Abstract
Dihydroorotate dehydrogenase (DHODH) catalyzes the oxidation of dihydroorotate to orotate during the fourth step of the de novo pyrimidine synthesis pathway. In rapidly proliferating mammalian cells, pyrimidine salvage pathway is insufficient to overcome deficiencies in that pathway for nucleotide synthesis. Moreover, as certain parasites lack salvage enzymes, relying solely on the de novo pathway, DHODH inhibition has turned out as an efficient way to block pyrimidine biosynthesis. Escherichia coli DHODH (EcDHODH) is a class 2 DHODH, found associated to cytosolic membranes through an N-terminal extension. We used electronic spin resonance (ESR) to study the interaction of EcDHODH with vesicles of 1,2-dioleoyl-sn-glycero-phosphatidylcholine/detergent. Changes in vesicle dynamic structure induced by the enzyme were monitored via spin labels located at different positions of phospholipid derivatives. Two-component ESR spectra are obtained for labels 5- and 10-phosphatidylcholine in presence of EcDHODH, whereas other probes show a single-component spectrum. The appearance of an additional spectral component with features related to fast-motion regime of the probe is attributed to the formation of a defect-like structure in the membrane hydrophobic region. This is probably the mechanism used by the protein to capture quinones used as electron acceptors during catalysis. The use of specific spectral simulation routines allows us to characterize the ESR spectra in terms of changes in polarity and mobility around the spin-labeled phospholipids. We believe this is the first report of direct evidences concerning the binding of class 2 DHODH to membrane systems.
Introduction
Growing organisms need nucleotides, the building blocks of DNA, RNA, and other biologically essential molecules. Inhibitors of nucleotide synthesis form an important group of chemotherapeutic agents, and cells that rely on this pathway are especially susceptible to such inhibitors. The de novo pyrimidine pathway seems to be significantly important. Besides RNA and DNA synthesis, pyrimidines are also needed for protein glycosylation, membrane lipid biosynthesis, and strand break repair (1–23). Several inhibitors of the enzyme involved in the only redox step of pyrimidines nucleotide pathway, dihydroorotate dehydrogenase (DHODH), have been in clinical trials and one inhibitor, Arava (leflunomide; Sigma, St. Louis, MO), has been approved for human use as rheumatoid arthritis agent (4,5). In addition, the increased interest in DHODH is due to its role as a target for a number of biologically active chemical or natural compounds. The enzyme has been identified as a pharmacological target for isoxazole, triazine, cinchoninic acid and (naphtha)quinone derivatives, which exerted antiproliferative, immunosuppressive, and antiparasitic effects (6–8). These compounds were found to interfere with aberrant imunological reactions, to combat parasitic protozoa infections, like malaria, to prevent the spreading of animal parasitic diseases, and to support antiviral therapies, by lowering the intracellular concentrations of pyrimidine nucleotides (9).
Dihydroorotate dehydrogenase catalyses the fourth sequential step in the de novo pyrimidine nucleotide synthesis pathway with the oxidation of dihydroorotate to orotate, the first aromatic intermediate in this biosynthetic pathway, with the aid of a flavin cofactor and an electron receptor (1). On the basis of sequence similarity, the DHODHs can be divided in two major classes (2). This division correlates with subcellular location of the proteins as well as their preferences for electron acceptors. Enzymes of class 1, found in Gram-positive bacteria and in the anaerobic yeast Saccharomyces cerevisiae, are located in the cytosol of the cell. They share <20% sequence identity with the membrane-bound enzymes of class 2, found in eukaryotes and in some prokaryotes such as the Gram-negative bacteria related to Escherichia coli. Class 1 enzymes can be further divided into two subtypes, namely 1A and 1B. The eukaryotic class 2 enzymes are located in the inner membrane of mitochondria and, in the case of E. coli enzyme, are associated with the cytoplasmatic membrane.
The structure of DHODH has been solved for four different organisms: Homo sapiens (HsDHODH) (10), E. coli (EcDHODH) (11), rat (12), Plasmodium falciparum (13), and Lactococcus lactis (3,14,15). The latter includes subtypes A and B of class 1 DHODHs. In all cases the structure of DHODH is an α/β barrel with eight parallel β strands forming the barrel and α helices wrapped around the outside. The orotate active site is at the top of the barrel where several additional strands form a binding pocket for the flavin cofactor and orotate. In addition to this main barrel, class 2 DHODH, such as HsDHODH and EcDHODH, contains a second domain situated at the N-terminus, which is supposed to be involved with membrane interaction (11).
Another remarkable difference between the two classes of DHODH is related to the mechanism used by them to complete the redox reaction. In class 1 DHODHs, the electron acceptors involved in the second half reaction of the redox process are either fumarate or NAD+ (1) whereas for class 2 DHOHDs this role is played by quinones present in the biological membranes (16,17). In the latter case, the N terminus has been proposed as the binding site for the electron acceptor (10). Thus, this N-terminal domain is supposedly responsible for both membrane association and binding of electron acceptor molecules.
Electron spin resonance (ESR) is a powerful technique that makes use of either transition metal ions or spin probes, usually involving stable nitroxide radicals bound to molecules such as phospholipids or cysteine residues in proteins, to monitor changes in the probe vicinity (18–21). Some advantages of spin-labeling ESR experiments are the possibility of using a selective probe that has a simple ESR spectra and their high sensitivity to the molecular motion of the spin-bearing moiety. The changes in the nitroxide surroundings can be related to a variety of biologically-relevant processes such as protein conformational changes (22–24), lipid-protein interactions (25–28), and the dynamic structure of biologic and model membranes (29–32).
