CoA to yield the phenolic lipids. The phenolic lipids in A. vinelandii were thus found to be synthesized solely from malonyl-CoA by the four members of the ars operon. This is the first demonstration that a type I FAS interacts directly with a type III PKS through substrate transfer.
Keywords: Azotobacter vinelandii, alkylresorcinol, alkylpyrone, long-chain fatty acid, cyst
Azotobacter vinelandii is a Gram-negative nitrogen-fixing soil bacterium that differentiates into metabolically dormant cysts under adverse environmental conditions (1). During encystment, a considerable portion of the membrane phospholipids are replaced by phenolic lipids, alkylresorcinols and alkylpyrones, which consist of polar aromatic rings and hydrophobic alkyl chains (Fig. 1A) (2, 3). The amphiphilic nature of the phenolic lipids contributes to the formation of stable monomolecular layers in vitro (4), and thus the phenolic lipids presumably allow the cysts to resist desiccation and heat.
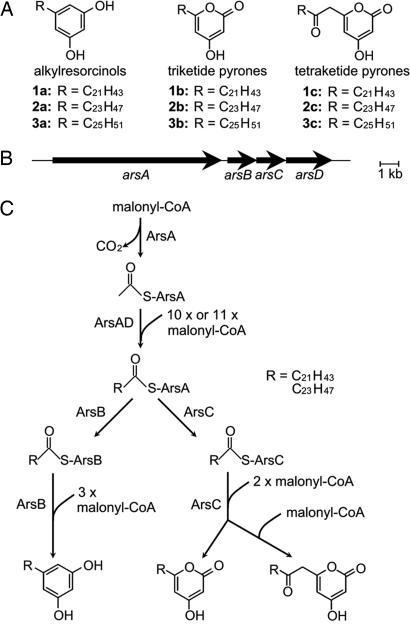
Fig. 1.
Phenolic lipid synthesis in A. vinelandii. (A) Structures of alkylresorcinols and alkylpyrones that accumulate in cysts. (B) Organization of the ars operon. (C) A proposed pathway for the biosynthesis of alkylresorcinols and alkylpyrones by the Ars proteins.
We previously revealed that the ars operon is essential for the biosynthesis of phenolic lipids in A. vinelandii (5). The ars operon directs the synthesis of two type III PKSs, ArsB and ArsC, and two putative type I FASs, ArsA and ArsD (Fig. 1B). Type III PKSs are simple homodimeric proteins that synthesize aromatic polyketides in plants, fungi, and bacteria (6, 7). ArsB and ArsC catalyze the synthesis of alkylresorcinols and alkylpyrones, respectively, from long-chain acyl-CoA and malonyl-CoA (Fig. 1A), indicating that the type III PKSs are responsible for the synthesis of the aromatic moiety of the phenolic lipids. However, details of the biosynthesis of the hydrophobic alkyl moiety of the phenolic lipids remained unknown. Because ArsA and ArsD are homologous to type I FASs and because functionally related genes in bacteria are often present in an operon, we postulated that ArsA and ArsD are responsible for the synthesis of the alkyl chains of the phenolic lipids. Type I FASs are large multifunctional enzymes that have a set of distinct catalytic domains and are distributed in mammals, fungi, and yeasts (8–12). In bacteria, however, the de novo synthesis of fatty acids is catalyzed by type II FASs, which are a group of monofunctional proteins with distinct properties (13, 14).
On the basis of the idea that ArsA and ArsD are involved in synthesis of the alkyl chain of the phenolic lipids, we reconstituted in vivo and in vitro phenolic lipid biosynthesis systems by using the Ars members, demonstrating that ArsA, ArsB, ArsC, and ArsD are required and sufficient for phenolic lipid biosynthesis. Another important finding was that the fatty acid products are covalently bound to ArsA, and the products are released by ArsB or ArsC in the presence of malonyl-CoA. Therefore, the fatty acids are transferred directly from ArsA to the type III PKSs as their starter substrates, resulting in phenolic lipid synthesis in A. vinelandii in the absence of acyl-CoAs intermediates. Thus, we propose a possible route for phenolic lipid biosynthesis solely from malonyl-CoA by the four enzymes in A. vinelandii (Fig. 1C).
Results
Domain Organization of ArsA and ArsD.
