Abstract
Interleukin-22 (IL-22) is a class 2 cytokine whose primary structure is similar to that of interleukin 10 (IL-10) and interferon-γ (IFN-γ). IL-22 induction during acute phase immune response indicates its involvement in mechanisms of inflammation. Structurally different from IL-10 and a number of other members of IL-10 family, which form intertwined inseparable V-shaped dimers of two identical polypeptide chains, a single polypeptide chain of IL-22 folds on itself in a relatively globular structure. Here we present evidence, based on native gel electrophoresis, glutaraldehyde cross-linking, dynamic light scattering, and small angle x-ray scattering experiments, that human IL-22 forms dimers and tetramers in solution under protein concentrations assessable by these experiments. Unexpectedly, low-resolution molecular shape of IL-22 dimers is strikingly similar to that of IL-10 and other intertwined cytokine dimeric forms. Furthermore, we determine an ab initio molecular shape of the IL-22/IL-22R1 complex which reveals the V-shaped IL-22 dimer interacting with two cognate IL-22R1 molecules. Based on this collective evidence, we argue that dimerization might be a common mechanism of all class 2 cytokines for the molecular recognition with their respective membrane receptor. We also speculate that the IL-22 tetramer formation could represent a way to store the cytokine in nonactive form at high concentrations that could be readily converted into functionally active monomers and dimers upon interaction with the cognate cellular receptors.
Introduction
Interleukin-22 (IL-22) is a novel cytokine primarily identified in murine CD4+ T cells upon induction with IL-9, and, later, in human cells (see (1–3) for reviews). IL-22 mediates acute-phase response signals in hepatocytes, eliciting the production of acute-phase reactive proteins. Its known target organs include pancreas, skin, liver, gastrointestinal tract, and kidneys. IL-22 induces pancreatic-associated protein, PAP1, in acinar cells and the antimicrobial proteins β-defensin 2 and 3 and psoriasin in human keratinocytes. The biological function of this cytokine, therefore, is to promote antimicrobial defense of the outer barriers (skin, lungs, gastrointestinal tract), directly stimulating the innate immune response of tissues (4). Furthermore, IL-22 acts as an effector cytokine and mediates communication between immune system and epithelial cells by modulating IL-23 response in physiological and pathological immune responses such as dermal inflammation and acanthosis (5).
Human and murine IL-22 orthologs encode 179 amino-acid proteins that are 79% identical to each other, bearing just 22% (human) and 25% (mouse) identity with their IL-10 paralogs. IL-22 is a member of the IL-10 family of cytokines (comprising also IL-19, -20, -24, -26, -28(A/B), and -29), proteins involved in cell signaling which triggers immune response (reviewed in (2)). IL-10 family belongs to Class 2 cytokine superfamily, which also includes interferons type I and type II. Interferon-γ (IFN-γ) is the only type II interferon, and both human and murine IL-22 genes are chromosomally located in a close proximity of the IFN-γ gene (6).
Class 2 cytokines bind to their specific receptors, which include tissue factor, IFN-γR1, IFN-γR2, IL-10R1, IL-10R2, IL-20R1, IL-20R2, IL-28R, and IL-22R1 (7). The extracellular cytokine-binding domain of these receptors is composed of two fibronectin type III domains, each consisting of two antiparallel β-sheets, formed by seven β-strands (8–12). On the cell surface, IL-22 binds to two receptors: an IL-22 specific receptor chain termed IL-22R1, and a second component of IL-10 receptor, IL-10R2, shared with IL-10, IL-26, IL28, and IL-29 (13,14). The IL-22R1 ligand-binding chain is mostly expressed in tissues of the digestive tract, whereas the IL-10R2 is a ubiquitously expressed signal-transducing chain (15,16). Enzyme-linked immunoabsorbant assay-based studies showed that IL-22 first binds to IL-22R1 and only afterwards does the IL-22/IL-22R1 complex expose an interface that is able to bind the IL-10R2 receptor chain (17). Supporting this model, another line of evidence showed that affinity interaction between IL-22 and IL-22R1 is in the nanomolar range, whereas that of IL-22 and IL-10R2 is in the millimolar range (18,19). Therefore, the assembly of the ternary IL-22/IL-22R1/IL-10R2 receptor complex shows sequential interactions dictated by the affinity of the pairing, before eliciting signaling on the cell membrane.
This simultaneous and complementary receptor recognition leads to activation of cytokine-specific signal transduction pathways. Upon binding to its receptors, IL-22 induces Jak1 and Tyk2 phosphorylation, activates three major MAP kinases pathways, and promotes activation of STAT (signal transducer and activation transcription factors) -1, -3, and -5 as measured by STAT-responsive promoter activation. STAT proteins translocate to the nucleus and activate the transcription of genes involved in IL-22 biological response (1,2).
Recombinant IL-22 has been produced both in Escherichia coli and Drosophila melanogaster S2 cells, purified to homogeneity and crystallized (20,21). Crystallographic studies of IL-22 revealed that the protein folds in an α-helical bundle with tertiary structure strikingly different from IL-10 architecture (11,18,22,23). Unlike IL-10, which exists as an intimate intertwined dimer forming a symmetric V-shaped molecule, bacterially expressed IL-22 is arranged in a crystal as an asymmetric dimer of two separate and independent molecules (24), and IL-22 crystals of protein produced in S2 insect cells contain six molecules in the asymmetric unit organized in three noncovalent dimers (21). Comparisons between IL-22/IL-22R1 homology model and equivalent residues involved in the cytokine-receptor binding of IL-10/IL-10R1 and IFN-γ/IFN-γR1 complexes suggest that IL-22 dimerization is probably an artifact of crystallization (21,24). Indeed, IL-22 amino-acid residues, from the putative IL-22R1 binding site, interact with other IL-22 molecules, and this incidental interaction is stabilized in the crystallographic arrangement. As a result, IL-22R1 binding site is hidden at the dimer molecular interfaces observed in crystal structures (21,24). If the structure of IL-22 adopted in solution is similar to that observed in crystallographic arrangements, it should hamper IL-22 recognition by the IL-22R1 receptor under physiological conditions.
To determine oligomeric state and to structurally characterize human IL-22 alone and in the complex with its high-affinity cognate receptor IL-22R1, we used dynamic light scattering, glutaraldehyde cross-linking, native gel electrophoresis, and small angle x-ray scattering (SAXS), a method of choice for experimental determination of protein quaternary structure and its molecular shape in solution (25,26). Our results reveal that under the experimental conditions IL-22 forms dimers and tetramers, and that SAXS-derived low resolution models of IL-22 dimers are surprisingly similar to the low resolution shape of the IL-10 intimate intertwined dimers. Furthermore, low resolution shape of IL-22 tetramers closely resembles tetrameric organization of IFN-γ crystallographic structure (10). Finally, we determined a low resolution ab initio molecular shape of the IL-22/IL-22R1 complex and demonstrated that it is composed of an IL-22 dimer recognized by two IL-22R1 receptors. Based on this collective evidence, we speculate that IL-22 interaction with its cognate receptors, similar to other class 2 cytokines, involves dimerization; and that the tetrameric form of the cytokine might represent inactive stock of IL-22, which dissociates into smaller multimeric forms upon interactions with its respective receptors on the cell surface.
Materials and methods
Protein expression and purification
Human IL-22 (residues 29–179) was purified from inclusion bodies as described previously (24). The purity of the IL-22 was estimated by SDS-PAGE and the concentration determined by UV absorbance. Protein bioactivity was assessed as described (27) and recombinant IL-22 protein was found to be fully active. Recombinant human IL-22R1 was produced in E. coli as follows. The IL-22R1 extracellular domain (corresponding to amino acids P18–T228) was cloned into a derivative of plasmid pET3A (Stratagene, La Jolla, CA). An additional glycine was added in front of P18 to avoid heterogenous processing of the N-terminal methionine. In addition, cysteine 204, which is not conserved in IL22R1 of other species, was mutated to leucine. Functional cellular assay of the mutated C204L receptor showed that it retains full activity. E. coli strain BL21-AI (Invitrogen Life Technologies, Paisley, UK) was used as the expression host. Mutated IL-22R1 was expressed as inclusion bodies. Inclusion bodies were purified as previously described (24,27) and solubilized in 6 M guanidine hydrochloride, 25 mM sodium acetate (pH 5.5). The purity of the IL-22R1 was estimated at 80% based on SDS-PAGE and Coomassie-blue staining analysis. The IL-22R1 protein was refolded by direct dilution of the solubilized inclusion bodies in the following folding mixture: 25 μg/ml IL-22R1, 50 mM Tris-HCl, 2 mM EDTA, 0.5 M L-arginine, 1 mM reduced glutathione, and 0.1 mM oxidized glutathione (pH 8.5). The solution was incubated for 1–2 weeks at 4°C. Then, glycerol and CHAPS (CALBIOCHEM, Darmstadt, Germany) were added at 10% (v/v) and 1 mM, respectively. The folding mixture was then concentrated by ultrafiltration and purified on a Superdex200 gel filtration column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). The protein was eluted with 25 mM Bicine, 150 mM NaCl, and 1 mM CHAPS (pH 9.5). IL2/IL22R1 complex was obtained by mixing of purified IL22 and IL22R1 at a molar ratio of ∼1.2 (IL22/IL22R1) in 25 mM Bicine, 150 mM NaCl, and CHAPS 1 mM (pH 9.5) and incubated overnight at 4°C. The complex was purified on a Superdex200 gel filtration column with 25 mM Bicine, 150 mM NaCl, and 1 mM CHAPS (pH 9.5) as elution buffer and concentrated by ultrafiltration to 5 mg/ml.