In this article, we use ESR to monitor EcDHODH-induced changes in the neighborhood of spin-labeled phospholipids incorporated into a membrane model system. We address the main goal of investigating the effect of EcDHODH binding to phospholipid vesicles. The use of specific spectral simulation routines allows us to fully characterize the ESR spectra in terms of changes in polarity and mobility in the surroundings of the spin-labeled phospholipid molecules. To the best of our knowledge, this is the first report showing direct evidences concerning the binding of class 2 DHODH to membrane systems and its implication in protein function.
Materials and methods
Expression and purification of EcDHODH
PAG1 plasmid and E. coli cell strains used for EcDHODH expression were kindly provided by Prof. K. F. Jensen (University of Copenhagen) (33). E. coli DHODH was overexpressed in S06645 E. coli cell strain grown in Luria-Broth medium. A cell pellet from 250 mL of cell culture was lysed in 10 mL of 50 mM sodium phosphate buffer pH 8.0 and 0.25 mM EDTA. To the lysate was added 5 mM magnesium chloride plus 0.2% Triton X-100 with subsequent centrifugation at 17,200 × g for 1 h. The supernatant was applied to a 20 mL DEAE-Sepharose column (Amersham Biosciences, Uppsala, Sweden) equilibrated with 50 mM sodium phosphate buffer pH 8.0 and 0.25 mM EDTA. The column was washed with 50 mL sodium phosphate buffer pH 8.0, 0.1 mM EDTA and 0.1% Triton X-100 and eluted with a linear gradient from 0 to 1 M NaCl. The fractions containing EcDHODH were combined in presence of 0.5% Triton X-100, followed by 1 M ammonium sulfate precipitation. The mixture was incubated for 1 h at 4°C and centrifuged at 20,000 × g for 1 h. The supernatant was applied to a 2 mL Phenyl-Sepharose column (Amersham Biosciences) equilibrated with 50 mM sodium phosphate buffer pH 7.0, 0.1 mM EDTA and 1.1 M ammonium sulfate. The column was washed with a linear gradient from 1.1 to 0 M ammonium sulfate. The protein is eluted with 50 mM sodium phosphate buffer pH 7.0, 0.1 mM EDTA and 0.5% Triton X-100.
EcDHODH/vesicles mixtures
EcDHODH is purified in the presence of the detergent Triton X-100, which is crucial for enzyme solubilization. The absence of the detergent leads to protein precipitation probably due to aggregation via its N-terminal domain. The solution containing EcDHODH in the presence of Triton X-100 is added to a dried phospholipid film formed on the wall of a glass tube from chloroform stock solutions of the lipid. It is well-known that the mixture of surfactants to phospholipids leads to alterations in the membrane structure, which depend basically on the ratio surfactant/phospholipids (34,35). A continuous increase in such a ratio is accompanied by a transition from a bilayer to a monolayer structure. In our case, the detergent Triton X-100 comes from the purification/solubilization process and its final concentration is hard to determine exactly. Several assays with different detergent concentration were carried out to assure that the amount of detergent present in the final samples was the minimum required for protein solubilization. In this work, final Triton X-100 concentration is above its critical micelle concentration and we estimated the surfactant/phospholipid ratio to be close to 1, which resulted in a mixture of mixed micelles and mixed vesicles described by López et al. (34) for a system constituted by Triton X-100 and phosphatidylcholine. To have a control experiment, enzyme-free samples containing mixtures of Triton X-100 and phospholipids at similar surfactant/phospholipid ratio as before were prepared and submitted to ESR analysis.
ESR spectroscopy
The headgroup spin label dipalmitoylphosphatidyl tempo (2,2,6,6,-tetramethyl-1-oxy) choline (DPPTC), the phospholipid labels 1-palmitoyl-2-(n-doxyl stearoyl) phosphatidylcholine (n = 5, 10, 12, 16-PC) and the lipid 1,2-dioleoyl-sn-glycero-phosphatidylcholine (DOPC) were purchased from Avanti Polar Lipids (Alabaster, AL). All labels and chemicals were used without further purification. Measured stock solutions of the lipid DOPC and the spin labels were mixed in a glass tube. The chloroform present in the stock solutions was removed by N2 flow followed by 1 h in a Speedvac system to ensure complete removal of the solvent. A measured amount of the buffered EcDHODH/Triton X-100 solution was added to the sample tube, and incubated for several minutes. A final volume of 100 μL of the samples containing mixtures of EcDHODH/Triton X-100/DOPC/spin label was drawn into a quartz flat cell, which was in turn placed in the ESR resonant cavity. Final enzyme concentration ranged from 89–103 μM. X-band ESR spectra of those samples were recorded on a Varian E109 spectrometer at room temperature. Acquisition conditions were: modulation amplitude, 1.0 G; modulation frequency, 100 kHz; microwave power, 10 mW; field range, 100 G.