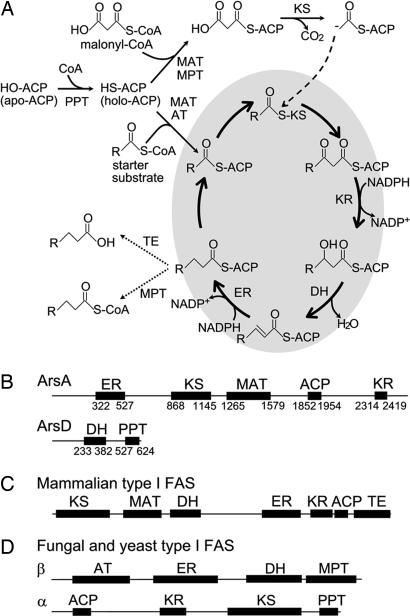
The biosynthesis pathway of fatty acids (14) is illustrated in Fig. 2A. The building blocks of fatty acid biosynthesis are transferred from CoA to the phosphopantetheine thiol of acyl carrier protein (ACP) by transesterification catalyzed by malonyl/acetyl transferase (MAT) or acetyl transferase (AT). A starter unit, mainly acetyl-ACP, is primed to ketosynthase (KS), and the resulting acyl-KS is condensed with an extender unit derived from malonyl-ACP by the action of KS. The newly formed β-keto group is reduced through sequential reactions catalyzed by ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER). Irrespective of the type of FAS, the growing fatty acids are attached to ACP. However, the mechanism of termination of the reaction differs in the different types of FASs. Mammalian type I FASs hydrolyze acyl-ACP to yield a free acid by thioesterase (TE) (15, 16), and fungal and yeast type I FASs transfer the acyl moiety to CoA by malonyl/palmitoyl transferase (MPT) (17) (Fig. 2A). In contrast, type II FASs release their products as ACP esters without cleaving the thioester bond (13).
Fig. 2.
Fatty acid biosynthesis and domain organization of FASs. (A) Fatty acid biosynthesis by FAS. (B) Domain organization of ArsA and ArsD. Numbers below the sequences indicate the amino acid residues, assigning the N-terminal Met as 1. (C) Domain organization of mammalian type I FASs. (D) Domain organization of α and β subunits of fungal and yeast type I FASs.
The domain organizations of ArsA and ArsD were determined through a BLAST search (18) and three-dimensional position-specific scoring matrix (3D-PSSM) analysis (19). Alignments of the amino acid sequences of ArsA, ArsD, and other type II FAS counterparts are shown in supporting information (SI) Fig. 6. Interestingly, the domain organizations of ArsA and ArsD are completely distinct from those of the mammalian and yeast type I FASs (Fig. 2 B–D). ArsA consists of 2,503 aa with a domain structure of ER–KS–MAT–ACP–KR (Fig. 2B). ArsD consists of 636 aa with a domain structure of DH and a phosphopantetheine transferase (PPT) domain (Fig. 2B). PPT catalyzes the posttranslational modification of ACP and transfers a 4′-phosphopantetheine group from CoA to a conserved serine residue of apo-ACP, producing holo-ACP. The most striking feature of ArsA and ArsD is the lack of the TE and MPT domains. The absence of these domains led us to speculate that the fatty acids produced by ArsA and ArsD remain attached to the ACP domain of ArsA and are transferred directly from ArsA to ArsB and ArsC. However, we previously showed that ArsB and ArsC use n-behenyl-CoA as a starter substrate at rates comparable with those of other type III PKSs (5). In addition, type III PKSs usually accept CoA esters as substrates, and the type III PKS reactions have been believed to be independent of ACP (6). As described below, however, we showed by producing ArsA, ArsB, ArsC, and ArsD in a heterologous host that these four enzymes are sufficient for phenolic lipid synthesis. Escherichia coli appeared to be a chemically and genetically clean host for analyzing the functions of ArsA and ArsD because it uses a type II FAS system to produce ACP esters of fatty acids, contains no type I FAS (13), and produces no phenolic lipids.
Heterologous Expression of ars Genes in E. coli.