Native polyacrylamide gel electrophoresis (native PAGE)
The electrophoresis was performed in 8–25% polyacrylamide gradient gel using the PhastSystem (Pharmacia, Peapack, NJ) according to the manufacturer's protocol. Molecular mass calibration kit containing the following proteins—97 kDa, phosphorylase b; 66 kDa, bovine serum albumin; 45 kDa, ovalbumin; 30 kDa, carbonic anhydrase; 20.1 kDa, soybean trypsin inhibitor; and 14.1 kDa, α-lactalbumin (Amersham Biosciences, Piscataway, NJ)—was run in parallel with sample to estimate molecular weights.
Glutaraldehyde cross-linking
IL-22 aliquots (2 μg) in 25 mM HEPES, 150 mM NaCl (pH 8) were incubated with 0.00–0.05% (v/v) glutaraldehyde (Merck, Whitehouse Station, NJ) in a total volume of 10 μl at 25°C for 30 min. Reactions were stopped by addition of SDS-PAGE loading dye and the products analyzed by SDS-PAGE.
Dynamic light scattering experiments
IL-22 oligomerization state was assessed by dynamic light scattering (DLS). DLS measurements were performed with a DynaPro MS200 instrument (Protein Solutions, Salt Lake City, UT) at 4°C using a 12 μl cuvette. Concentrated IL-22 protein (10 mg/ml) was sequentially diluted to 5, 2.5, 1.25, 0.625, and 0.325 mg/ml in 10 mM Tris-HCl, 10 mM NaCl (pH 8) before measurements.
Small-angle x-ray scattering data acquisition
SAXS data were measured at the small-angle scattering beamline of the National Synchrotron Light Laboratory (Campinas, Brazil) using a one-dimensional position-sensitive detector (28) as described previously (29). IL-22 scattering was measured at concentrations of 10, 5, and 2.5 mg/mL in 10 mM Tris-HCl, and 10 mM NaCl, pH 8, and IL-22/IL-22R1 at 3.5 mg/mL and 1.0 mg/mL, in 20 mM Bicine, 150 mM NaCl, and 1 mM CHAPS, pH 9.5 (Fig. 1). Since our SAXS curves in all cases exhibit a wide range of Guinier behavior, without any significant positive deviation at very small q and with radii of gyration derived from Guinier plots that are compatible with those of the proposed models, we can safely conclude that no effects of protein aggregation were occurring under experimental conditions. Samples were measured at the wavelength λ = 0.1488 nm, with detector/sample distance of 1033 mm, covering the momentum transfer range 0.15 < q < 3.5 nm−1 (q = 4πsinθ/λ, where 2θ is the scattering angle). The protein solutions and buffer were exposed in 1-min frames to monitor radiation damage and beam stability. Scattering from the buffer was subtracted after the data being normalized to the intensity of the incident beam and corrected for detector response, and the difference curves were scaled for concentration.
Figure 1.
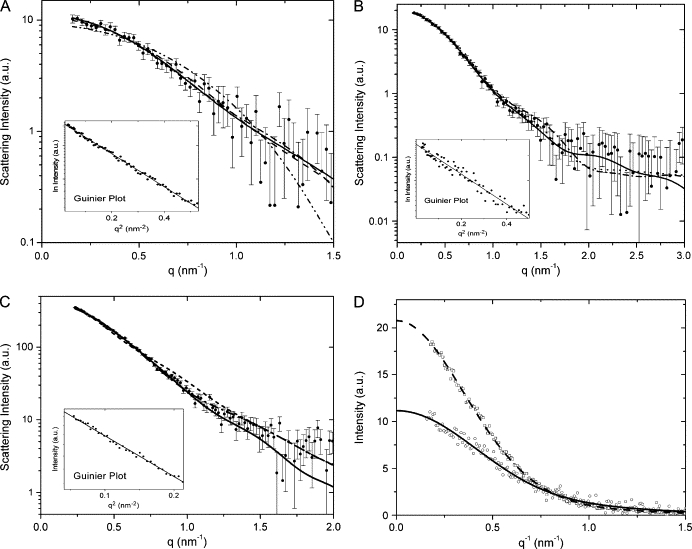
Experimental solution scattering curves of IL-22 (log I versus q) and results of the fitting procedures for (A) IL-22 dimer, experimental curve after subtraction of a constant value as described in Materials and Methods (solid spheres with error bars); scattering intensity from the DAMs (DAMMIN) (solid line); from the proposed high-resolution model (dash-dot); from IL-10 (dash); and from the crystallographically observed IL-22 dimer (30) (dash-dot-dot). (B) IL-22 tetramer, experimental curve (solid spheres with error bars); scattering intensity from the DAMs (DAMMIN) (solid line); from the proposed high-resolution model (dash-dot); and from IFN-γ tetramer (dots). (C) IL-22/IL-22R1, experimental scattering curve (solid spheres with error bars); scattering intensity from the DAMs (DAMMIN) (solid line); from the proposed high-resolution model (dash-dot); and from IL-10/IL-10R1 (short dash). Insets display the correspondent Guinier plots (log I versus q2). (D) Experimental intensities I(q)/c versus q for IL-22 dimer (open circles) and IL-22 tetramer (open squares) are superposed with I(q)/c extrapolated to zero scattering angle for IL-22 dimer (solid line) and IL-22 tetramer (dash line).
Scattering data analysis
SAXS analysis was carried out using the ATSAS program package (30). The distance distribution functions p(r) and the radii of gyration (Rg) were evaluated by the indirect Fourier transform program GNOM (30). Low resolution particle shapes of IL-22 multimers were restored from the experimental SAXS data using two ab initio procedures. In the first method, the shape is described by an angular envelope function, parameterized in terms of spherical harmonics, using multipole expansion techniques. The maximum number of the spherical harmonics is selected to keep the number of free parameters close to the number of Shannon channels in the experimental data (31). The envelopes were obtained with the SASHA program (30). In the second method, the low resolution shapes were restored from the experimental data using a separate ab initio procedure implemented in DAMMIN (30). In this program, the particle is represented by the dummy atom model (DAM) and a simulated annealing protocol is used to search for a compact interconnected model compatible with the data.
If I(q) is given in absolute units, the extrapolated SAXS intensity, I(0), corresponding to a dilute and monodisperse set of N scattering objects per unit volume, can be written as (26)
| (1) |
where Δρ is the difference in electron density between the protein and the surrounding solvent and v is the volume of the protein assembly. The product N.v and the difference Δρ in Eq. 1 remain approximately constant and independent of the degree of protein association and thus the quotient I(0)/c is given by
| (2) |
k being a constant. Equation 2 tells us that I(0)/c is proportional to the monomer/multimer volume. Thus, even if the SAXS intensity is known in relative units, the experimental values of I(0) for different protein concentration can be used to determine the presence of transitions involving different types of multimers and eventually to check the validity of independent findings about the presence and nature of these transitions. For example, for a dimer-to-tetramer transition for which , the experimental value of I(0)/c for tetramers is expected to be two times I(0)/c for dimers.