Results and discussions
ESR spectra and nonlinear least-squares analysis
The ESR spectra of the headgroup spin probe (DPPTC) as well as acyl chain labels (5-, 10-, 12- and 16-PC) incorporated into DOPC/Triton X-100 mixed vesicles in the presence and absence of EcDHODH are shown in Fig. 1. Only minor changes between samples with and without EcDHODH are detectable for DPPTC, 12-, and 16-PC probes, whereas the appearance of a second component (sharp peak in the low-field resonance) is observed for probes 5- and 10-PC in the presence of the titled enzyme. In these two-component spectra, the major contribution is attributed to the bulk labeled phospholipids whereas the second one is assigned to label molecules in close contact with the protein (the so-called boundary lipid (36–38)). A qualitative analysis of the extra component suggests that a spectrum constituted by sharp lines like those observed in Fig. 1 B and C should be the result of a much less hindered motion of the spin probes when in the vicinity of the enzyme. The faster motion experienced by the boundary labels averages out the anisotropy of the magnetic interactions (hyperfine and Zeeman) leading to narrower resonance lines than usually observed for spin probes experiencing slow and anisotropic motion. A control experiment using class 1 Trypanosoma cruzi DHODH (TcDHODH), which misses the N-terminal domain (cloning, expression, and purification of TcDHODH followed a protocol adapted from (39), in the presence of the same membrane model system described above yielded no alterations whatsoever in the ESR spectra of either the headgroup or a carbon-chain spin probe (data not shown).
Figure 1.
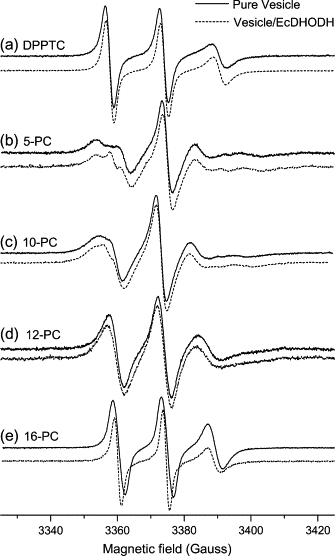
ESR spectra of spin labels (a) DPPTC, (b) 5-, (c) 10-, (d) 12-, and (e) 16-PC incorporated into vesicles of DOPC/Triton X-100 in the absence (solid line) and in the presence of EcDHODH (dashed line). Experimental conditions: microwave frequency 9.5 GHz; modulation amplitude 1.0 G; modulation frequency 100 kHz; microwave power 10 mW.
To fully characterize the modifications measured in the presence of EcDHODH, the ESR spectra of the spin probes in mixtures of vesicle/enzyme were simulated by means of a nonlinear least-squares program developed by Freed et al. (40–42). The parameters involved in the fitting procedure were as follows: hyperfine tensor components (Axx,Ayy,Azz), rotational diffusion rates (R⊥ and R//), and a lorentzian () inhomogeneous broadening. The dynamics of the spin probe is characterized by R⊥ and R//, which represent the rotational diffusion rates of the nitroxide radical around the axes perpendicular and parallel to the mean symmetry axis for the rotation. This symmetry axis is also the direction of preferential orientation of the spin label moiety (41). For n-PC chain labels, R⊥ accounts for the wagging motion of the long axis of the carbon chain (Fig. 2 A). As for the label DPPTC, it represents the wagging motion of the headgroup region (Fig. 2 B). To avoid local minima the simulation process was restarted from different sets of seed values.
Figure 2.
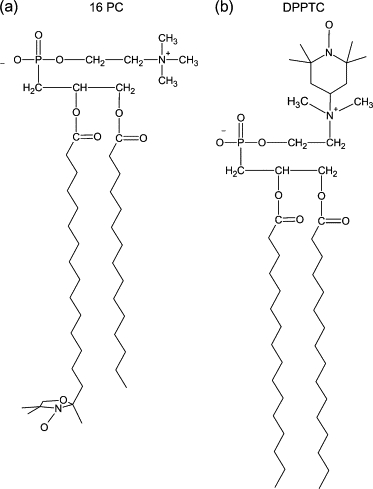
Chemical structures of the (a) acyl-chain (16-PC) and (b) headgroup (DPPTC) spin labels showing the principal magnetic axes (xm, ym, zm).
The ESR spectra of the spin probes incorporated into the model membrane system (Fig. 3) can be divided, for simulation purposes, in two categories. One comprises the one-component spectra of DPPTC, 5-, 10-, 12-, and 16-PC in pure vesicles (without EcDHODH) and of DPPTC, 12-, and 16-PC in EcDHODH-containing samples, which were then treated first. Seed values for the magnetic parameters (Axx, Ayy, Azz, gxx, gyy, gzz) were obtained from Ge et al (1990) (43). During the simulations process, the magnetic parameters were initially kept fixed, and the rotational diffusion tensor component R⊥ was varied. After that, variations in the hyperfine and g-tensor components were carried out separately to avoid high correlation values between those parameters that can come up when they are varied together. Once a reasonable fit was obtained for the mixtures containing only vesicles of DOPC/Triton X-100, the calculated parameters thus obtained were used as starting values for the fits of EcDHODH-containing samples. The best-fit values calculated for the one-component ESR spectra are presented in Table 1 and the best spectral fits are shown in Figs. 3, 4 A, and 5 A along with the respective experimental data.
Figure 3.
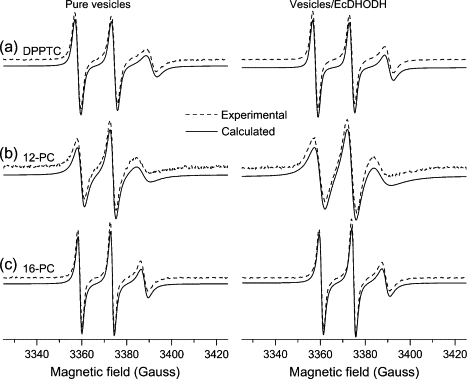
Experimental (dashed line) and simulated (solid line) ESR spectra from (a) DPPTC, (b) 12-, and (c) 16-PC labels in mixtures of DOPC/Triton X-100 (left column) and DOPC/Triton X-100/EcDHODH (right column).
