Abstract
Monovalent cation binding by DNA A-tracts, runs of four or more contiguous adenine or thymine residues, has been determined for two curved ∼200 basepair (bp) restriction fragments, one taken from the M13 origin of replication and the other from the VP1 gene of SV40. These two fragments have previously been shown to contain stable, centrally located bends of 44° and 46°, respectively, located within ∼60 bp “curvature modules” containing four or five irregularly spaced A-tracts. Transient electric birefringence measurements of these two fragments, sequence variants containing reduced numbers of A-tracts in the SV40 curvature module or changes in the residues flanking the A-tracts in the M13 curvature module, have been combined with the free solution electrophoretic mobilities of the same fragments using known equations to estimate the effective charge of each fragment. The effective charge is reduced, on average, by one-third charge for each A-tract in the curvature module, suggesting that each A-tract binds a monovalent cation approximately one-third of the time. Monovalent cation binding to two or more A-tracts is required to observe significant curvature of the DNA helix axis.
Introduction
The binding of monovalent cations in the minor groove of DNA A-tracts, runs of 4–6 contiguous adenine (or thymine) residues, is a topic of continuing interest (1,2). The localization of cations in the A-tract minor groove would lead to asymmetric neutralization of the charged phosphate residues (3–5), giving directionality to the bending fluctuations of the helix backbone (6–10) and providing a molecular explanation for the A-tract-induced curvature of DNA (11). Recent x-ray diffraction and NMR studies have indicated that the A-tract minor groove is occupied ∼10% of the time by Na+ ions (12), ∼40% of the time by Rb+ ions (12), ∼50% of the time by Cs+ ions (13), and ∼10% of the time by Tl+ ions (14). Tl+ ions also appear to be localized ∼30% of the time in the major groove of G/C-tracts adjacent to A-tracts (14). The bound cations are readily exchangeable with each other and with water molecules in the solvent; water molecules occupy the ion binding sites when cations are not present (15).
X-ray diffraction and NMR measurements are indirect methods of determining cation binding to DNA. A more direct method of measuring cation binding is free solution capillary electrophoresis (CE) (16–18). The mobility of an analyte is determined by the ratio between its effective charge, Q, and its frictional coefficient, f. Therefore, according to the Nernst-Einstein equation
| (1) |
where μ is the electrophoretic mobility, Dt is the translational diffusion coefficient, kB is Boltzmann's constant, T is the absolute temperature, zeff is the effective number of charged phosphate residues, and eo is the (unsigned) charge of the electron (19,20). If the translational diffusion coefficient is independent of sequence, as it is for small DNA oligomers (21), the free solution mobility becomes a direct measure of the effective charge (16,18). In our previous study of monovalent cation binding to DNA A-tracts, we showed that 20-basepair (bp) oligomers containing phased A- or T-tracts migrate more slowly than an oligomer of the same size without A-tracts, indicating that A-tracts bind monovalent cations, reducing the effective net charge of the oligomer (16). A subsequent study showed that the free solution mobility of a stably curved, 199-bp restriction fragment taken from the VP1 gene in simian virus 40 (SV40) increased progressively as the number of A-tracts in the “curvature module” in the center of the fragment was decreased (17), again indicating that DNA A-tracts bind monovalent cations.
More quantitative analysis of monovalent cation binding to the A-tracts in the SV40 sequence variants requires knowledge of the translational diffusion coefficients of the various mutants. The translational diffusion coefficients can be calculated from the corresponding rotational diffusion coefficients using known equations (22) if the DNA molecules can be approximated as rigid rods. Previous studies have shown that this approximation is reasonable for DNA molecules in the size range considered here (23–25). From the calculated translational diffusion coefficients and the measured free solution mobilities, the effective charge of various DNA molecules can be estimated from Eq. 1. Comparison of the effective charge of DNA molecules containing the same number of bps but different numbers of A-tracts then provides a quantitative measure of monovalent cation binding to DNA A-tracts.
The curved DNA molecules used in this study were a 199-bp fragment taken from the VP1 gene in SV40 and a 217-bp fragment taken from the M13 origin of replication that was cloned into the plasmid Litmus 28. Transient electric birefringence (TEB) measurements, using the method of overlapping fragments (26), have shown that the SV40 and M13 fragments contain stable, centrally located bends of 44° ± 2° and 46° ± 2°, respectively (26–28). The apparent bend angle in the M13 fragment has been confirmed by atomic force microscopy (29).