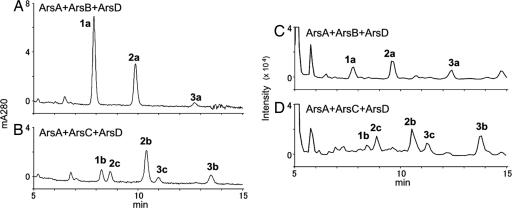
For expression of the Ars proteins in E. coli, pETDuet–ArsA, pACYC–ArsB, pCDF–ArsC, pETDuet–ArsD, and pETDuet–ArsAD were constructed. In addition to these plasmids, pRSF–ACC, which carries the genes encoding the two subunits of acetyl-CoA carboxylase of Corynebacterium glutamicum, was used to increase the intracellular pool of malonyl-CoA (20, 21). The vector plasmids pETDuet, pACYC, pCDF, and pRSF, which contain different replication origins and different selective markers, can be maintained together in the same E. coli cell. E. coli cells harboring combinations of the plasmids were prepared, and lipid extracts of the cells were analyzed by HPLC. E. coli harboring pETDuet–ArsAD, pACYC–ArsB, and pRSF–ACC produced three products (Fig. 3A). A major peak eluting at 7.9 min was determined to contain 5-heneicosylresorcinol (1a) (Fig. 1A) by comparing its liquid chromatography (LC) atmospheric pressure chemical ionization (APCI) tandem mass spectometry (MS/MS) spectrum with that of an authentic sample (data not shown). Similarly, other peaks were determined to contain 5-tricosylresorcinol (2a) and 5-hexacosylresorcinol (3a). E. coli harboring pETDuet–ArsAD, pCDF–ArsC, and pRSF-ACC yielded five products (Fig. 3B). Comparison of their LC-APCIMS/MS spectra with those of authentic samples identified these products as triketide pyrones (1b to 3b) and tetraketide pyrones (2c and 3c) (Figs. 1A and 3B). In contrast, no polyketides were produced when pETDuet–ArsAD was replaced with pETDuet–ArsA or pETDuet–ArsD (data not shown). Interestingly, although we previously revealed that ArsB and ArsC accept a broad range of starter substrates in vitro (5), the expression of ArsB or ArsC alone did not yield any phenolic lipids (data not shown). All of these data suggest that ArsA and ArsD are both required and sufficient for the synthesis of the starter substrates for the type III PKSs, ArsB and ArsC, in phenolic lipid synthesis.
Fig. 3.
Chromatography analysis of the products of Ars reactions. (A and B) HPLC chromatograms of lipid extracts prepared from E. coli harboring pETDuet–ArsAD and pACYC–ArsB (A) or pETDuet–ArsAD and pCDF–ArsC (B). (C and D) Negative extracted ion LC-APCIMS chromatograms of in vitro reactions containing ArsA, ArsB, and ArsD (C) and ArsA, ArsC, and ArsD (D).
Analysis of the ArsAD Reaction in Vitro.
We purified recombinant ArsA, ArsB, ArsC, and ArsD proteins produced in E. coli to investigate their reactions in vitro. The proteins were prepared separately as His-tagged proteins, using the pET and pCold systems (SI Fig. 7). ArsB (43 kDa), ArsC (44 kDa), and ArsD (70 kDa) gave a major single protein band on SDS/PAGE, but ArsA migrated at two positions of ≈220 and 190 kDa. The 190-kDa protein was probably an N-terminally truncated form of ArsA, because the His tag was attached to its C terminus.
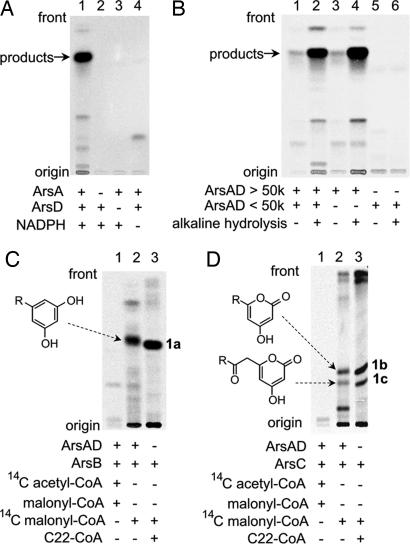
All of the in vitro reactions contained free CoA and MgCl2 to allow phosphopantetheinylation of the ACP domain of ArsA by the PPT domain of ArsD. We first analyzed the reactions of ArsA and ArsD by radio thin-layer chromatography (TLC), using [14C]malonyl-CoA as a substrate. The products were detected as free acids by treatment of the reaction mixture with alkali. An apparent radiolabeled product was detected only in the reaction that contained both ArsA and ArsD in the presence of NADPH (Fig. 4A, lane 1). The [14C]-product was found to actually contain C22–C26 fatty acids, as indicated by comigration of the radioactivity with authentic samples in LC-APCIMS (SI Fig. 8). The ratio of the amounts of the C22, C24, and C26 fatty acids was ≈1:2:1. This production ratio was inconsistent with that of the lipids isolated from the A. vinelandii cyst membrane (2, 3). A major lipid of the cyst membrane was 1a, which was derived from C22 fatty acid. This difference may have been due to the absence of ArsB or ArsC in the reactions of ArsA and ArsD. Neither [14C]acetyl-CoA nor [14C]palmitoyl-CoA was incorporated into the products (SI Fig. 9). This result was not entirely surprising, because most type I FASs are able to initiate fatty acid biosynthesis even in the absence of a starter substrate such as acetyl-CoA, although their reaction rates are low (22, 23). This capacity is attributable to the inherent malonyl decarboxylase activity of FAS (22). These results clearly indicate that ArsA and ArsD are previously uncharacterized FASs that catalyze the synthesis of C22–C26 fatty acids solely from malonyl-CoA.