Table 1.
Best-fit parameters from NLSL simulations of the ESR spectra obtained from spin labels (headgroup DPPTC and n-PC) incorporated into mixtures of DOPC/Triton X-100 and DOPC/Triton X-100/EcDHODH
| Sample | Component | gxx | gyy | gzz | Axx | Ayy | Azz | A0 | R⊥ (×108 s−1) |
|---|---|---|---|---|---|---|---|---|---|
| No EcDHODH | |||||||||
| DPPTC | 1 | 2.0078 | 2.0046 | 2.0022 | 6.0 | 5.9 | 37.5 | 16.5 | 0.58 |
| 5-PC | 1 | 2.0075 | 2.0049 | 2.0020 | 7.4 | 6.5 | 30.2 | 14.7 | 0.22 |
| 10-PC | 1 | 2.0084 | 2.0063 | 2.0033 | 6.5 | 6.0 | 31.4 | 14.6 | 0.36 |
| 12-PC | 1 | 2.0089 | 2.0063 | 2.0033 | 5.3 | 4.9 | 33.2 | 14.5 | 0.79 |
| 16-PC | 1 | 2.0102 | 2.0063 | 2.0033 | 5.3 | 4.9 | 33.2 | 14.5 | 1.94 |
| EcDHODH | |||||||||
| DPPTC | 1 | 2.0078 | 2.0047 | 2.0022 | 6.0 | 4.9 | 37.8 | 16.2 | 0.78 |
| 5-PC | 1 | 2.0075 | 2.0049 | 2.0020 | 7.4 | 6.5 | 30.2 | 14.7 | 0.22 |
| 2 | 2.0068 | 2.0047 | 2.0015 | 6.5 | 5.2 | 36.3 | 16.0 | 1.47 | |
| 12-PC | 1 | 2.0090 | 2.0063 | 2.0033 | 5.3 | 4.9 | 33.3 | 14.5 | 0.66 |
| 16-PC | 1 | 2.0102 | 2.0063 | 2.0033 | 5.3 | 4.9 | 33.3 | 14.5 | 1.69 |
A-tensor components are in Gauss. A0=(Axx+Ayy+Azz)/3. Estimated errors: R⊥ (5%), Axx and Ayy (10%), Azz (5%).
Figure 4.
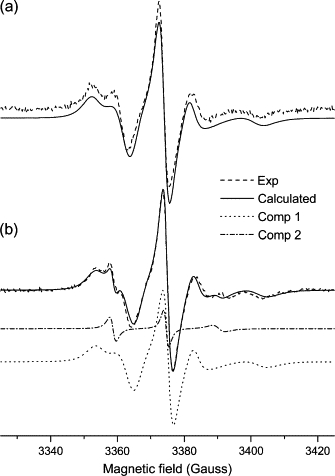
Experimental (dashed line) and calculated (solid line) ESR spectra from 5-PC spin probe in mixtures of (a) DOPC/Triton X-100 and (b) DOPC/Triton X-100/EcDHODH. b shows the individual components: 1 (bulk lipid, dotted line) and 2 (boundary lipid, dash-dotted line) obtained by means of NLSL simulations.
Figure 5.
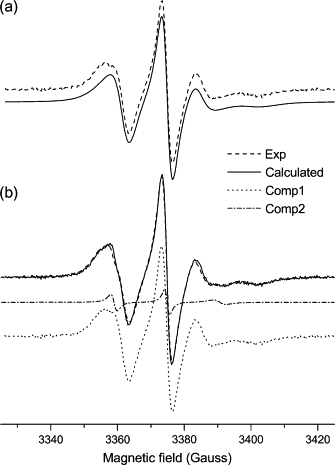
Experimental (dashed line) and calculated (solid line) ESR spectra from 10-PC spin probe in mixtures of (a) DOPC/Triton X-100 and (b) DOPC/Triton X-100/EcDHODH. b shows the individual components: 1 (bulk lipid, dotted line) and 2 (boundary lipid, dash-dotted line) obtained as described in the text.
As for 5-PC two-component spectral simulation, we used separate sets of parameters for each component. For the bulk component, the parameters determined previously for the one-component spectra (Table 1) were used to calculate the spectrum of what we denoted as component 1 in Table 1. These values were kept fixed and only the parameters for the boundary probes (component 2 in Table 1) were allowed to vary. The best-fit parameters thus obtained are shown in Table 1 and the calculated spectra in Fig. 4 B, where it is also shown the individual components calculated by the NLSL program. As pointed out above, component 2 gives rise to parameters attributable to a probe molecule experiencing a fast-motion regime.
The spectrum of the spin probe 10-PC in mixtures containing the enzyme was treated differently than the 5-PC spectrum described above. The NLSL program could not satisfactorily handle such a weak contribution to the overall spectrum because, in this case, the extra component is seen as a minor bump in the low-field resonance (Fig. 5 B). Even using two sets of parameters for the simulations, one of them was repeatedly set as a null contribution to the overall spectrum. A careful analysis of the individual spectrum determined for component 2 of the 5-PC probe in EcDHODH-containing model membranes (Fig. 4 B) suggested that the general features of the sharp-line spectrum in the 10-PC case were not dramatically different from those observed for component 2 of the 5-PC probe. To verify whether this was a reasonable assumption, we manually added the experimental spectrum of 10-PC in pure vesicles (Fig. 5 A) with the calculated spectrum for 5-PC boundary probe (component 2 in Table 1 and sharp-line spectrum in Fig. 4 B). The intensities of these individual spectra were adjusted to achieve the best reproduction of the two-component experimental spectrum of 10-PC in EcDHODH/DOPC/Triton X-100. A very good agreement between the final sum spectrum and the experimental 10-PC spectrum from EcDHODH-containing vesicles was obtained as can be seen in Fig. 5 B. This indicates that 5- and 10-PC probe molecules in contact with EcDHODH experience similar microenvironments.