The apparent bend centers in the SV40 and M13 fragments are located within 50–60 bp “curvature modules” containing four or five irregularly spaced A- or T-tracts. The sequences of the curvature modules are given in Scheme 1; the locations of the apparent bend centers are indicated by double vertical lines. The curvature modules are necessary and sufficient to cause stable bending of the DNA helix axis; replacing the curvature modules by random inserts of the same size eliminates the curvature (26–28).
Scheme 1.

Sequences of the curvature modules in the VP1 gene of SV40 and the M13 origin of replication
The A-tracts are highlighted in bold; the locations of the apparent bend centers are indicated by double vertical lines.
Sequence variants of the SV40 fragment were constructed in which the A- and T-tracts, singly and together, were mutated to other sequences, generating 30 fragments containing 0–5 permuted A-tracts. Sequence variants of the M13 curvature module had the GT and CA dinucleotides flanking the A3T5 and T7 groups mutated to other residues; in another construct, the A3T motif near the apparent bend center was replaced by a T4 group. The sequences of the mutated SV40 and M13 curvature modules are given in Tables S1 and S2 of the Supplementary Material. The M13 mutants were studied to indicate the generality of the results obtained with the SV40 sequence variants and to explore the effect of the flanking nucleotides on monovalent cation binding. Several polyacrylamide gel electrophoresis studies have suggested that the A-tract-induced curvature of DNA is modulated by the flanking nucleotides (30–36).
The results obtained with the parent SV40 and M13 fragments and their sequence variants indicate that the effective charge of curved, A-tract-containing DNAs is reduced, on average, by one-third charge per A-tract. Hence, each A-tract in a curvature module binds a monovalent cation approximately one-third of the time. Monovalent counterion binding by DNA A-tracts does not appear to be influenced by the flanking sequences; rather it is influenced only by the number of A-tracts in the curvature module. Significant curvature of the DNA helix axis requires monovalent cation binding by two or more closely spaced A-tracts.
Materials and methods
DNA samples
Restriction fragments from the M13 origin of replication, which has been cloned into the plasmid Litmus 28 (coordinates 1047–1254), and the VP1 gene in SV40 (coordinates 1826–2017) were prepared by methods described previously (26–28). The A- and T-tracts in the curvature module of the SV40 fragment and the CA and TG dinucleotides flanking the A-tracts in the M13 curvature module were modified by site-directed mutagenesis, as described previously (17). The final purification step in all cases was adsorption of the DNA onto small diethylaminoethyl cellulose (D52, Whatman) columns; the fragments were then eluted into the desired buffer and stored at −20°C. All fragments were sequenced to confirm their identities. The sequences of the SV40 and M13 curvature modules and the various mutants are given in Tables S1 and S2 of the Supplementary Material. The various fragments were also characterized by polyacrylamide gel electrophoresis, using methods described previously (37). The anomalously slow mobilities of the curved fragments were analyzed by calculating μ-decrements, defined as Δμ/μ = [(μnormal – μcurved)/μnormal] × 100. The μ-decrements observed for the parent SV40 and M13 fragments and their curvature module variants are given in Tables S1 and S2.
Buffers
The buffer usually used for the TEB experiments was 1 mM Tris-Cl, pH 8.0, plus 0.1 mM EDTA, called buffer B in previous work (26,27,38). Equivalent results were obtained using 0.2 mM Tris-Cl buffer (buffer C). CE experiments were carried out using Tris-acetate-EDTA (TAE) buffers (13 or 40 mM Tris base titrated to pH 8.0 with glacial acetic acid, plus 2 mM EDTA). Equivalent results were obtained in the two buffers.
Transient electric birefringence measurements
The apparatus and procedures used to carry out the TEB measurements have been described previously (26–28,38,39). Each DNA solution was pulsed 15 times in the single-shot mode under a given set of experimental conditions, and the field-free decay of each birefringence signal was analyzed using a nonlinear least-squares fitting program (CURVEFIT) adapted from the program designed by Bevington (40). The decay curves were fit with a single relaxation time, τ. Attempts to analyze the decay curves with two relaxation times led to significant increases in the uncertainties of the fitted τ-values. The relaxation times observed for the various fragments were constant within ±2%, even when different, independently prepared DNA stock solutions were used. Control experiments showed that the relaxation times were independent of DNA concentration, pulse length, and electric field strength. Therefore, a standard set of operating conditions—DNA concentration = 14 μg/mL (21 μM bp), electric field strength = 5 kV/cm, and pulse length = 8 μs—was used for all measurements. The temperature was 20°C ± 1°C.