Fig. 4.
Radio-TLC analysis of in vitro reaction products. (A) Analysis of products produced from malonyl-CoA by ArsA and ArsD. The production of radiolabeled products was observed in the presence of [2-14C]malonyl-CoA, NADPH, ArsA, and ArsD (lane 1). No radioactivity was seen in the absence of ArsA (lane 2), ArsD (lane 3), or NADPH (lane 4). (B) Analysis of a hydrolyzed reaction mixture containing ArsA and ArsD. After incubation of malonyl-CoA in the presence of ArsA and ArsD, the reaction mixture was hydrolyzed by alkali, extracted with ethyl acetate, and subjected to TLC. Lane 1 is a negative control containing a complete reaction mixture that was not subjected to hydrolysis. [14C] attached to ArsA was fractionated into the water layer and did not appear in the TLC analysis. Lane 2 contains a complete reaction mixture that was hydrolyzed and extracted with ethyl acetate. Lanes 3 and 4 contain a macromolecular fraction of the reaction mixture that was hydrolyzed (lane 4) or not hydrolyzed (lane 3). Lanes 5 and 6 contain a low-molecular-weight fraction that was hydrolyzed (lane 6) or not hydrolyzed (lane 5). (C) Analysis of the products of ArsA, ArsB, and ArsD from two different starter substrates. The starter substrates used were [2-14C]acetyl-CoA (lane 1) and [2-14C]malonyl-CoA (lane 2). Lane 3 contains an authentic sample 1a prepared from behenyl-CoA (C22-CoA) and malonyl-CoA by ArsB. (D) Analysis of the products of ArsA, ArsC, and ArsD from two different starter substrates. The starter substrates used were [2-14C]acetyl-CoA (lane 1) and [2-14C]malonyl-CoA (lane 2). Lane 3 contains authentic samples 1b and 1c prepared from behenyl-CoA (C22-CoA) and malonyl-CoA by ArsC.
Because ArsA and ArsD lack TE and MPT domains, the C22–C26 fatty acids were expected to remain attached to the phosphopantetheinyl thiol of the ACP domain in ArsA. Therefore, we separated low-molecular-weight compounds from macromolecules in the reaction mixture by ultrafiltration. As expected, fatty acids were detected in the macromolecular fraction, which was hydrolyzed by alkali to cleave the thioester bond between the fatty acids and the phosphopantetheine thiol of ACP (Fig. 4B, lane 4). A very small amount of fatty acids was detected in the macromolecular fraction that had not been subjected to alkali hydrolysis (Fig. 4B, lane 3). These products presumably resulted from nonenzymatic hydrolysis that occurred after the separation, because no such products were found in the fraction of low-molecular-weight compounds (Fig. 4B, lanes 5 and 6).
In Vitro Analysis of the ArsABCD Reaction.
We next reconstituted an in vitro phenolic lipid synthesis system by adding the type III PKSs to the reaction of ArsA and ArsD. Incubation of ArsA, ArsB, and ArsD in the presence of [14C]malonyl-CoA, CoA, MgCl2, and NADPH yielded a radiolabeled band, as revealed by TLC (Fig. 4C, lane 2). Further LC-MS analysis revealed that the product contained alkylresorcinols (1a to 3a) derived from the C22–C26 fatty acids produced by the actions of ArsA and ArsD (Fig. 3C). Similarly, the reaction of ArsA, ArsC, and ArsD gave a radiolabeled band (Fig. 4D, lane 2), shown by LC-MS analysis to contain triketide pyrones (1b to 3b) and tetraketide pyrones (2c and 3c) (Fig. 3D). These in vitro results were consistent with those obtained from the above in vivo study (Fig. 3 A and B). Again, [14C]acetyl-CoA was not incorporated into the products (Fig. 4 C and D, lane 1), indicating that malonyl-CoA is the only carbon source required for phenolic lipid synthesis by the Ars enzymes.
Hand-to-Hand Transfer of Starter Substrate from ArsA to Type III PKSs.
We monitored the localization of radioactivity derived from [14C]malonyl-CoA in the sequential reaction catalyzed by ArsA, ArsB, ArsC, and ArsD. After the incubation of ArsA and ArsD with [14C]malonyl-CoA, CoA, MgCl2, and NADPH, we removed low-molecular-weight compounds, including [14C]malonyl-CoA that remained intact, by ultrafiltration. The resulting macromolecular fraction was subjected to SDS/PAGE followed by autoradiography. Radioactivity was detected on ArsA (Fig. 5A, lane 1), indicating that the fatty acids were attached to ArsA, probably via a thioester bond with its ACP domain. The attachment of the fatty acids to ArsA was in agreement with the domain architecture of ArsA and ArsD, which contain no TE or MPT domain (Fig. 2B).