Binding mechanism and enzyme catalysis
The isotropic hyperfine parameter A0 (A0 = Axx + Ayy + Azz) is a well-known measure of the relative polarity around the nitroxide moiety (18,30,44). The higher the A0 value, the more polar is that environment. Hence, from Table 1, we can see that the headgroup spin label DPPTC is, as expected, in a much more hydrophilic environment than the chain labels showing A0 value (16.5 G) comparable to the values observed for spin labels free in aqueous solution (ca. 16.9 G). From the headgroup region toward the vesicle interior, the A0 values drop down to 14.5 G for the 16-PC probe (Table 1). In the hydrophobic part of the model membrane, A0 parameter does not change significantly, thus suggesting that the polarity inside the vesicle does not show abrupt alterations. The same pattern for A0 is also obtained for the bulk labels in samples containing the enzyme (component 1 in Table 1), which indicates that EcDHODH does not modify the solvent (or other polar molecules) accessibility to the hydrophobic carbon chains of the vesicles.
On the other hand, a considerable increase in polarity (from 14.7 G to 16.0 G) is observed for the component 2 in the 5- and 10-PC spectra (Table 1) in the presence of EcDHODH. This result can be rationalized in terms of the residue composition of the EcDHODH N-terminal domain. The hydrophobic pattern for the residues in the two α-helices and one 310 helix that constitute the N-terminal domain determined by ProtScale software (45) (Fig. 6) allows us to infer that such a region shows an amphipathic character with alternating hydrophobic and hydrophilic regions. The existence of a significant number of polar residues could account for the increase in polarity observed for the component 2 of 5-PC label in the presence of the enzyme. Norager et al. (11) suggested that this residue distribution in the N-terminal domain would make it possible for the enzyme to adhere to the membrane, but not as an integral membrane protein. Our ESR data supports this peripheral docking of EcDHODH to membranes because major changes are observed no further down the acyl chain than position n = 10. However, we should bear in mind that, because of the low-ordered structure of the model membrane core, the protein penetration depth cannot be rigorously determined by our experiments. Nevertheless we can conclude that the modifications induced by the presence of the enzyme do take place in a somehow localized manner. Furthermore, the use of class 1 TcDHODH in similar ESR experiments resulted in no spectral changes, thus suggesting that it is the N-terminal extension the domain responsible for protein/membrane interaction.
Figure 6.
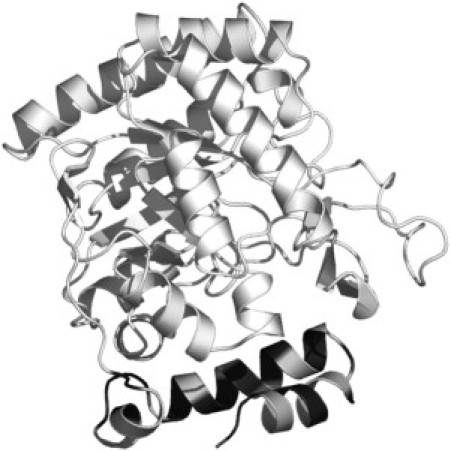
Ribbon representation of EcDHODH structure emphasizing the hydrophobicity pattern for N-terminal domain (comprising residues in the two α-helices and one 310 helix) as determined by ProtScale software. The dehydrogenase-active domain is shown in cyan. Hydrophobic and hydrophilic residues in the N-terminal domain are colored in dark and light gray, respectively.
The dynamics of the several spin probes can be discussed in terms of R⊥ and R// parameters. As observed previously in other articles that made use of the NLSL program, we also found that our simulations were insensitive to R//. Hence, this value was kept fixed at R// = 10 R⊥ during all simulations (27,46,47). In the absence of EcDHODH, R⊥ values of the chain labels followed an increasing gradient when one goes down along the acyl chain of the spin probe molecule (Table 1), which is compatible with the low-ordered and highly flexible organization of molecules inside mixed vesicles of phospholipid/detergent. The headgroup label DPPTC presents faster motion due to its exposition to the solvent, resulting in a less immobilized regime of motion.
In the presence of EcDHODH, we observed a greater R⊥ value for the DPPTC probe and a decrease in R⊥ values for the bulk labels (component 1 in Table 1) positioned in the hydrophobic part of the membrane as compared to the vesicles without the protein (Table 1). The enzyme induces lower fluidity of the carbon chains, while increasing the mobility of the headgroup probe probably due to the breakage of hydrogen bonds that would otherwise be formed by the headgroup. As for component 2 of the 5-PC ESR spectrum (Table 1), its R⊥ value (1.47 × 108 s−1) increases significantly when compared to the R⊥ parameter (0.22 × 108 s−1) obtained for the same 5-PC localized in the vesicle bulk (component 1), reaching rotational diffusion rates close to the ones observed for 16-PC labels.