The relaxation times of normal fragments containing the same number of bps as the curved fragments were calculated from standard curves of the logarithm of the relaxation times plotted as a function of the logarithm of the number of bps in each fragment (26,27,38). The τ-values of curved and normal fragments containing the same number of bps were used to calculate τ-decrements, defined as Δτ/τ = [(τnormal – τcurved)/τnormal] × 100.
Capillary electrophoresis measurements
Capillary zone electrophoresis measurements were carried out using a P/ACE MDQ Capillary Electrophoresis System (Beckman Coulter, Fullerton, CA) operated in the reverse polarity mode (detector at the anode) with ultraviolet detection at 254 nm, using methods described previously (17,41). The capillaries (Bio-Rad, Hercules, CA) were typically 40 cm in length (29.7 cm to the detector) and were coated internally with polyacrylamide to minimize the electroosmotic flow (EOF) of the solvent. Previous studies have shown that polyacrylamide coatings do not affect the observed mobilities (41). DNA samples containing ∼50 μg/mL were injected by pressure into the capillary; the sample plug occupied ∼1% of the capillary length. The applied electric field strength was 200 V/cm. Control experiments showed that the observed mobilities were independent of DNA concentration, sample plug length, and applied electric field strength. All measurements were made at 20°C ± 1°C.
The migration times of the parent SV40 and M13 fragments and their sequence variants were measured at least two to three times under a given set of conditions. Because the EOF mobility was negligible, the electrophoretic mobilities, μ, could be calculated directly from the migration times, t, according to Eq. 2:
| (2) |
where Ld is the length of the capillary to the detector and E is the electric field strength (19,20). Replicate measurements made on the same day were constant within ±0.04%; the average day-to-day variation in the mobilities was ±0.2%.
Theory
The rotational diffusion coefficient of a rigid, rod-like macromolecule is related to its size and shape as shown in Eq. 3:
| (3) |
where Dr is the rotational diffusion coefficient, kB is Boltzmann's constant, T is the absolute temperature, p is the axial ratio (p = L/d, where L is the end-to-end length of the rod and d is the diameter), η is the viscosity of the solvent, and δ is a correction for end effects, which can be expressed as δ = −0.662 + 0.917/p – 0.050/p2 (22). The rotational diffusion constant is calculated from the birefringence relaxation time, τ, using the well-known relationship: Dr = 1/6τ (42).
Eq. 3 contains two unknowns, the length and the diameter (or axial ratio) of the macromolecule. For small DNA restriction fragments, d is often taken to be 26 Å, somewhat larger than the crystallographic diameter, to allow for hydration effects and/or moderate coiling of the helix backbone (39,43). Since the diameter (or axial ratio) enters only through a logarithmic term in Eq. 3, reasonable variations in d have little effect on the calculated length.
The translational diffusion coefficient, Dt, of a rigid rod-like molecule is also related to its size and shape, according to Eq. 4:
| (4) |
where v is a correction for end effects, defined as ν = 0.312 + 0.565/p – 0.100/p2 (22). Combining Eqs. 3 and 4 relates the translational diffusion constant of a rod to its rotational diffusion coefficient, according to Eq. 5:
| (5) |
Inspection of Eq. 5 shows that the second term in parentheses is essentially constant for sequence isomers containing the same number of bps. Inserting the appropriate value for the 199-bp SV40 sequence isomers, evaluating the various constants in Eqs. 1 and 5, remembering that eo/kBT = 39.6 V−1 at 20°C, and converting to birefringence relaxation times instead of rotational diffusion coefficients leads to a simple expression for the effective charge of each of the SV40 sequence variants:
| (6) |
The constant in Eq. 6 is based on the assumption that the temperature is 20°C, the mobility is expressed as cm2/Vs, and the relaxation time is given in seconds. The constant is also weakly dependent on the DNA axial ratio, as can be seen from Eq. 5. For the 217-bp M13 sequence variants, the appropriate value of the constant is 6.54 × 106.