High-resolution modeling of IL-22
The coordinate set for IL-22 monomer was obtained from the Protein Data Bank (PDB ID: 1YKB and 1M4R) (21,24). IFN-γ and IL-10 coordinates (PDB ID: 1FYH and 1ILK, respectively) were similarly obtained (10,23). The crystallographic models of monomeric IL-22 could be unambiguously positioned inside the DAM for the IL-22 dimer. Relative positions of IL-22 were found by iterative rotation of its envelope functions to minimize the discrepancy with the ab initio low-resolution structure, using an automated procedure. Two models of the solution IL-22 dimers were placed within the tetramer ab initio envelopes allowing for their relative rotation and translation around a common twofold axis. The models were displayed using the program MASSHA (30). Radii of gyration, maximum intraparticle distances (Dmax), envelope functions, and scattering curves were calculated from these atomic coordinates with use of the program CRYSOL (30) taking into account the influence of the hydration shell. SUPCOMB (30) was used to superimpose ab initio low-resolution models with the crystallographic structure. Figures for the SAXS IL-22 dimer and tetramer were generated in the program PyMOL (32).
Rigid-body modeling of the IL-22/IL-22R1 complex
One IL-22 monomer was superimposed with each domain of IL-10 dimer in the IL-10/IL-10R1 template structure (PDB ID: 1J7V) (11) using CCP4 superpose (33). IL-22R1 was obtained by homology modeling using IL-10R1 starting model and also superimposed with the same IL-10/IL-10R1 template. This search structure, as well as the IL-10/IL-10R1 complex, did not fit well the experimental SAXS data. Therefore, it was further separated in two equal subunits containing one IL-22 and one IL-22R1 each, and their relative position and orientation were minimized using SASREF program against SAXS curve (30). The intermolecular contacts mediated by DE loop were maintained during the minimization procedure. CRYSOL (30) was used to evaluate the high-resolution model parameters and SUPCOMB (30) was used to superimpose the model with ab initio DAM. Figures for the SAXS IL-22/IL-22R1 complex were generated in the program PyMOL (32).
Sequence alignment
IL-22 homologs protein sequences were retrieved from the nonredundant database using PSI-BLAST (34) and aligned with the program ClustalW (35).
Results
SAXS, dynamic light scattering, native page, and cross-linking experiments reveal IL -22 dimers and tetramers in solution
By comparison with a reference solution of bovine serum albumin, SAXS studies showed that IL-22 is present predominantly as dimers of 32 ± 3 kDa, at concentrations below 2.5 mg/ml, whereas the protein forms mostly tetramers of molecular weight 63 ± 5 kDa, at concentrations above 5 mg/ml. The experimental intensities extrapolated to zero angle normalized by the concentration, I(0)/c, derived from the SAXS curves, are plotted in Fig. 1 D. The I(0)/c for a protein solution with the concentration c = 5.0 mg/ml is approximately two times that corresponding to c = 2.5 mg/ml, ([I(0)/c]5.0mg/ml =1.85 ± 0.1 [I(0)/c]2,5mg/ml). As explained in Scattering Data Analysis (Eq. 2), this finding confirms the presence of a dimer-to-tetramer transition for protein solutions with concentrations changing from c = 2.5 mg/ml to c = 5 mg/ml, independently deduced from GNOM and DAMMIN analyses. Further evidence of IL-22 oligomerization in solution comes from dynamic light scattering measurements. As determined by DLS IL-22, the protein Stokes radius varies from 3.5 nm at high concentrations to 2.9 nm at lower concentrations. These results favorably agree with the Stokes radii calculated on the basis of the ab initio low-resolution SAXS models of the IL-22 dimer and tetramer, which are 2.9 nm and 3.3 nm, respectively (36).
The experimental value of Dmax and Rg, respectively, 8.0 nm and 2.74 nm for the dimer, and 11 nm and 3.25 nm for the IL-22 tetramer, suggest that the protein is of approximately globular shape in both oligomerization states (Table 1). This notion is also supported by the profiles of the distance distribution functions p(r) (Fig. 2), which are typical of globular prolate particles (37).
Table 1.
SAXS parameters for IL-22 dimer and tetramer and IL-22/IL-22R1
| IL-22 dimer |
IL-22 tetramer |
IL-22/IL-22R1 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter | Exp.* | Mod† | DAM‡/Env§ | Exp.* | Mod† | DAM‡/Env§ | Exp.* | Mod† | DAM‡/Env§ |
| Dmax (nm) | 8.0 ± 0.5 | 8.2 | 8.2/10.4 | 11.0 ± 0.5 | 10.5 | 10.8/11.0 | 11.5 ± 0.5 | 13.0 | 11.9/13.5 |
| Rg (nm) | 2.74 ± 0.06 | 2.83 | 2.78/2.94 | 3.25 ± 0.04 | 3.32 | 3.32/3.51 | 3.56 ± 0.02 | 3.34 | 3.48/3.59 |
| Discrepancy χ | — | 1.80 | 1.81/1.41 | — | 1.75 | 1.73/1.62 | — | 1.31 | 1.20/1.23 |
| Resolution (nm) | 4.2 | — | 4.2/4.2 | 1.8 | — | 1.8/1.8 | 3.1 | — | 3.1/3.1 |
Calculated from the experimental data.
Parameters of the proposed high-resolution structures.
Parameters of the dummy atoms models averaged over 20 models.
Parameters of the envelope models obtained from SASHA.
Figure 2.
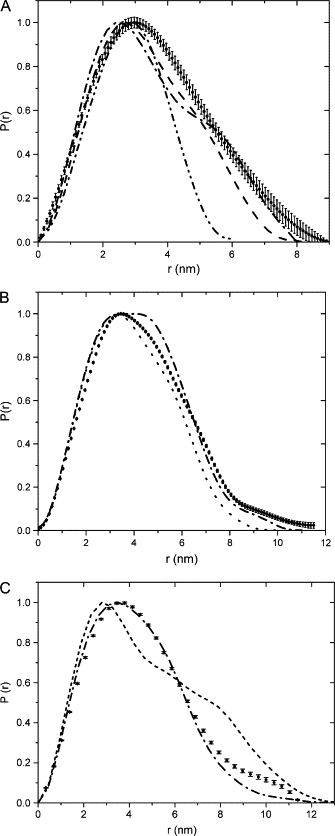
Distance distribution functions of the IL-22 dimer (A), distance distribution functions: from the experimental x-ray scattering data (solid spheres with error bars); from the proposed high-resolution model (dash-dot); and from the IL-22 dimer observed in crystal structure (PDB ID 1M4R) (dash-dot-dot) and from IL-10 (dash). IL-22 tetramer (B), distance distribution functions: from the experimental x-ray scattering data (solid spheres with error bars); from the proposed from the proposed high-resolution model (dash-dot); and from IFN-γ tetramer (dots). IL-22/IL-22R1 (C), distance distribution functions: from the experimental x-ray scattering data for (solid spheres with error bars); from the proposed from the proposed high-resolution model (dash-dot); and from IL-10/IL-10R1 (short dash).
Moreover, separation of IL-22 on an 8–25% native PAGE gradient gel showed a predominant band of 65 kDa, corresponding to the size of the tetrameric form and another faint band with a molecular weight of 33 kDa, which corresponds to the IL-22 dimer (Fig. 3 A). These oligomeric forms of IL-22 were also observed by glutaraldehyde cross-linking experiments; the presence of three bands, located at monomer (17 kDa), dimer (35 kDa), and tetramer (70 kDa) positions, indicates the ability of IL-22 to assemble as dimers and tetramers (Fig. 3 B). These results are in agreement with the observations of SAXS (Table 1) and DLS experiments, and indicate a thermodynamic equilibrium between IL-22 oligomerization states. Taken together these studies strongly suggest that IL-22 protein is present as dimers and tetramers in solution.
Figure 3.
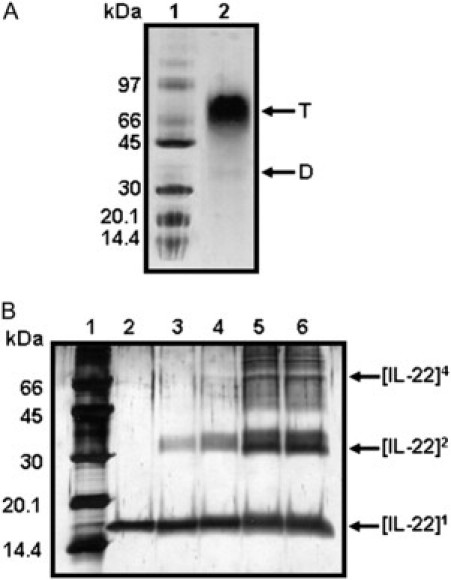
IL-22 forms dimers and tetramers in solution. (A) Electrophoresis pattern of purified IL-22 revealed by native gel electrophoresis. Lane 1, molecular weight calibration standards; Lane 2, IL-22 (0.5 mg/ml). Samples were run on 8–25% gradient gel using the PhastSystem and bands were visualized by Coomassie-blue staining. (B) Glutaraldehyde cross-linking of IL-22. The IL-22 aliquots (2 μg) were cross-linked with 0.000% (lane 2), 0.005% (lane 3), 0.010% (lane 4), 0.020% (lane 5), and 0.050% (lane 6) glutaraldehyde for 30 min at 25°C. Reactions were stopped by addition of SDS-PAGE loading dye and the products analyzed by SDS-PAGE and silver staining. Dimer (D) and Tetramer (T) bands are indicated. Molecular markers and their weights are given (lane 1, A and B).