Fig. 5.
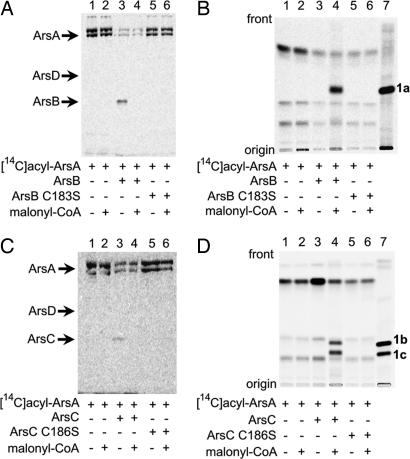
Radiolabeling analysis of the direct transfer of acyl products from ArsA to ArsB (A and B) or ArsC (C and D) by SDS/PAGE (A and C) and radio-TLC (B and D). The substrates used were [14C]acyl-ArsA (lane 1), [14C]acyl-ArsA + malonyl-CoA (lane 2), [14C]acyl-ArsA + ArsB or ArsC (lane 3), [14C]acyl-ArsA + ArsB or ArsC + malonyl-CoA (lane 4), [14C]acyl-ArsA + ArsB or ArsC mutants (lane 5), and [14C]acyl-ArsA + ArsB or ArsC mutants + malonyl-CoA (lane 6). Lane 7 contains 1a (B) or 1b and 1c (D). Production of radiolabeled alkylresorcinol (B) and alkylpyrone (D) was observed (lane 4). The chemical structures of these compounds are shown in Fig. 1A.
We next incubated ArsB with the radiolabeled ArsA. As expected, the radioactivity on ArsA decreased with a concomitant transfer of radioactivity to ArsB (Fig. 5A, lane 3), indicating that ArsB received the fatty acids directly from ArsA. Adding nonlabeled malonyl-CoA to this reaction resulted in the removal of radioactivity from ArsB (Fig. 5A, lane 4) and the detection of the synthesis of alkylresorcinols by radio-TLC analysis (Fig. 5B, lane 4). The C183S mutant of ArsB did not have the ability to accept fatty acids from ArsA (Fig. 5A, lane 5), suggesting that the active-site cysteine 183 bound the fatty acids, as in the case of the priming of other type III PKSs with acyl-CoA (6). These results showed that the fatty acids produced by ArsA and ArsD were directly transferred to ArsB and served as starter substrates for the synthesis of alkylresorcinols from malonyl-CoA as an extender substrate. We were able to duplicate the results by replacing ArsB with ArsC; the fatty acids on ArsA were transferred directly to ArsC (Fig. 5 C and D).
Discussion
The in vivo and in vitro reconstitution of the phenolic lipid biosynthesis systems have clearly demonstrated that alkylresorcinols and alkylpyrones are synthesized from malonyl-CoA by the actions of the four Ars enzymes. Of these enzymes, ArsB and ArsC have been shown to be type III PKSs that synthesize alkylresorcinols and alkylpyrones, respectively, from several acyl-CoAs with various side-chain lengths as starter substrates (5). However, the present study confirmed that the active-site cysteine residues of ArsB and ArsC receive C22–C26 fatty acids, the products of the actions of ArsA and ArsD on malonyl-CoA, directly from ArsA, which probably has the fatty acids attached to its ACP domain. To our knowledge, this is the first experimental demonstration of the direct transfer of the product of a type I FAS to a type III PKS. The fatty acids received by ArsB and ArsC serve as starter substrates for the synthesis of alkylresorcinols and alkylpyrones, respectively, by two or three successive extensions with malonyl-CoA. Thus, we have revealed the whole biosynthesis pathway of the phenolic lipids that are essential for cyst formation in A. vinelandii (Fig. 1C).