Eq. 6 also assumes that equations that have been derived for straight rods (22) can be used to describe the relationship between Dt and Dr for moderately curved molecules, such as the SV40 and M13 sequence mutants studied here, keeping the value of p constant for the various sequence mutants. This assumption was evaluated by calculating zeff for several of the SV40 sequence mutants using the actual values of p calculated from the birefringence relaxation times, instead of using a constant p for all mutants. No significant differences in zeff were obtained by the two methods of calculation.
The quantity of interest in this study is not zeff but the decrease in zeff as a function of the number of A-tracts in the curvature module. Equating the decrease in the effective charge to the increase in the number of bound cations, ΔN, leads to
| (7) |
The normal fragment used as the control for the SV40 mutants was 199K, which has no A-tracts in the curvature module. The normal fragment used for the M13 mutants was a 219 bp fragment taken from plasmid Litmus 28. Although this control fragment has two extra bps, its birefringence relaxation time and free solution mobility are very close to the values expected for a normal fragment containing 217 bp (17,38), as shown below by the last two entries in Table 2.
Table 2.
Birefringence relaxation times and free solution mobilities observed for M13 curvature module mutants
Results and discussion
Characterization of SV40 and M13 curvature module sequence variants
A typical birefringence signal observed for the parent 199-bp SV40 fragment is shown in the inset of Fig. 1. The decay of the birefringence, shown in the main part of the figure, is monoexponential, with a relaxation time of 3.74 ± 0.06 μs. Similar monoexponential decay curves were observed for all mutants of the SV40 and M13 curvature modules and for normal fragments ranging in size from 79 to 250 bps (38). Typical birefringence relaxation times observed for the parent SV40 fragment and its curvature module sequence variants are given in Table 1; the relaxation times of the M13 mutants and controls are given in Table 2. Previous studies have shown that the τ-decrements observed for the curved SV40 and M13 molecules and their sequence variants are essentially independent of the buffer in which the measurements are carried out, indicating that the parent fragments and their sequence variants are stably curved, not anisotropically flexible (26–28).
Figure 1.
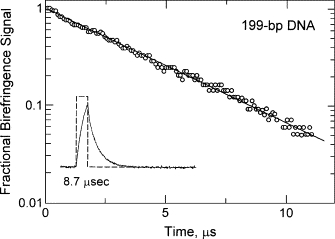
Analysis of the birefringence decay curve observed for the parent 199-bp SV40 fragment in 1 mM Tris-Cl buffer. The logarithm of the fractional birefringence signal remaining after time, t, is plotted as a function of the time after removal of the electric field. The birefringence decay curve is monoexponential, as shown by the straight line through the data points; the relaxation time is 3.74 ± 0.06 μs. The inset shows the applied 8.7 μs square-wave pulse (dashed line) and the corresponding birefringence signal (noisier curved line). Adapted from Lu et al. (26) with permission.
Table 1.
Birefringence relaxation times and free solution mobilities observed for SV40 curvature module variants
| Name | Number of A-tracts in curvature module | τ × 106, s* | μ × 104, cm2/Vs† |
|---|---|---|---|
| 199 | 5 | 3.71 | 3.1087 |
| 198A | 4 | 3.73 | 3.1120 |
| 199B | 4 | 3.87 | 3.1190 |
| 199E | 4 | 3.77 | 3.1209 |
| 199F | 4 | 4.11 | 3.1125 |
| 198C | 3 | 4.02 | 3.1375 |
| 199Q | 3 | 4.08 | 3.1332 |
| 199U | 3 | 4.05 | 3.1358 |
| 199G | 3 | 4.03 | 3.1308 |
| 199P | 3 | 3.99 | 3.1279 |
| 199D | 3 | 4.08 | 3.1254 |
| 199R | 3 | 3.88 | 3.1225 |
| 198B | 3 | 3.87 | 3.1245 |
| 199V | 3 | 3.91 | 3.1208 |
| 199O | 2 | 4.11 | 3.1443 |
| 199H | 2 | 4.00 | 3.1339 |
| 198I | 2 | 4.13 | 3.1353 |
| 198J | 2 | 4.16 | 3.1520 |
| 198H | 2 | 4.15 | 3.1429 |
| 198G | 2 | 3.98 | 3.1263 |
| 199T | 2 | 4.10 | 3.1432 |
| 198D | 2 | 3.99 | 3.1392 |
| 199S | 2 | 4.17 | 3.1425 |
| 199J | 1 | 4.13 | 3.1369 |
| 199N | 1 | 4.18 | 3.1487 |
| 199I | 1 | 4.12 | 3.1435 |
| 199E | 1 | 3.96 | 3.1209 |
| 198F | 1 | 3.99 | 3.1379 |
| 199K | 0 | 4.24 | 3.1445 |
In buffer B. The average standard deviation of triplicate measurements of each sample was ±0.04.