Our results allow us to conclude that the presence of EcDHODH leads to a spacer effect between n = 5 and n = 10 carbon atoms of the DOPC vesicles. The determination of the size of this region is not accurate because a system composed of mixed vesicles does not present an ordering of the carbon chains as high as phospholipid bilayers. Nonetheless, this is strong evidence that a peripheral docking of the protein is taking place. The high A0 and R⊥ values (16.0 G and 1.47 × 10 s−1, respectively) for component 2 of the 5- and 10-PC probes indicate that a defect-like structure is formed by the adhesion of the EcDHODH N-terminal domain to the vesicle. The formation of defects in bilayers has been detected previously by ESR (48,49). Kleinschmidt et al. reported the existence of a sharp component in the ESR spectrum from a spin-labeled stearic acid incorporated in DTPG bilayers after addition of melittin (48). Ge et al. showed that binding of ADP ribosylation factor 6 (ARF6), an activator of phospholipase D, to phosphatidylinositol 4,5-bisphosphate (PIP2)-containing vesicles creates defects in the bilayer structure at the headgroup and/or near the n = 8–10 position of the carbon acyl chain (26). Moreover, Ge et al. (49) concluded that the sharp component can be significantly enhanced or reduced through specific lipid/lipid or lipid/protein interactions.
The formation of such defect may play a fundamental role in the catalytic cycle of the enzyme because the N-terminal region is also responsible for regulating the access to the protein active site. The crystal structure of HsDHODH in the presence of several inhibitors showed that the N-terminal contains the site for quinone binding (10). The N-terminal domain, along with the active site loop, acts as a cleft, shielding the FMN cofactor and orotate from the solvent (11) When bound to the membrane, the N-terminal induces the appearance of the defect (spacer effect) so that quinones dispersed in the membrane, which act as electron acceptors in the second half of the redox reaction catalyzed by EcDHODH, can now bind to the protein. The appearance of such defect is thus crucial for enzyme catalysis to take place.
Acknowledgments
The authors thank the Brazilian agencies Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for financially supporting this work. M.C.N. thanks a Young Scientist Fellowship from FAPESP (Grant No. 01/14583-0). A.J.C.F. thanks CNPq for a Research Fellowship (307102/2006-8). This work is part of a joint program Programa Núcleo de Excelência (PRONEX)/FAPESP/CNPq (Grant No. 03/09859-2). The authors are also grateful to Prof. K. Jensen who kindly provided the PAG1 plasmid and E. coli cell strains S06645 and S06740 used for EcDHODH expression.
Footnotes
Editor: David D. Thomas.
References
- 1.Jones M.E. Pyrimidine nucleotide biosynthesis in animals: Genes, enzymes, and regulation of UMP biosynthesis. Annu. Rev. Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- 2.Bjornberg O., Rowland P., Larsen S., Jensen K.F. Active site of dihydroorotate dehydrogenase A from Lactococcus lactis investigated by chemical modification and mutagenesis. Biochemistry. 1997;36:16197–16205. doi: 10.1021/bi971628y. [DOI] [PubMed] [Google Scholar]
- 3.Rowland P., Nielsen F.S., Jensen K.F., Larse S. The crystal structure of the flavin containing enzyme dihydroorotate dehydrogenase A from Lactococcus lactis. Structure. 1997;5:239–252. doi: 10.1016/s0969-2126(97)00182-2. [DOI] [PubMed] [Google Scholar]
- 4.Dimitrijevic M., Bartlett R.R. Leflunomide, a novel immunomodulating drug, inhibits homotypic adhesion of peripheral blood and synovial fluid mononuclear cells in rheumatoid arthritis. Inflamm. Res. 1996;45:550–556. doi: 10.1007/BF02342226. [DOI] [PubMed] [Google Scholar]
- 5.Smolen J.S., Kalden J.R., Scott D.L., Rozman B., Kvien T.K., Larsen A., Loew-Friedrich I., Oed C., Rosenburg R. Efficacy and safety of leflunomide compared with placebo and sulphasalazine in active rheumatoid arthritis: a double-blind, randomised, multicentre trial. Lancet. 1999;353:259–266. doi: 10.1016/s0140-6736(98)09403-3. [DOI] [PubMed] [Google Scholar]
- 6.Ruckemann K., Fairbanks L.D., Carrey E.A., Hawrylowicz C.M., Richards D.F., Kirschbaum B., Simmonds H.A. Leflunomide inhibits pyrimidine de novo synthesis in mitogen-stimulated T-lymphocytes from healthy humans. J. Biol. Chem. 1998;273:21682–21691. doi: 10.1074/jbc.273.34.21682. [DOI] [PubMed] [Google Scholar]
- 7.Davis J.P., Cain G.A., Pitts W.J., Magolda R.L., Copeland R.A. The immunosuppressive metabolite of leflunomide is a potent inhibitor of human dihydroorotate dehydrogenase. Biochemistry. 1996;35:1270–1273. doi: 10.1021/bi952168g. [DOI] [PubMed] [Google Scholar]
- 8.Loffler M., Grein K., Knecht W., Klein A., Bergjohann U. Dihydroorotate dehydrogenase - Profile of a novel target for antiproliferative and immunosuppressive drugs. Adv. Exp. Med. Biol. 1998;431:507–513. [PubMed] [Google Scholar]