Ab initio IL-22 and IL-22/IL-22R1 molecular shape determination
First, shape determinations of the IL-22 dimers, IL-22 tetramers, and IL-22/IL-22R1 complex were performed using multipole expansion methods. The experimental data were fitted ab initio by the scattering from an envelope function starting from a spherical initial approximation. The molecular envelope of the IL-22 dimer, obtained by this method, is V-shaped and has two well-developed lobes (Fig. 4 A), whereas the molecular envelope for IL-22 tetramer is X-shaped and has four separate domains (Fig. 5 A). Molecular envelope of the IL-22/IL-22R1 complex is prolate and elongated (Fig. 6 A).
Figure 4.
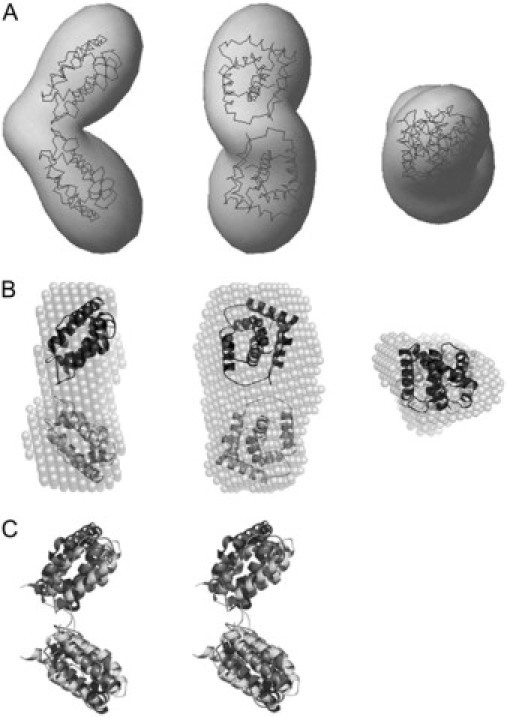
SAXS ab initio envelope for IL-22 dimer structure and proposed high-resolution model based on IL-10 dimer. (A) Three orthogonal views of the superposition of the envelope model obtained by SASHA (30) (shaded envelope) with the model based on two IL-22 high-resolution structures (shaded lines) (24). (B) Three orthogonal views of the superposition of the dummy residues model obtained by DAMMIN (30), as an average of 20 independent simulations (shaded spheres), with the model based on two IL-22 high-resolution monomer structures (dark shaded ribbons) (24). (C) Stereoview of the IL-22 dimer model (dark shaded ribbons) superimposed on the IL-10 intertwined dimer (light shaded ribbons).
Figure 5.
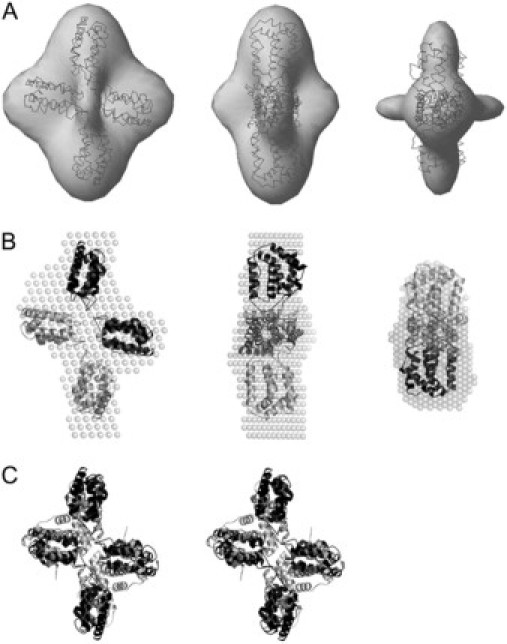
SAXS ab initio envelope for IL-22 tetramer and proposed high-resolution model based on IFN-γ tetramer. (A) Three orthogonal views of the superposition of the envelope model obtained by SASHA (30) and shown as a shaded envelope with the model based on two IL-22 dimer models (shaded lines) (24). (B) Three orthogonal views of the superposition of the dummy residues model obtained by DAMMIN (30), as an average of 20 independent simulations (shaded spheres), with the model based on two IL-22 high-resolution dimeric structures (dark and light ribbons) (24). (C) Stereoview of the IL-22 tetramer model (solid ribbons) superimposed on the IFN-γ tetramer (shaded ribbons).
Figure 6.
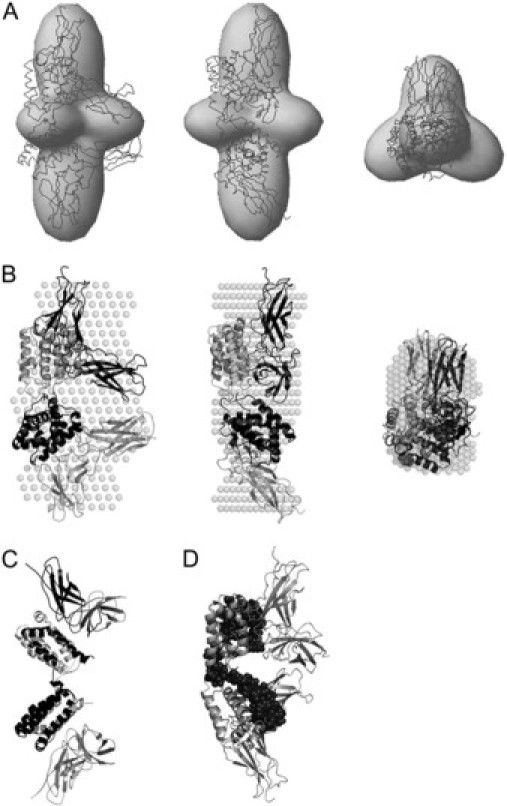
SAXS ab initio envelope for IL-22/IL-22R1 dimer and proposed high-resolution model based on IL-10/IL-10R1. (A) Three orthogonal views of the superposition of the envelope model obtained by SASHA (30) displayed as a shaded envelope, with IL-22/IL-22R1 high-resolution complex (shaded lines). (B) Three orthogonal views of the superposition of the dummy residues model obtained by DAMMIN (30), as an average of 20 independent simulations (shaded spheres), with IL-22/IL22R1 high-resolution complex (cartoons). (C) Stereoview of the IL-10/IL-10R1 structure (PDB ID: 1J7V) (cartoons). (D) Putative IL-22/IL-22R1/IL-22R2 molecular interfaces. The amino-acid residues implicated in interactions with IL-22R2 receptor (50) are shown in solid spheres and are confined in the vicinity of the IL-22/IL-22R1 molecular interface.
The particle shapes were also computed from the IL-22 and IL-22/IL-22R1 experimental data by the ab initio procedure implemented in the DAMMIN program (30). No symmetry restraints were applied. Twenty independent ab initio simulations were performed, and 760 ± 20 dummy atoms were attributed to the final model of IL-22 dimer, as well as 690 ± 30 atoms were assigned to the final model of the IL-22 tetramer and 1286 ± 24 atoms were assigned to the final model of the IL-22/IL-22R1 complex.
To verify the uniqueness of the shape restoration using DAMs (Figs. 4 B, 5 B, and 6 B), several independent restorations were performed using different starting conditions, which all converged to similar molecular shapes. Maximum concentrations of the protein used for SAXS solution studies of the IL-22 dimers, tetramers, and the IL-22/IL-22R1 complex limited the maximum resolution of the final models to 4.2 nm, 1.8 nm, and 3.1 nm, respectively (Table 1). This resolution does not permit unambiguous determination of the spatial positions of their secondary structure elements, but allowed us to obtain the overall shapes of the IL-22 and IL-22/IL-22R1 complexes and relative positions of their subunits. It is important to stress that the use of two independent ab initio reconstructions of SAXS data for the IL-22 and IL-22/IL-22R1 molecular envelopes yielded very consistent and coherent results.