In 40 mM TAE. The average standard deviation of triplicate measurements of each sample was ±0.0008.
Typical electropherograms observed by free solution CE for the 199-bp SV40 fragment and a mutant, 199K, in which all the A-tracts in the curvature module have been mutated to other sequences are illustrated in the inset of Fig. 2. Both peaks are nearly Gaussian in shape, but the parent 199-bp fragment migrates significantly more slowly than oligomer 199K. Similar electropherograms were observed for the other fragments used in this study (not shown).
Figure 2.
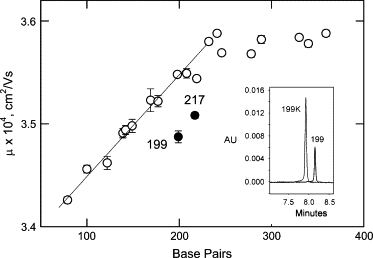
Comparison of the free solution mobility of the parent SV40 (199-bp) and M13 (217-bp) restriction fragments (●) with the mobilities of normal, non-A-tract-containing fragments of various sizes (○), measured in 13 mM TAE buffer. The error bars represent the standard deviation of triplicate measurements; the straight line is drawn to guide the eye. Inset: Typical electropherograms observed for the curved 199-bp SV40 fragment (right peak) and oligomer 199K, which had all five A-tracts in the curvature module mutated to other sequences (left peak). The optical density, in arbitrary units, is plotted as a function of migration time.
The dependence of the free solution mobility of normal, non-A-tract-containing DNA restriction fragments on molecular weight is illustrated in the main part of Fig. 2 (open circles). The mobilities increase approximately linearly with molecular weight until leveling off at a constant plateau value for fragments larger than ∼250 bp. Similar results have been observed for other DNA restriction fragments in the same molecular weight range (17,41). The mobilities of the curved SV40 (199-bp) and M13 (217-bp) fragments are significantly lower than expected on the basis of their molecular weights, as shown by the solid circles in the same figure. The free solution mobilities of the various SV40 and M13 sequence mutants are given in the last columns of Tables 1 and 2.
Preferential counterion binding to DNA A-tracts
The effective charge of the curved 199-bp SV40 fragment and mutants containing reduced numbers of A-tracts in the curvature module was calculated from Eq. 6; the apparent number of cations bound to the A-tracts, ΔN, was then calculated from Eq. 7. The parent SV40 fragment contains five A-tracts in the curvature module. Five sequence variants contained one A-tract, nine mutants contained two A-tracts in various positions, nine mutants contained three permuted A-tracts, five mutants contained four permuted A-tracts, and one mutant, 199K, had all five A-tracts mutated to other sequences. To focus on the generality of the results, and not on cation binding to specific A-tracts, the ΔN values obtained for all mutants containing the same number of A-tracts in the curvature module were averaged. The average ΔN value is plotted in Fig. 3 as a function of the number of A-tracts in the curvature module (open circles).
Figure 3.
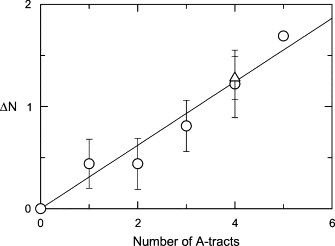
Dependence of the change in the average number of bound cations, ΔN, on the number of A-tracts in the curvature module. (○) SV40 sequence variants; (▵), M13 sequence variants. The error bars correspond to the standard deviation of the ΔN values observed for sequence variants containing the same number of A-tracts. The line was drawn by linear regression (r2 = 0.96); the slope of the line is 0.31.