- 9.Knecht W., Henseling J., Loffler M. Kinetics of inhibition of human and rat dihydroorotate dehydrogenase by atovaquone, lawsone derivatives, brequinar sodium and polyporic acid. Chem. Biol. Interact. 2000;124:61–76. doi: 10.1016/s0009-2797(99)00144-1. [DOI] [PubMed] [Google Scholar]
- 10.Liu S.P., Neidhardt E.A., Grossman T.H., Ocain T., Clardy J. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure. 2000;8:25–33. doi: 10.1016/s0969-2126(00)00077-0. [DOI] [PubMed] [Google Scholar]
- 11.Norager S., Jensen K.F., Bjömberg O., Larsen S. E. coli dihydroorotate dehydrogenase reveals structural and functional distinctions between different classes of dihydroorotate dehydrogenase. Structure. 2002;10:1211–1223. doi: 10.1016/s0969-2126(02)00831-6. [DOI] [PubMed] [Google Scholar]
- 12.Hansen M., Le Nours J., Johansson E., Antal T., Ullrich A., Loffler M., Larsen S. Inhibitor binding in a class 2 dihydroorotate dehydrogenase causes variations in the membrane-associated N-terminal domain. Protein Sci. 2004;13:1031–1042. doi: 10.1110/ps.03533004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurt D.E., Widom J., Clardy J. Structure of Plasmodium falciparum dihydroorotate dehydrogenase with a bound inhibitor. Acta Crystallogr. D. Biol. Crystallogr. 2006;62:312–323. doi: 10.1107/S0907444905042642. [DOI] [PubMed] [Google Scholar]
- 14.Rowland P., Bjornberg O., Nielsen F.S., Jensen K.F., Larsen S. The crystal structure of Lactococcus lactis dihydroorotate dehydrogenase A complexed with the enzyme reaction product throws light on its enzymatic function. Protein Sci. 1998;7:1269–1279. doi: 10.1002/pro.5560070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowland P., Norager S., Jensen K.F., Larsen S. Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster. Struct. Fold. Des. 2000;8:1227–1238. doi: 10.1016/s0969-2126(00)00530-x. [DOI] [PubMed] [Google Scholar]
- 16.Karibian D. Dihydroorotate dehydrogenase (Escherichia coli) Methods Enzymol. 1978;51:58–63. doi: 10.1016/s0076-6879(78)51010-0. [DOI] [PubMed] [Google Scholar]
- 17.Knecht W., Kohler R., Minet M., Loffler M. Anti-peptide immunoglobulins from rabbit and chicken eggs recognise recombinant human dihydroorotate dehydrogenase and a 44-kDa protein from rat liver mitochondria. Eur. J. Biochem. 1996;236:609–613. doi: 10.1111/j.1432-1033.1996.00609.x. [DOI] [PubMed] [Google Scholar]
- 18.Hubbell W.L., McConnell H.M. Molecular motion in spin-labeled phospholipids and membranes. J. Am. Chem. Soc. 1971;93:314–326. doi: 10.1021/ja00731a005. [DOI] [PubMed] [Google Scholar]
- 19.Berliner L.J. Academic Press; New York: 1979. Spin Labeling Theory and Applications II. [Google Scholar]
- 20.Hubbell W.L., Mchaourab H.S., Altenbach C., Lietzow M.A. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 21.Borbat P.P., Costa-Filho A.J., Earle K.A., Moscicki J.K., Freed J.H. Electron spin resonance in studies of membranes and proteins. Science. 2001;291:266–269. doi: 10.1126/science.291.5502.266. [DOI] [PubMed] [Google Scholar]
- 22.Dong J.H., Yang G.Y., Mchaourab H.S. Structural basis of energy transduction in the transport cycle of Msb. Science. 2005;308:1023–1028. doi: 10.1126/science.1106592. [DOI] [PubMed] [Google Scholar]
- 23.Park S.Y., Borbat P.P., Gonzalez-Bonet G., Bhatnagar J., Pollard A.M., Freed J.H., Bilwes A.M., Crane B.R. Reconstruction of the chemotaxis receptor-kinase assembly. Nat. Struct. Mol. Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 24.Eps N.V., Oldham W.M., Hamm H.E., Hubbell W.L. Structural and dynamical changes in an α-subunit of a heterotrimeric G protein along the activation pathway. Proc. Natl. Acad. Sci. USA. 2006;103:16194–16199. doi: 10.1073/pnas.0607972103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh D., Horväth L.I. Structure, dynamics and composition of the lipid-protein interface. Perspectives from spin-labelling. Biochim. Biophys. Acta. 1998;1376:267–296. doi: 10.1016/s0304-4157(98)00009-4. [DOI] [PubMed] [Google Scholar]
- 26.Ge M., Cohen J.S., Brown H.A., Freed J.H. ADP ribosylation factor 6 binding to phosphatidylinositol 4,5-bisphosphate-containing vesicles creates defects in the bilayer structure: an electron spin resonance study. Biophys. J. 2001;81:994–1005. doi: 10.1016/S0006-3495(01)75757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Citadini A.P.S., Pinto A.P.A., Araújo A.P.U., Nascimento O.R., Costa-Filho 1 A.J. EPR studies of chlorocatechol 1,2-dioxygenase: evidences of iron reduction during catalysis and of the binding of amphipatic molecules. Biophys. J. 2005;88:3502–3508. doi: 10.1529/biophysj.104.055251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuello L.G., Cortes D.M., Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 29.Ge M.T., Field K.A., Aneja R., Holowka D., Baird B., Freed J.H. Electron spin resonance characterization of liquid ordered phase of detergent-resistant membranes from RBL-2H3 cells. Biophys. J. 1999;77:925–933. doi: 10.1016/S0006-3495(99)76943-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh D. Polarity and permeation profiles in lipid membranes. Proc. Natl. Acad. Sci. USA. 2001;98:7777–7782. doi: 10.1073/pnas.131023798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa-Filho A.J., Shimoyama Y., Freed J.H. A 2D-ELDOR study of the liquid ordered phase in multilamellar vesicle membranes. Biophys. J. 2003;84:2619–2633. doi: 10.1016/S0006-3495(03)75067-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collado M.I., Goni F.M., Alonso A., Marsh D. Domain formation in sphingomyelin/cholesterol mixed membranes studied by spin-label electron spin resonance spectroscopy. Biochemistry. 