IL-22 dimers mimic IL-10 quaternary arrangement
The SAXS-derived IL-22 dimer model is an open V-shaped molecule with two clearly developed substructures each comprising one IL-22 monomer (Figs. 4 A and 5 B). Attempts to fit this model with IL-22 crystallographic dimers described previously (21,24) were proved to be unsuccessful (Fig. 1 A). Moreover, solution IL-22 dimer distance-distribution function is grossly different from that of IL-22 dimer observed in crystals (Fig. 2 A). Finally, IL-22 crystallographic dimer could not be convincingly superimposed with the IL-22 low-resolution structure. Failure to fit ab initio, low-resolution IL-22 structure with IL-22 dimers observed in crystals, prompted us to analyze IL-22 multimers in more detail and to compare them with the multimeric assemblies of other class-2 cytokines. Strikingly, we found that IL-22 dimer structure is very similar to the IL-10 intertwined dimer (22–24,38). The V-opening angle is ∼90° for both IL-22 and IL-10, and the relative positions and orientations of the IL-10 domains are almost identical to those of IL-22 monomers forming a dimer. This is clearly observed by the superposition of both structures presented in Fig. 4 C. Collectively, analysis of IL-22 SAXS models suggest a completely different structural organization for IL-22 molecules in solution which mimic IL-10 intertwined dimer.
IL-22 tetramers are similar to IFN-γ crystallographic tetramer
The IL-22 tetramer is an oblate and somewhat twisted X-shaped molecule (Fig. 5, A and B). Similarly to the IL-22 dimer, ab initio tetramer low-resolution structure contains well-developed lobes, or domains, that correspond to the individual monomers. The tetramer can be thought of as a dimer of V-shaped dimers bound through molecular interactions at the base of the V. Neither E. coli-expressed nor insect-cell-derived crystallographic IL-22 dimer can convincingly explain our experimental SAXS data, because simulated x-ray scattering curves for these assemblies do not fit well experimental low-resolution scattering intensities. In addition, their distance distribution functions are very different from the experimentally derived one. Furthermore, these high-resolution structures cannot be adequately superimposed with the ab initio low-resolution IL-22 tetramer model (results not shown). Since none of the crystallographic assemblies of IL-22 proved to be suitable for modeling the low-resolution envelope of the tetramer, we systematically tested multimers of other class-2 cytokines observed in x-ray crystal structures. By fitting of high-resolution crystallographic models into the low-resolution molecular envelope, we found a high similarity of the ab initio low-resolution structure of IL-22 tetramer and the packing of the crystal homodimeric structure of single chain monovalent human interferon-γ (ScIFN-γ), described by Randal and Kossiakoff (10). The SAXS-derived high resolution IL-22 tetramer model, built from two IL-22 high-resolution dimers models assembled in accordance with scIFN-γ quaternary structure arrangement, fit well both experimental x-ray scattering data (Fig. 1 B) and the distance distribution function (Fig. 2 B). Furthermore, it is also consistent with two independently generated ab initio low resolution structures of IL-22 tetramer, as shown in the superposition of Fig. 5, A and B. Thus, it seems clear that solution IL-22 tetramer structure is similar to the tetrameric organization of IFN- γ crystallographic structure (Fig. 5 C).
IL-22/IL-22R1 solution structure is similar but not identical to crystallographic IL-10/IL10-R1 complex
To gain more insights into the physiological relevance of the observed IL-22 multimeric forms, we decided to study a macromolecular complex between IL-22 with its high-affinity cellular receptor IL-22R1. Since IL-22 recognition by IL-22R1 is directly relevant to a signaling by this class-2 cytokine on a cellular level, we generated the low resolution shape of the IL-22/IL-22R1 complex using SAXS technique as described above. The obtained low-resolution structure of IL-22/IL22R1 leaves no doubt regarding its oligomerization state (Fig. 6, A and B). IL-22/IL-22R1 complex is composed by an IL-22 dimer and two IL-22R1 receptors, each receptor interacting with one IL-22 monomer. The intramolecular interface of this complex is composed of the loop DE interactions mediating interactions between two IL-22 monomers in the dimer, and the interactions between two IL-22R1 receptors. IL-22/IL22R1 complex architecture is similar, but not identical to that of IL-10/IL-10R1 crystallographic model (Fig. 6 C), in which the later interactions are absent. Furthermore, the angle between monomer in the IL-22 dimer is more open due to the interactions between N-terminal regions of IL-22R1s. These conformational changes are not surprising given a high flexibility of the loop DE described by Nagem and co-workers (24) resulting in a much weaker dimerization interface between two IL-22 molecules in the dimer, as opposed to the relatively rigid structure of the intimate intertwined IL-10 dimer. Despite aforementioned differences in the relative orientations between IL-22/IL-22R1 and IL-10/IL-10R1 complexes, a similarity in their quaternary architecture is evident (Fig. 6, C and D).
Discussion
In this work we report the ab initio, low-resolution models of IL-22 alone and as a complex with its cognate receptor, IL-22R1, generated from the synchrotron small angle x-ray scattering data. IL-22 forms dimers or tetramers in solution, depending on the protein concentration. The dimer is an open V-shaped molecule and the tetramer adopts a partially twisted X-shaped conformation. Our experimental x-ray scattering data can be fitted very well with the V-shaped IL-22 high-resolution model (Fig. 1 A). The distance distribution function based on V-shaped dimer is also consistent with the experimentally derived one (Fig. 2 A). Moreover, there is a very good agreement between the low-resolution shapes of the IL-22 dimers and the proposed V-shaped quaternary arrangement, which we found to be similar to the IL-10 cytokine structure (Fig. 4 C). However, IL-10 dimerization is mediated by the swapping of the helices C and D of its α-helix bundle (22), whereas IL-22 dimerization interface is mediated by residues on the surface of the protein. This is surprising because surface residues are generally the first to be mutated under evolutionary pressure, since such substitutions usually do not interfere with the protein fold and overall three-dimensional structure. It is important to stress that the asymmetric dimer arrangement observed in IL-22 crystal structure (24) is not consistent with our solution SAXS data. Based on our low-resolution model we propose an explanation for the V-shaped dimeric association of IL-22 in solution as follows.
The IL-22 rigid body model obtained with MASHA (30) is a V-shaped molecule in which the loop DE composes the interface of dimerization (Fig. 7 A, left panel). At this interface, protein-protein association might be mediated by three types of interactions:
-
1.
Central hydrophobic interactions via Ile-134.
-
2.
Electrostatic binding interaction of His-133 and negatively charged Glu-135 side chains.
-
3.
A hydrogen-bond type interaction which might be mediated by Thr-131 and Asp-137 (Fig. 7 A, right panel). Given the twofold symmetry of the dimer, both loops are in juxtaposition, allowing for their mutual interactions.
Figure 7.
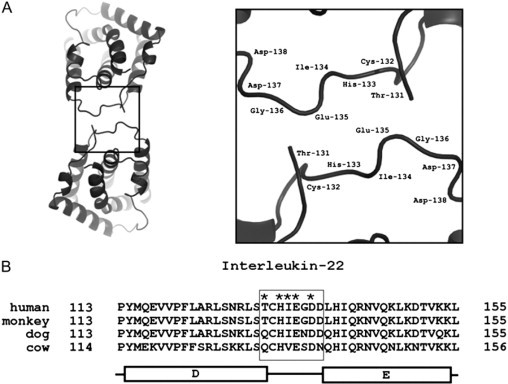
Putative IL-22 dimerization interface. (A) Top view of IL-22 dimer (left panel). Interaction between two DE loops (solid box on the left panel) are shown in detail in the right panel. (B) Alignment of IL-22 amino-acid sequences of helix D, loop DE, and helix E of human (Genbank entry AJ277247), monkey (XP_001117159), dog (XP_538274), and cow (XP_584453) homologs. The conserved potential residues involved in DE loop interaction are indicated (asterisks inside the box).
Crystal structures of IL-22 expressed in E. coli and S2 insect cells were obtained, respectively, at pH values close to 7 and 6. At such pH values, the His-133 side chain is charged, which might weaken IL-22 dimers, possibly leading to their dissociation, and formation of alternative asymmetric dimeric forms observed in crystals. In line with this hypothesis, alignment of helix D, loop DE, and helix E amino acids of IL-22 from human, monkey, dog, and cow, shows conservation for the potential residues involved in dimerization contacts in these species (Fig. 7 B). To examine this hypothesis, mutagenesis studies of key interface residues are currently under way.
There is well-documented evidence of a number of proteins whose multimeric arrangements are different in crystals and in solution, similarly to the differences between solution and crystallographic IL-22 structures. Indeed, x-ray diffraction data provide a wealth of high-resolution information about proteins secondary and tertiary structures, but are not necessarily effective for their quaternary structural analysis. These molecular arrangements are susceptible to appearance of spurious multimeric forms which can be observed in a crystal as a result of high precipitant, protein concentrations, and nonphysiological pHs used under crystallization conditions. For example, ferredoxin reductases were also crystallized as dimers (39,40), but are known to act as monomers in solution (41). Conversely, both yeast hexokinase and bovine β-lactoglobulin, which are capable of forming dimers in solution (42–44), were observed as monomers in a crystal (45–48). These evidences, together with IL-22 structural differences between its solution and crystal organizations, indicate that the protein structure observed in solution should be more closely related to the functional form in vivo.