The results in Fig. 3 indicate that the average number of bound cations increases approximately linearly with the number of A-tracts in the curvature module. The slope of the line is 0.31, indicating that a Tris+ ion is bound to an A-tract approximately one-third of the time. Stated differently, approximately one-third of the A-tracts in the SV40 curvature module bind a Tris+ ion at any given time. Preferential cation binding to the A-tracts appears to be independent of A-tract length and polarity, since there is no correlation between the number of bound cations and the presence or absence of a particular A-tract in the curvature module (data not shown). The average occupancy rate of ∼30% indicates that the preferential binding of Tris+ ions to DNA A-tracts is within the range of values observed for the binding of other monovalent cations to the AATT motif in the Dickerson-Drew dodecamer (12–14). The consistency of monovalent cation binding to DNA A-tracts observed by x-ray diffraction, NMR, and birefringence/electrophoresis methods lends confidence to the interpretation of the results obtained by all three methods.
The apparent number of cations bound to sequence variants of the M13 curvature module agrees with that observed for the SV40 mutants, as shown by the open triangle in Fig. 3. Hence, preferential cation binding to DNA A-tracts is not limited to the particular A-tracts in the SV40 curvature module. In addition, the apparent number of cations bound by the A-tracts in the M13 curvature module is approximately independent of the presence or absence of CA or TG dinucleotides adjacent to the A3T5 and T7 groups, indicating that the flanking dinucleotides do not affect monovalent cation binding to DNA A-tracts. Replacing the A3T sequence near the apparent bend center by a T4 group also does not affect monovalent cation binding, suggesting that these two sequence motifs bind Tris+ ions to a similar extent. A comparison of the binding of monovalent cations to A-tracts of different types will be described in a future communication (Q. Dong, E. Stellwagen, and N. C. Stellwagen, manuscript in preparation).
The results in Fig. 3 do not address the issue of whether monovalent cations are bound in the major or minor groove of DNA A-tracts; they address only the fact that A-tracts bind more monovalent cations than random sequence DNA. Regardless of the binding site, the juxtaposition of several A-tracts within a relatively short sequence span, as observed in the SV40 and M13 curvature modules, would lead to a patch of reduced charge density along the helix backbone. This patch of reduced charge density, with or without electrostatic collapse of the helix backbone toward the minor groove (6,7,10,36,44), could be part of the recognition signal used by transcription factors and other proteins to find their targets on the DNA surface (15,44–46) and/or may contribute to indirect readout (47) when the residues in an A-tract are not directly contacted by the protein side chains.
Monovalent cation binding to the A-tracts in the SV40 and M13 curvature modules is directly correlated with the anomalous electrophoretic mobilities observed in polyacrylamide gels, as shown in Fig. 4 A, where the average number of cations bound to sequence variants containing the same number of A-tracts is plotted as a function of the average gel mobility decrements, Δμ/μ, observed for the same fragments. The linear correlation indicates that a primary determinant of the anomalously slow electrophoretic mobilities observed for A-tract-containing DNAs in polyacrylamide gels is their reduced effective charge density. The differences in the effective charge are amplified by the sieving effects of the polyacrylamide gel matrix, explaining why polyacrylamide gel electrophoresis is such a good reporter of DNA curvature (48).
Figure 4.
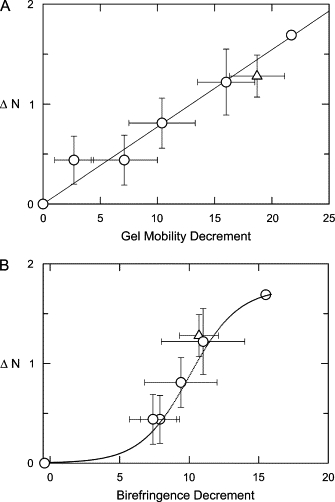
The average number of cations bound by SV40 (○) and M13 (▵) sequence variants containing the same number of A-tracts is plotted as a function of (A) the average polyacrylamide gel mobility decrements, Δμ/μ, and (B) the average τ-decrements, Δτ/τ, observed for the same fragments. The error bars represent the average deviation of the ΔN, Δμ/μ, and Δτ/τ values observed for sequence variants containing the same number of A-tracts. The straight line in (A) was drawn by linear regression (r2 = 0.96). The dotted line in (B) is a sigmoidal fit of the data points.