The direct transfer of the fatty acids from ArsA to ArsB and ArsC was expected from the domain architecture of ArsA and ArsD, which belong to the type I FAS family. ArsA has the structure of ER–KS–MAT–ACP–KR, and ArsD has the structure of DH–PPT (Fig. 2B). Mammalian type I FASs form an α2 homodimer with a domain structure of KS–MAT–DH–ER–KR–ACP–TE (Fig. 2C) and release fatty acids by cleavage of a thioester linkage between the fatty acids and ACP by the thioesterase activity of the TE domain. Fungal and yeast type I FASs form an α6β6 heterododecamer with a domain structure of AT–ER–DH–MPT/ACP–KR–KS–PPT (Fig. 2D) and transfer their product from ACP to CoA through the malonyl/palmitoyl transferase activity of the MPT domain. The absence of TE and MPT domains in ArsA and ArsD led us to speculate that the fatty-acid products would remain attached to the ACP domain of ArsA and would be transferred directly to ArsB and ArsC. In fact, the fatty acids produced by ArsA and ArsD are directly transferred from the ACP domain to the active-site cysteine residues of ArsB and ArsC, where they serve as starter substrates for phenolic lipid synthesis.
ArsA and ArsD cooperatively synthesize C22–C26 fatty acids solely from malonyl-CoA. The phospholipids produced by a type II FAS in A. vinelandii mainly have chain lengths of C16 and C18 (13, 24), which suggests that ArsB and ArsC use the C22–C26 fatty acids produced by ArsA and ArsD of type I FASs. The hand-to-hand transfer of the fatty acids from ArsA to the type III PKSs ArsB and ArsC presumably facilitates efficient catalysis by these PKSs.
A BLAST search, using the ArsA and ArsD sequences, revealed that some actinomycetes and cyanobacteria, including Streptomyces avermitilis and Gloeobacter violaceus, also possess a type I FAS containing domains similar to ArsA and ArsD sequences (SI Fig. 10). Therefore, the ArsAD-like system is not unique to A. vinelandii. In S. avermitilis and G. violaceus, an ArsD-like protein and a putative type I PKS are fused into one polypeptide (SI Fig. 10). In these cases, the product(s) synthesized by the ArsAD-like systems could be passed to the fused type I PKS portion as a substrate by a mechanism similar to that observed for ArsA and the type III PKSs.
We previously showed that ArsB and ArsC use behenyl-CoA as a starter substrate and malonyl-CoA as an extender substrate to synthesize alkylresorcinols and alkylpyrones in vitro (5), in agreement with the idea that type III PKSs use acyl-CoA thioesters as starter substrates. The transfer of the fatty acids from the ACP domain of ArsA to ArsB and ArsC implies that these type III PKSs can accept the starter fatty acids not only from CoA but also from ACP. Very recently, SCO7671, a type III PKS in Streptomyces coelicolor A3(2), has been shown to accept acyl-ACP as a starter substrate in vitro (25), although its biological role is unknown. These intriguing findings warrant future structural and mechanistic studies.
Recently, enzymatic reactions other than thioesterification or transesterification between ACP and CoA to release fatty acids from type I FASs were found in some organisms. For example, Pap5A catalyzes the transfer of mycocerosic acid analogs to an alcohol, phthiocerol, from the ACP domain of a type I FAS, Mas, in Mycobacterium tuberculosis (26). In addition, direct transfer of C6 fatty acid is predicted to occur between HexAB (type I FASs) and PksA (a type I PKS) in Aspergillus parasiticus (27, 28). It is also hypothesized that steely, a fusion protein of a type I FAS and a type III PKS, directly transfers acyl products attached to the ACP domain of the type I FAS to the active site of the type III PKS in Dictyostelium discoideum (29). In the present study, we showed that the radiolabeled fatty acids attached to the ACP domain of ArsA are directly transferred to the catalytic cysteines of the type III PKSs ArsB and ArsC to yield phenolic lipids. To our knowledge, this is the first demonstration that a type I FAS directly interacts with a type III PKS through substrate transfer.
Materials and Methods
Construction of Plasmids and Mutagenesis.
See SI Text.
Heterologous Expression of Ars Enzymes in E. coli.
pETDuet–ArsA, pETDuet–ArsD, pETDuet–ArsAD, pACYC–ArsB, pCDF–ArsC, and pRSF–ACC (21) were used for the coexpression of ArsABCD in E. coli BLR (DE3); 100 μg/ml ampicillin, 34 μg/ml chloramphenicol and 20 μg/ml kanamycin, and 50 μg/ml streptomycin were used when appropriate. Recombinant strains were grown at 26°C in 3 ml of Luria–Bertani broth. When the optical density at 600 nm reached 0.5, heterologous expression in the cells was induced by the addition of 0.25 mM isopropyl thio-β-d-galactoside. The cells were then cultured for an additional 24 h at 15°C. The cells were harvested by centrifugation and extracted with chloroform/methanol (2:1, vol/vol). After the cell debris had been filtered off, the filtrate was evaporated to dryness. The residual material was dissolved in methanol for LC-APCI-MS and HPLC analysis. LC-APCI-MS analysis was carried out by using the esquire high-capacity trap plus system (Bruker Daltonics) with a Pegasil-B C4 reversed-phase column (4.6 × 250 mm) (Shenshu Scientific) and 90% acetonitrile in water containing 0.1% trifluoroacetic acid as an eluent at a flow rate of 1 ml/min. HPLC analysis was performed by using the LaChrom Elite system (Hitachi) with the same column, eluent, and flow rate as for the LC-APCI-MS analysis. UV spectra were detected by using the Hitachi diode array detector L-2450.