Therefore, the fact that the IL-22 monomers arrange themselves in solution as dimers, which closely resembles intertwined IL-10s, might have functional implications. Indeed, comparison of the IL-22 solution model with IL-10/IL-10R1 high-affinity receptor complex (11) demonstrates that the IL-22 dimer adopts position and orientation which could be functionally productive for IL-22R1 receptor recognition. Such IL-22/IL-22R1 complex would create molecular interfaces similar to those of IL-10/IL-10R1 complex (11,17–19,49,50), which could be recognized by the extracellular cytokine-binding domain of IL-10R2. This molecule is a common receptor chain shared by both cytokines (IL-22 and IL-10), and this further strengthens our hypothesis of the putative physiological relevance of the observed IL-22 dimer.
To obtain compelling evidence to support our hypothesis of the physiological importance of the IL-22 dimers, we determined the solution low-resolution structure of the complex IL-22/IL-22R1. The model given in Fig. 6, A and B, shows dimeric IL-22 bound to two IL-22R1 receptor chains. The V-angle observed for soluble IL-22 dimers becomes more open when the cytokines bind to IL-22R1s, due to the interactions between N-terminal regions of IL-22R1s and as a result of the weaker IL-22 dimer interface mediated by the flexible loop DE. However, the identity in stoichiometry and similarity of the quaternary structures between IL-22/IL-22R1 and IL-10/IL-10R1 complexes are evident, further supporting a notion of the physiological relevance of the IL-22 dimers.
Low-resolution structure of IL-22/IL22-R1 complex helps to shed light on a molecular assembly of IL-22 with its soluble receptor, IL-22BP. IL-22BP, and IL-22R1 have ∼34% of amino-acid sequence identity. Moreover, IL-22BP also competes with IL-22R1 for IL-22, which is indicative of least partial overlap in their IL-22 binding sites (3). The collective evidence points toward putative structural similarities in IL-22/IL-22BP and IL-22/IL-22R1 quaternary organizations, and allows one to argue that IL-22/IL-22BP architecture should be grossly similar to that of IL-22/IL-22R1 (Fig. 6 A). Furthermore, the present IL-22/IL-22R1 model permits us to speculate about IL-22/IL-22R1/IL-22R2 ternary complex assembly. It is known that IL-22R2 recognizes specific peptides in helices A and D of IL-22 (50). Mapping of the correspondent residues on our low-resolution model of IL-22/IL-22R1 binary complex reveals that in agreement with previous studies (19,50) these peptides are at, or close to, the interface between IL-22 and IL-22R1 (Fig. 6 D). Hence, it is probable that the surface areas of IL-22R1 in the vicinity of the binary complex interface might also participate in IL-22R2 recognition and the ternary complex formation. This model is consistent with the sequential mechanism of IL-22 ternary complex formation (17–19). However, much more thorough studies are required to elucidate structural details of the binary (IL-22/IL-22BP) and ternary (IL-22/IL-22R1/IL-22R2) complex assemblies.
The twisted X-shaped conformation of IL-22 tetramer is another novel finding from our study. The low-resolution scattering showed that its quaternary structure is composed of two V-shaped IL-22 dimers (Fig. 5, A and B). We found that this arrangement is similar to that of crystallographic single-chain monovalent human interferon-γ, described previously (10) (Fig. 5 C). The ScIFN-γ was made by duplication of IFN-γ sequence and linking the C-terminal end of one copy to the N-terminus of the second one. Such hybrid molecule folds similarly to the IFN-γ intimate intertwined dimer (with the difference that both IFN-γ polypeptide sequences belong now to the same polypeptide chain), but recognizes only one IFN-γR1 high-affinity receptor chain though the intermolecular interactions mostly involving helices A, B, and the loop connecting them (residues 1–34) (10). The second binding site of such chimera becomes obstructed most probably by restraints imposed by the linker peptide on the conformation of helices A, B, and the connecting loop. A similarity between the molecular shapes of the IL-22 tetramer and the ScIFN-γ is indeed striking (Fig. 5 C).
Comparison of the IL-22 tetramers (Fig. 5) with the IL-22/IL-22R1 molecular complexes (Fig. 6) shows that the interactions of IL-22s with IL-22R1s lead to the major rearrangements in the relative conformation of the IL-22 monomers within a dimer and in significant increase in the V-angle of the IL-22 dimer observed in the absence of the cognate receptor. Furthermore, the molecular interfaces of two dimers forming a tetramer and of the IL-22/IL-22R1 complex partially overlap. Therefore, molecular interactions of IL-22 with the cognate receptor must lead to the dissociation of the tetrameric form of IL-22. A similar self-association of proteins into higher multimeric forms has been described previously. For example, a retinoid X receptor forms tetramers, which dissociate into functionally active dimers upon interaction with ligands and/or DNA response elements (51,52). Retinoid X receptor tetramers are considered to function as a reservoir of inactive protein which becomes readily available in an active, dimeric, form on request from the physiological stimuli (53). Recently another member of the nuclear receptor family, thyroid hormone nuclear receptor, was also found to form tetramers, which dissociate into dimers on hormone or cognate DNA signal (54,55). Thus, it is tempting to speculate that IL-22 tetramers might in fact represent a nonactive form of the cytokine which is formed at high protein concentrations and dissociate into the active, lower multimeric forms upon interaction with the cognate cellular receptors.
IL-22 functional form was predicted to be a monomer, a result which was later experimentally supported by surface plasmon resonance experiments (18,19). The fact that IL-22 is capable of monomer formation prompted speculations that IL-22 recruits one IL-22R1 and one IL-10R2 to form a functional IL-22/receptor complex, unlike IL-10, which binds two IL-10R1s and two IL-10R2s to a functional intertwined dimer (18). It is important to mention that surface plasmon resonance experiments have been conducted with IL-22 protein that was immobilized at a concentration of 5 mg/ml in 10 mM sodium acetate buffer pH 5.0 (18). Under this pH, His-133 is uncharged—a condition which, according to our model for dimer interaction, prevents IL-22 dimerization. We conducted a number of biochemical and biophysical experiments at pH 8 and propose SAXS models which show that IL-22 forms dimers and tetramers in solution. The V-shaped dimer and the partially twisted X-shaped molecular arrangements were also observed biochemically. The molecular weights of such species were independently obtained by DLS, glutaraldehyde cross-linking, and native PAGE (Fig. 3), supporting the SAXS models, and indicating that dimer and tetramer species are in thermodynamic equilibrium. Our data are also coherent with the previous biochemical reports, based on cross-linking studies, showing that IL-22 forms oligomers with its receptors in hamster cells (13). We also determined the solution structure of the IL-22/IL-22R1 complex, which reveals the IL-22 dimer being recognized by two IL-22R1 high-affinity receptor chains. This molecular assembly lends support to a hypothesis that the IL-22 dimeric state has a physiological relevance and that this quaternary arrangement could be physiologically significant at high cytokine concentrations and under cellular settings in which an amplified molecular response is required. The differences in the observed IL-22 multimeric forms with the previous plasmon resonance studies presumably stem from the lower concentrations of the cytokine accessible under such experiments, which are likely to represent conditions of IL-22 interactions with the cognate receptors under very low cytokine concentrations.
Furthermore, our results propose a model which unifies interleukin/receptor stoichiometry models. It was shown previously that IL-22 (and related cytokines IL-19 and IL-20) form 1:1:1 stoichiometric complexes with correspondent receptor chains, whereas IL-10 (as well as IFN-γ) intimate intertwined dimers are recognized by their cognate membrane receptors in 1:2:2 stoichiometry (1–3,10,11,13,14,17–19). V-shaped dimers of IL-22 offer a possibility to bind IL-22R1 and IL-10R2 receptors in 1:2:2 stoichiometry, while preserving a 1:1:1 molar ratio for the IL-22 monomer.