Preferential cation binding to DNA A-tracts is not directly correlated with the curvature of the helix backbone, as shown in Fig. 4 B where the average number of cations bound to sequence variants containing the same number of A-tracts is plotted as a function of the average τ-decrements observed for the same fragments. The data appear to be well fitted by a sigmoidal function, suggesting that stable curvature of the DNA helix axis requires the cooperative binding of monovalent cations to two or more closely spaced A-tracts. Studies of the fluorescence anisotropy of cationic residues tethered in the A-tract minor groove have also indicated that curvature of the DNA helix backbone requires the presence of adjacent positive charges (44). The combined results suggest that fractional occupancy of the A-tract major/minor groove(s) by the first cation facilitates occupancy by additional cation(s) in adjacent A-tracts. Hence, the number of cations bound in the SV40 curvature module increases linearly with the number of A-tracts (Figs. 3 and 4 A), whereas curvature depends on the cooperative binding of two or more cations to adjacent A-tracts (Fig. 4 B).
Conclusion
The results in this study have shown that DNA A-tracts bind monovalent cations about one-third of the time, reducing the effective charge of A-tract-containing DNAs by about one-third charge per A-tract. Preferential cation binding to the A-tracts is linearly correlated with the gel mobility decrements observed for the same fragments (Fig. 4 A); however, stable curvature of the helix axis requires the cooperative binding of monovalent cation(s) to two or more closely spaced A-tracts (Fig. 4 B). The exact sequence and spacing of the A-tracts affects preferential counterion binding to some extent, leading to the error bars reported in Figs. 3 and 4. Quantitative studies of monovalent cation binding to various types of A-tracts will be described in a future communication (Q. Dong, E. Stellwagen, and N. C. Stellwagen, manuscript in preparation).
Acknowledgments
The expert technical assistance of Brock Weers in preparing the restriction fragments used in this study is gratefully acknowledged.
Partial financial support of this work by grant GM610090 from the National Institute of General Medical Sciences (to N.C.S.) is also acknowledged.
Footnotes
Yongjun Lu's present address is Dept. of Internal Medicine, University of Iowa, Iowa City, IA 52242.
Editor: Jonathan B. Chaires.
Appendix A.
Data Supplement 1
References
- 1.Olson W.K., Zhurkin V.B. Twenty years of DNA bending. In: Sarma R.H., Sarma M.H., editors. Biological Structure and Dynamics. Adenine Press; Schenectady, NY: 1996. pp. 341–370. [Google Scholar]
- 2.Beveridge D.L., Dixit S.B., Barreiro G., Thayer K.M. Molecular dynamics simulations of DNA curvature and flexibility: helix phasing and premelting. Biopolymers. 2004;73:380–403. doi: 10.1002/bip.20019. [DOI] [PubMed] [Google Scholar]
- 3.Strauss J.K., Maher L.J., III DNA bending by asymmetric phosphate neutralization. Science. 1994;266:1829–1834. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- 4.Strauss J.K., Roberts C., Nelson M.G., Switzer C., Maher L.J., III DNA bending by hexamethylene-tethered ammonium ions. Proc. Natl. Acad. Sci. USA. 1996;93:9515–9520. doi: 10.1073/pnas.93.18.9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young M.A., Jayaram B., Beveridge D.L. Intrusion of counterions into the spine of hydration in the minor groove of B-DNA: fractional occupancy of electronegative pockets. J. Am. Chem. Soc. 1997;119:59–69. [Google Scholar]