Expression and Purification of Ars Proteins.
ArsB and ArsC were purified as described in ref. 5. ArsA with a histidine tag at its C terminus and ArsD with a histidine tag at its N terminus were produced in E. coli BL21 (DE3). E. coli BL21 (DE3) harboring pET26b–ArsA was grown at 26°C in 200 ml of Luria–Bertani broth containing 20 μg/ml kanamycin. When the optical density at 600 nm reached 0.5, heterologous expression in the cells was induced by the addition of 0.25 mM isopropyl thio-β-d-galactoside. The cells were then cultured for an additional 24 h at 15°C. E. coli BL21 (DE3) harboring pCold I–ArsD was grown at 37°C in 200 ml of Luria–Bertani broth containing 100 μg/ml ampicillin. When the optical density at 600 nm reached 0.5, the cells were incubated for 30 min at 15°C and then heterologous expression was induced by the addition of 0.025 mM isopropyl thio-β-d-galactoside. The cells were then cultured for an additional 24 h at 15°C, harvested by centrifugation, and resuspended in buffer containing 100 mM potassium phosphate (pH 7.7), 500 mM KCl, 10% glycerol, and 1 mM Tris[2-carboxyethyl]phosphine. From a cell-free extract prepared by sonication, each protein was purified on a His-trap HP column (GE Healthcare Biosciences). The purified proteins were dialyzed against a buffer containing 100 mM potassium phosphate (pH 7.7), 150 mM KCl, 10% glycerol, and 1 mM Tris[2-carboxyethyl]phosphine.
FAS Assay.
The standard reaction mixture contained 10 μM [2-14C]malonyl-CoA, 100 mM potassium phosphate (pH 7.7), 10% glycerol, 1 mM Tris[2-carboxyethyl]phosphine, 10 mM MgCl2, 100 μM CoA, 5 mM NADPH, 1 μM ArsA, and 1 μM ArsD in a volume of 200 μl. After incubation at 30°C for 1 h, the reactions were quenched by the addition of 200 μl of ice-cold 20% trichloroacetic acid (wt/vol). The pellet was resuspended in 50 μl of 350 mM KOH and incubated at 65°C for 20 min to release covalently bound products. After acidification with 20 μl of 6 M HCl, the products were extracted with 400 μl of ethyl acetate and the organic layer was evaporated to dryness. The residual material was dissolved in 15 μl of methanol for TLC. A silica-gel 60 WF254 TLC plate (Merck) was developed in benzene/acetone/acetic acid (85:15:1, vol/vol/v), and the 14C-labeled compounds were detected by using a Bio-imaging Analyzer System (BAS)-MS imaging plate (Fujifilm Life Science).
FAS-PKS Assay.
The standard reaction mixture contained 10 μM [2-14C]malonyl-CoA, 100 mM potassium phosphate (pH 7.7), 10% glycerol, 1 mM Tris[2-carboxyethyl]phosphine, 10 mM MgCl2, 100 μM CoA, 5 mM NADPH, 1 μM ArsA, 1 μM ArsB, 1 μM ArsC, and 1 μM ArsD in a volume of 200 μl. After incubation at 30°C for 1 h, the reactions were quenched by the addition of 20 μl of 6 M HCl. The products were extracted with 400 μl of ethyl acetate, and the organic layer was evaporated to dryness. The residual material was dissolved in 15 μl of methanol for TLC and LC-APCI-MS analysis, carried out as described above.
Direct Transfer of Acyl Substrates from ArsA to Type III PKSs.
The reaction of ArsA and ArsD was carried out as described in the FAS assay section to produce [14C]acyl-ArsA. [14C]acyl-ArsA from which low-molecular-weight compounds, such as [2-14C]malonyl-CoA and NADPH, were removed by ultrafiltration, using a 50,000-MW cutoff membrane (Amicon Ultra-4; Millipore), was used as a substrate for the ArsB and ArsC reactions. Solutions of 1 μM ArsB or ArsC and 100 μM malonyl-CoA were added to the [14C]acyl-ArsA solution and incubated at 30°C for 20 min. For SDS/PAGE analysis on 10% (wt/vol) polyacrylamide gels, the reactions were quenched by boiling before electrophoresis. The 14C-labeled proteins were detected by using a BAS-MS imaging plate. For TLC analysis, the reactions were quenched by adding 20 μl of 6 M HCl, and the products were extracted and analyzed as described above.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by the New Energy and Industrial Technology Development Organization of Japan, a Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics” from Monkasho, and a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709819105/DC1.