The concentration of IL-22 was already determined for physiological and pathological conditions. In patients with psoriasis, a chronic inflammatory skin disease, the IL-22 serum concentration is ∼80-fold higher than the concentration observed for healthy donors (56). This elevated IL-22 plasma level correlates with the disease severity. In particular, patients with severe psoriasis have blood IL-22 concentrations of ∼76 pg/ml (56). Such low concentrations of the protein are beyond the sensitivity of our experimental techniques. At these concentrations, when IL-22 acts as an endocrine factor, IL-22 would be expected to be monomeric. However, local concentration of this cytokine in inflammatory focus may be increased in several orders of magnitude when compared to serum concentrations driving the thermodynamic equilibrium to form dimeric and/or tetrameric species. Given the practical limitation in the determination of protein concentration in situ, we speculate that the oligomeric species demonstrated in this work can be present in inflammatory focus, e.g., dermatoses, where protein IL-22 concentration is increased. In line with our proposal, we observed, in native gel electrophoresis, IL-22 dimers at concentrations as low as 1 μg/ml (data not shown). The local cytokine concentration close to the cell surface and the dimer-dimer affinities could be significantly higher for the functional cytokine dimers bound to the extracellular domains of class-2 cytokine receptors. It was shown, for example, that the binding affinities of the molecules in the two-dimensional diffusion case are significantly (orders-of-magnitude) higher than that observed in solution (57). Therefore, affinities of multimers association are expected to be substantially enhanced when cytokines are bound to the receptors anchored in the membrane, similarly to what is observed in the follicle-stimulating hormone-receptor interactions (58). We expect, therefore, that IL-22 might be present in multimeric forms in diseases, particularly in peripheral tissues, generated in the chronic psoriatic plaques.
Furthermore, physiological effect of cytokine/receptors recognition seems to be cumulative. ScIFN-γ, for example, which contains only one productive cognate receptor binding site, retains only 1% of the physiological activity of the wild-type IFN-γ (10). We hypothesize that the ability of IL-22 to form dimers bound simultaneously to two IL-22R1 and two IL-10R2 receptor chains (1:2:2 stoichiometry) could be used by the cell to amplify molecular signals conducted by this cytokine.
All these data collectively suggest that the concentration-dependent dimerization of monomeric cytokines might be a common theme in their molecular recognition by respective cellular receptors, and that dimeric quaternary structures of these cytokines, along with the monomeric forms, are likely to be of functional importance.
Acknowledgments
We thank Laboratório Nacional de Luz Síncrotron for access to SAXS beamline and other facilities.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil, via grants No. 02/14041-6 and No. 06/00182-2, and by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil.
Footnotes
Editor: Jill Trewhella.
References
- 1.Wolk K., Sabat R. Interleukin-22: a novel T- and NK-cell derived cytokine that regulates the biology of tissue cells. Cytokine Growth Factor Rev. 2006;17:367–380. doi: 10.1016/j.cytogfr.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Petska S., Krause C.D., Sarkar D., Walter M.R., Shi Y., Fisher P.B. Interleukin-10 and related cytokines and receptors. Annu. Rev. Immunol. 2004;22:929–979. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- 3.Nagem R.A., Ferreira Junior J.R., Dumoutier L., Renauld J.C., Polikarpov I. Interleukin-22 and its crystal structure. Vitam. Horm. 2006;74:77–103. doi: 10.1016/S0083-6729(06)74004-3. [DOI] [PubMed] [Google Scholar]
- 4.Wolk K., Kunz S., Witte E., Friedrich M., Asadullah K., Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Zheng Y., Danilenko D.M., Valdez P., Kasman I., Eastham-Anderson J., Wu J., Ouyang W. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 6.Dumoutier L., Van Roost E., Ameye G., Michaux L., Renauld J.C. IL-TIF/IL-22: genomic organization and mapping of the human and mouse genes. Genes Immun. 2000;1:488–494. doi: 10.1038/sj.gene.6363716. [DOI] [PubMed] [Google Scholar]
- 7.Langer J.A., Cutrone E.C., Kotenko S. The Class II cytokine receptor (CRF2) family: overview and patterns of receptor-ligand interactions. Cytokine Growth Factor Rev. 2004;15:33–48. doi: 10.1016/j.cytogfr.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Harlos K., Martin D.M., O’Brien D.P., Jones E.Y., Stuart D.I., Polikarpov I., Miller A., Tuddenham E.G., Boys C.W. Crystal structure of the extracellular region of human tissue factor. Nature. 1994;370:662–666. doi: 10.1038/370662a0. [DOI] [PubMed] [Google Scholar]
- 9.Thiel D.J., le Du M.H., Walter R.L., D’Arcy A., Chene C., Fountoulakis M., Garotta G., Winkler F.K., Ealick S.E. Observation of an unexpected third receptor molecule in the crystal structure of human interferon-γ receptor complex. Structure Fold Des. 2000;8:927–936. doi: 10.1016/s0969-2126(00)00184-2. [DOI] [PubMed] [Google Scholar]
- 10.Randal M., Kossiakoff A.A. The structure and activity of a monomeric interferon-γ:α-chain receptor signaling complex. Structure. 2001;9:155–163. doi: 10.1016/s0969-2126(01)00567-6. [DOI] [PubMed] [Google Scholar]
- 11.Josephson K., Logsdon N.J., Walter M.R. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity. 2001;15:35–46. doi: 10.1016/s1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- 12.Walter M.R., Windsor W.T., Nagabhushan T.L., Lundell D.J., Lunn C.A., Zauodny P.J., Narula S.K. Crystal structure of a complex between interferon-γ and its soluble high-affinity receptor. Nature. 1995;376:230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- 13.Kotenko S.V., Izotova L.S., Mirochnitchenko O.V., Esterova E., Dickensheets H., Donnelly R.P., Petska S. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rβ) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes. J. Biol. Chem. 2001;276:2725–2732. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- 14.Xie M.H., Aggarwal S., Ho W.H., Foster J., Zhang Z., Stinson J., Wood W.I., Goddard A.D., Gurney A.L. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2–4 and IL-22R. J. Biol. Chem. 2000;275:31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- 15.Nagalakshmi M.L., Murphy E., McClanahan T., de Waal Malefyt R. Expression patterns of IL-10 ligand and receptor gene families provide leads for biological characterization. Int. Immunopharmacol. 2004;4:577–592. doi: 10.1016/j.intimp.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Nagalakshmi M.L., Rascle A., Zurawski S., Menon S., de Waal Malefyt R. Interleukin-22 activates STAT3 and induces IL-10 by colon epithelial cells. Int. Immunopharmacol. 2004;4:679–691. doi: 10.1016/j.intimp.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Li J., Tomkinson K.N., Tan X.Y., Wu P., Yan G., Spaulding V., Deng B., Annis-Freeman B., Heveron K., Zollner R., De Zutter G., Wright J.F., Crawford T.K., Liu W., Jacobs K.A., Wolfman N.M., Ling V., Pittman D.D., Veldman G.M., Fouser L.A. Temporal associations between interleukin 22 and the extracellular domains of IL-22R and IL-10R2. Int. Immunopharmacol. 2004;4:693–708. doi: 10.1016/j.intimp.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Logsdon N.J., Jones B.C., Josephson K., Cook J., Walter M.R. Comparison of interleukin-22 and interleukin-10 soluble receptor complexes. J. Interferon Cytokine Res. 2002;22:1099–1112. doi: 10.1089/10799900260442520. [DOI] [PubMed] [Google Scholar]
- 19.Logsdon N.J., Jones B.C., Allman J.C., Izotova L., Schwartz B., Pestka S., Walter M.R. The IL-10R2 binding hot spot on IL-22 is located on the N-terminal helix and is dependent on N-linked glycosylation. J. Mol. Biol. 2004;342:503–514. doi: 10.1016/j.jmb.2004.07.069. [DOI] [PubMed] [Google Scholar]