2005;44:4911–4918. doi: 10.1021/bi0474970. [DOI] [PubMed] [Google Scholar]
- 33.Bjornberg O., Gruner A.C., Roepstorff P., Jensen K.F. The activity of Escherichia coli dihydroorotate dehydrogenase is dependent on a conserved loop identified by sequence homology, mutagenesis, and limited proteolysis. Biochemistry. 1999;38:2899–2908. doi: 10.1021/bi982352c. [DOI] [PubMed] [Google Scholar]
- 34.López O., Maza A., Coderch L., López-Iglesia C., Wherli E., Parra J.L. Direct formation of mixed micelles in the solubilization of phospholipid liposomes by Triton X-100. FEBS Lett. 1998;426:314–318. doi: 10.1016/s0014-5793(98)00363-9. [DOI] [PubMed] [Google Scholar]
- 35.Kragh-Hansen U., Maire M., Moller J.V. The mechanism of detergent solubilization of liposomes and protein-containing membranes. Biophys. J. 1998;75:2932–2946. doi: 10.1016/S0006-3495(98)77735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jost P.C., Griffith O.H., Capaldi R.A., Vanderkooi G.A. Evidence for boundary lipid in membranes. Proc. Natl. Acad. Sci. USA. 1973;70:480–484. doi: 10.1073/pnas.70.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang S.Y., Gutowsky H.S., Hsung J.C., Jacobs R., King T.E., Rice D., Oldfield E. Nuclear magnetic resonance investigations of the cytochrome oxidase-phospholipid interaction: a new model for boundary lipid. Biochemistry. 1979;18:3257–3267. doi: 10.1021/bi00582a010. [DOI] [PubMed] [Google Scholar]
- 38.Marsh D., Watts A. Spin labeling and lipid-protein interactions in membranes. In: Jost P.C., Griffith O.H., editors. Lipid-Protein Interactions. John Wiley & Sons; New York: 1982. p. 53. [Google Scholar]
- 39.Takashima E., Inaoka D.K., Osanai A., Nara T., Odaka M., Aoki T., Inaka K., Harada S., Kita K. Characterization of the dihydroorotate dehydrogenase as a soluble fumarete reductase in Trypanosoma cruzi. Mol. Biochem. Parasitol. 2002;122:189–200. doi: 10.1016/s0166-6851(02)00100-7. [DOI] [PubMed] [Google Scholar]
- 40.Meirovitch E., Nayeem A., Freed J.H. An analysis of protein-lipid interactions based on model simulations of ESR spectra. J. Phys. Chem. 1984;88:3454–3465. [Google Scholar]
- 41.Schneider D.J., Freed J.H. Calculating slow motional magnetic resonance spectra: a user's guide. In: Berliner L.J., Reuben J., editors. Biological Magnetic Resonance. Vol. 8. Plenum Publishing; New York: 1989. pp. 1–76. [Google Scholar]
- 42.Budil D.E., Lee S., Saxena S., Freed J.H. Nonlinear-least squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J. Magn. Reson. A. 1996;120:155–189. [Google Scholar]
- 43.Ge M., Rananavare S.B., Freed J.H. ESR studies of stearic-acid binding to bovine serum-albumin. Biochim. Biophys. Acta. 1990;1036:228–236. doi: 10.1016/0304-4165(90)90039-y. [DOI] [PubMed] [Google Scholar]
- 44.Griffith O.H., Jost P.A. Lipid spin labels in biological membranes. In: Berliner L.J., editor. Spin Labeling Theory and Applications. Academic Press; New York: 1976. pp. 453–523. [Google Scholar]
- 45.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press, Totowa; NJ: 2005. pp. 571–607. [Google Scholar]
- 46.Kar L., Ney-Igner E., Freed J.H. Electron spin resonance and electron-spin-echo study of oriented multilayers of Lα-dipalmitoylphosphatidylcholine water system. Biophys. J. 1985;48:569–595. doi: 10.1016/S0006-3495(85)83814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ge M., Freed J.H. Electron-spin resonance study of aggregation of Gramicidin in dipalmitoylphosphatidylcholine bilayers and hydrophobic mismatch. Biophys. J. 1999;76:264–280. doi: 10.1016/S0006-3495(99)77195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleinschmidt J.H., Mahaney J.E., Thomas D.D., Marsh D. Interaction of bee venom melittin with zwitterionic and negatively charged phospholipid bilayers: a spin-label electron spin resonance study. Biophys. J. 1997;72:767–778. doi: 10.1016/s0006-3495(97)78711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge M.T., Gidwani A., Brown H.A., Holowka D., Baird B., Freed J.H. Ordered and disordered phases coexist in plasma membrane vesicles of RBL-2H3 mast cells. An ESR study. Biophys. J. 2003;85:1278–1288. doi: 10.1016/S0006-3495(03)74563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