References
- 1.Lin LP, Sadoff HL. J Bacteriol. 1968;95:2336–2343. doi: 10.1128/jb.95.6.2336-2343.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su CJ, Reusch RN, Sadoff HL. J Bacteriol. 1981;147:80–90. doi: 10.1128/jb.147.1.80-90.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reusch RN, Sadoff HL. Nature. 1983;302:268–270. doi: 10.1038/302268a0. [DOI] [PubMed] [Google Scholar]
- 4.Kozubek A, Tyman JH. Chem Rev. 1999;99:1–26. doi: 10.1021/cr970464o. [DOI] [PubMed] [Google Scholar]
- 5.Funa N, Ozawa H, Hirata A, Horinouchi S. Proc Natl Acad Sci USA. 2006;103:6356–6361. doi: 10.1073/pnas.0511227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin MB, Noel JP. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 7.Shen B. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 8.Schweizer E, Hofmann J. Microbiol Mol Biol Rev. 2004;68:501–517. doi: 10.1128/MMBR.68.3.501-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maier T, Jenni S, Ban N. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 10.Leibundgut M, Jenni S, Frick C, Ban N. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 11.Jenni S, Leibundgut M, Boehringer D, Frick C, Mikolasek B, Ban N. Science. 2007;316:254–261. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- 12.Lomakin IB, Xiong Y, Steitz TA. Cell. 2007;129:319–332. doi: 10.1016/j.cell.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Rock CO, Cronan JE. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 14.White SW, Zheng J, Zhang Y-M, Rock CO. Annu Rev Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 15.Smith S, Witkowski A, Joshi AK. Prog Lipid Res. 2003;42:289–317. doi: 10.1016/s0163-7827(02)00067-x. [DOI] [PubMed] [Google Scholar]
- 16.Chakravarty B, Gu Z, Chirala SS, Wakil SJ, Quiocho FA. Proc Natl Acad Sci USA. 2004;101:15567–15572. doi: 10.1073/pnas.0406901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynen F. Eur J Biochem. 1980;112:431–442. doi: 10.1111/j.1432-1033.1980.tb06105.x. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Kelley LA, MacCallum RM, Sternberg MJ. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 20.Davis MS, Solbiati J, Cronan JE., Jr J Biol Chem. 2000;275:28593–28598. doi: 10.1074/jbc.M004756200. [DOI] [PubMed] [Google Scholar]
- 21.Miyahisa I, Kaneko M, Funa N, Kawasaki H, Kojima H, Ohnishi Y, Horinouchi S. Appl Microbiol Biotechnol. 2005;68:498–504. doi: 10.1007/s00253-005-1916-3. [DOI] [PubMed] [Google Scholar]
- 22.Kresze GB, Steber L, Oesterhelt D, Lynen F. Eur J Biochem. 1977;79:191–199. doi: 10.1111/j.1432-1033.1977.tb11797.x. [DOI] [PubMed] [Google Scholar]
- 23.Arai K, Kawaguchi A, Saito Y, Koike N, Seyama Y, Yamakawa T, Okuda S. J Biochem (Tokyo) 1982;91:11–18. doi: 10.1093/oxfordjournals.jbchem.a133667. [DOI] [PubMed] [Google Scholar]
- 24.Su CJ, Reusch R, Sadoff HL. J Bacteriol. 1979;137:1434–1436. doi: 10.1128/jb.137.3.1434-1436.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruschow S, Buchholz TJ, Seufert W, Dordick JS, Sherman DH. ChemBioChem. 2007;8:863–868. doi: 10.1002/cbic.200700026. [DOI] [PubMed] [Google Scholar]
- 26.Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, Mohanty D, Gokhale RS. Mol Cell. 2005;17:631–643. doi: 10.1016/j.molcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe CM, Townsend CA. Chem Biol. 2002;9:981–988. doi: 10.1016/s1074-5521(02)00213-2. [DOI] [PubMed] [Google Scholar]
- 28.Crawford JM, Dancy BC, Hill EA, Udwary DW, Townsend CA. Proc Natl Acad Sci USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Austin MB, Saito T, Bowman ME, Haydock S, Kato A, Moore BS, Kay RR, Noel JP. Nat Chem Biol. 2006;2:494–502. doi: 10.1038/nchembio811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.