- 20.Nagem R.A., Lucchesi K.W., Colau D., Dumoutier L., Renauld J.C., Polikarpov I. Crystallization and synchrotron x-ray diffraction studies of human interleukin-22. Acta Crystallogr. D Biol. Crystallogr. 2002;58:529–530. doi: 10.1107/s0907444902001063. [DOI] [PubMed] [Google Scholar]
- 21.Xu T., Logsdon N.J., Walter M.R. Structure of insect-cell-derived IL-22. Acta Crystallogr. D Biol. Crystallogr. 2005;61:942–950. doi: 10.1107/S0907444905009601. [DOI] [PubMed] [Google Scholar]
- 22.Zdanov A., Schalk-Hihi C., Wlodawer A. Crystal structure of human interleukin-10 at 1.6 Å resolution and a model of a complex with its soluble receptor. Protein Sci. 1996;5:1955–1962. doi: 10.1002/pro.5560051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zdanov A., Schalk-Hihi C., Gustchina A., Tsang M., Weatherbee J., Wlodawer A. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon gamma. Structure. 1995;3:591–601. doi: 10.1016/s0969-2126(01)00193-9. [DOI] [PubMed] [Google Scholar]
- 24.Nagem R.A., Colau D., Dumoutier L., Renauld J.C., Ogata C., Polikarpov I. Crystal structure of recombinant human interleukin-22. Structure. 2002;10:1051–1062. doi: 10.1016/s0969-2126(02)00797-9. [DOI] [PubMed] [Google Scholar]
- 25.Koch M.H., Vachette P., Svergun D.I. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 26.Svergun D.I., Koch M.H. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 2003;66:1735–1782. [Google Scholar]
- 27.Dumoutier L., Lejeune D., Colau D., Renauld J.C. Cloning and characterization of IL-22 binding protein, a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J. Immunol. 2001;166:7090–7095. doi: 10.4049/jimmunol.166.12.7090. [DOI] [PubMed] [Google Scholar]
- 28.Kellerman G., Vicentin F., Tamura E., Rocha M., Tolentino H., Barbosa A., Craievich A., Torriani I. The small-angle x-ray scattering beamline at the Brazilian synchrotron light laboratory. J. Appl. Crystallogr. 1997;30:880–883. [Google Scholar]
- 29.Fischer H., Dias S.M., Santos M.A., Alves A.C., Zanchin N., Craievich A.F., Apriletti J.W., Baxter J.D., Webb P., Neves F.A., Ribeiro R.C., Polikarpov I. Low resolution structures of the retinoid X receptor DNA-binding and ligand-binding domains revealed by synchrotron x-ray solution scattering. J. Biol. Chem. 2003;278:16030–16038. doi: 10.1074/jbc.M206953200. [DOI] [PubMed] [Google Scholar]
- 30.Konarev P.V., Petoukhov M.V., Volkov V.V., Svergun D.I. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon C.E., Weaver W. University of Illinois Press; Urbana, IL: 1949. The Mathematical Theory of Communication. [Google Scholar]
- 32.DeLano W.L. DeLano Scientific; San Carlos, CA: 2002. The PyMol Molecular Graphics System. [Google Scholar]
- 33.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 34.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia De La Torre J., Huertas M.L., Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feign L.A., Svergun D.I. Plenum Press; New York: 1987. Structure Analysis by Small-Angle X-Ray and Neutron Scattering. [Google Scholar]
- 38.Walter M.R., Nagabhushan T.L. Crystal structure of interleukin 10 reveals an interferon γ-like fold. Biochemistry. 1995;34:12118–12125. doi: 10.1021/bi00038a004. [DOI] [PubMed] [Google Scholar]
- 39.Deng Z., Aliverti A., Zanetti G., Arakaki A.K., Ottado J., Orellano E.G., Calcaterra N.B., Ceccarelli E.A., Carrillo N., Karplus P.A. A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies. Nat. Struct. Biol. 1999;6:847–853. doi: 10.1038/12307. [DOI] [PubMed] [Google Scholar]
- 40.Morales R., Charon M.H., Kachalova G., Serre L., Medina M., Gomez-Moreno C., Frey M. A redox-dependent interaction between two electron-transfer partners involved in photosynthesis. EMBO Rep. 2000;1:271–276. doi: 10.1093/embo-reports/kvd057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ceccarelli E.A., Arakaki A.K., Cortez N., Carrillo N. Functional plasticity and catalytic efficiency in plant and bacterial ferredoxin-NADP(H) reductases. Biochim. Biophys. Acta. 2004;1698:155–165. doi: 10.1016/j.bbapap.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Schulze I.T., Colowick S.P. The modification of yeast hexokinases by proteases and its relationship to the dissociation of hexokinase into subunits. J. Biol. Chem. 1969;244:2306–2316. [PubMed] [Google Scholar]
- 43.Hoggett J.G., Kellett G.L. Kinetics of the monomer-dimer reaction of yeast hexokinase PI. Biochem. J. 1992;287:567–572. doi: 10.1042/bj2870567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kontopidis G., Holt C., Sawyer L. Invited review: β-lactoglobulin: binding properties, structure, and function. J. Dairy Sci. 2004;87:785–796. doi: 10.3168/jds.S0022-0302(04)73222-1. [DOI] [PubMed] [Google Scholar]
- 45.Bennett W.S., Jr., Steitz T.A. Structure of a complex between yeast hexokinase A and glucose. I. Structure determination and refinement at 3.5 Å resolution. J. Mol. Biol. 1980;140:183–209. doi: 10.1016/0022-2836(80)90102-3. [DOI] [PubMed] [Google Scholar]
- 46.Kuser P.R., Krauchenco S., Antunes O.A., Polikarpov I. The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action. J. Biol. Chem. 2000;275:20814–20821. doi: 10.1074/jbc.M910412199. [DOI] [PubMed] [Google Scholar]
- 47.Brownlow S., Morais Cabral J.H., Cooper R., Flower D.R., Yewdall S.J., Polikarpov I., North A.C., Sawyer L. Bovine β-lactoglobulin at 1.8 Å resolution—still an enigmatic lipocalin. Structure. 1997;5:481–495. doi: 10.1016/s0969-2126(97)00205-0. [DOI] [PubMed] [Google Scholar]
- 48.Oliveira K.M., Valente-Mesquita V.L., Botelho M.M., Sawyer L., Ferreira S.T., Polikarpov I. Crystal structures of bovine β-lactoglobulin in the orthorhombic space group C222(1). Structural differences between genetic variants A and B and features of the Tanford transition. Eur. J. Biochem. 2001;268:477–483. doi: 10.1046/j.1432-1033.2001.01918.x. [DOI] [PubMed] [Google Scholar]
- 49.Yoon S.I., Logsdon N.J., Sheikh F., Donnelly R.P., Walter M.R. Conformational changes mediate IL-10R2 binding to IL-10 and assembly of the signaling complex. J. Biol. Chem. 2006;281:35088–35096. doi: 10.1074/jbc.M606791200. [DOI] [PubMed] [Google Scholar]
- 50.Wolk K., Witte E., Reineke U., Witte K., Friedrich M., Sterry W., Asadullah K., Volk H.-D., Sabat R. Is there an interaction between interleukin-10 and interleukin-22? Genes Immun. 2005;6:8–18. doi: 10.1038/sj.gene.6364144. [DOI] [PubMed] [Google Scholar]
- 51.Kersten S., Kelleher D., Chambon P., Gronemeyer H., Noy N. Retinoid X receptor-α forms tetramers in solution. Proc. Natl. Acad. Sci. USA. 1995;92:8645–8649. doi: 10.1073/pnas.92.19.8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kersten S., Pan L., Chambon P., Gronemeyer H., Noy N. Role of ligand in retinoid signaling. 9-cis-retinoic acid modulates the oligomeric state of the retinoid X receptor. Biochemistry. 1995;34:13717–13721. doi: 10.1021/bi00042a001. [DOI] [PubMed] [Google Scholar]
- 53.Kersten S., Dong D., Lee W., Reczek P.R., Noy N. Auto-silencing by the retinoid X receptor. J. Mol. Biol. 1998;284:21–32. doi: 10.1006/jmbi.1998.2168. [DOI] [PubMed] [Google Scholar]
- 54.Figueira A.C., Dias S.M., Santos M.A., Apriletti J.W., Baxter J.D., Webb P., Neves F.A., Simeoni L.A., Ribeiro R.C., Polikarpov I. Human thyroid receptor forms tetramers in solution, which dissociate into dimers upon ligand binding. Cell Biochem. Biophys. 2006;44:453–462. doi: 10.1385/CBB:44:3:453. [DOI] [PubMed] [Google Scholar]
- 55.Figueira A.C., Neto M.O., Bernardes A., Dias S.M., Craievich A.F., Baxter J.D., Webb P., Polikarpov I. Low-resolution structures of thyroid hormone receptor dimers and tetramers in solution. Biochemistry. 2007;46:1273–1283. doi: 10.1021/bi061698h. [DOI] [PubMed] [Google Scholar]
- 56.Wolk K., Witte E., Wallace E., Docke W.D., Kunz S., Asadullah K., Volk H.D., Sterry W., Sabat R. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur. J. Immunol. 2006;36:1309–1323. [Google Scholar]
- 57.Grasberger B., Minton A.P., DeLisi C., Metzger H. Interaction between proteins localized in membranes. Proc. Natl. Acad. Sci. USA. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan Q.R., Hendrickson W.A. Structure of human follicle-stimulating hormone in complex with its receptor. Nature. 2005;433:269–277. doi: 10.1038/nature03206. [DOI] [PMC free article] [PubMed] [Google Scholar]