- 6.Stefl R., Wu H., Ravindranathan S., Sklenář V., Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc. Natl. Acad. Sci. USA. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Range K., Mayaan E., Maher L.J., III, York D.M. The contribution of phosphate-phosphate repulsions to the free energy of DNA bending. Nucleic Acids Res. 2005;33:1257–1268. doi: 10.1093/nar/gki272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manning G.S. The contribution of transient counterion imbalances to DNA bending fluctuations. Biophys. J. 2006;90:3208–3215. doi: 10.1529/biophysj.105.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hud N.V., Sklenář V., Feigon J. Localization of ammonium ions in the minor groove of DNA duplexes in solution and the origin of DNA A-tract bending. J. Mol. Biol. 1999;286:651–660. doi: 10.1006/jmbi.1998.2513. [DOI] [PubMed] [Google Scholar]
- 10.McDonald R.J., Dragan A.I., Kirk W.R., Neff K.L., Privalov P.L., Maher L.J., III DNA bending by charged peptides: electrophoretic and spectroscopic analyses. Biochemistry. 2007;46:2306–2316. doi: 10.1021/bi061921a. [DOI] [PubMed] [Google Scholar]
- 11.Hud N.V., Plavec J. A unified model for the origin of DNA sequence-directed curvature. Biopolymers. 2003;69:144–159. doi: 10.1002/bip.10364. [DOI] [PubMed] [Google Scholar]
- 12.Marincola F.C., Denisov V.P., Halle B. Competitive Na+ and Rb+ binding in the minor groove of DNA. J. Am. Chem. Soc. 2004;126:6739–6750. doi: 10.1021/ja049930z. [DOI] [PubMed] [Google Scholar]
- 13.Tereshko V., Minasov G., Egli M. A “hydrat-ion” spine in a B-DNA minor groove. J. Am. Chem. Soc. 1999;121:3590–3595. [Google Scholar]
- 14.Howerton S.B., Sines C.C., VanDerveer D., Williams L.D. Locating monovalent cations in the grooves of B-DNA. Biochemistry. 2001;40:10023–10031. doi: 10.1021/bi010391+. [DOI] [PubMed] [Google Scholar]
- 15.Subirana J.A., Soler-López M. Cations as hydrogen bond donors: a view of electrostatic interactions in DNA. Annu. Rev. Biophys. Biomol. Struct. 2003;32:27–45. doi: 10.1146/annurev.biophys.32.110601.141726. [DOI] [PubMed] [Google Scholar]
- 16.Stellwagen N.C., Magnusdóttir S., Gelfi C., Righetti P.G. Preferential counterion binding to A-tract DNA oligomers. J. Mol. Biol. 2001;305:1025–1033. doi: 10.1006/jmbi.2000.4362. [DOI] [PubMed] [Google Scholar]
- 17.Stellwagen E., Lu Y.J., Stellwagen N.C. Curved DNA molecules migrate anomalously slowly in free solution. Nucleic Acids Res. 2005;33:4425–4432. doi: 10.1093/nar/gki748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stellwagen E., Dong Q., Stellwagen N.C. Quantitative analysis of monovalent counterion binding to random-sequence, double-stranded DNA using the replacement ion method. Biochemistry. 2007;46:2050–2058. doi: 10.1021/bi062132w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bockris J.O’.M., Reddy A.K.N. 2nd ed. Plenum Press; New York: 1998. Modern Electrochemistry 1: Ionics. [Google Scholar]
- 20.Grossman P.D. Factors affecting the performance of capillary electrophoresis separations: Joule heating, electroosmosis, and zone dispersion. In: Grossman P.D., Colburn J.E., editors. Capillary Electrophoresis. Academic Press; San Diego: 1992. pp. 3–43. [Google Scholar]
- 21.Stellwagen N.C., Magnusdóttir S., Gelfi C., Righetti P.G. Measuring the translational diffusion coefficients of small DNA molecules by capillary electrophoresis. Biopolymers. 2001;58:390–397. doi: 10.1002/1097-0282(20010405)58:4<390::AID-BIP1015>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.García de la Torre J., Lopez Martinez M.C., Tirado M.M. Dimensions of short, rodlike macromolecules from translational and rotational diffusion coefficients. Study of the Gramicidin dimer. Biopolymers. 1984;23:611–615. [Google Scholar]
- 23.Nicolai T., Mandel M. Dynamic light scattering by aqueous solutions of low molar mass DNA fragments in the presence of NaCl. Macromolecules. 1989;24:6128–6138. [Google Scholar]
- 24.Goïnga H.Tj., Pecora R. Dynamics of low molar weight DNA fragments in dilute and semidilute solutions. Macromolecules. 1991;24:6128–6138. [Google Scholar]
- 25.Tirado M.M., López Martínez C., García de la Torre J. Comparison of theories for the translational and rotational diffusion coefficients of rod-like molecules. Application to short DNA fragments. J. Chem. Phys. 1984;81:2047–2052. [Google Scholar]
- 26.Lu Y.J., Weers B.D., Stellwagen N.C. Analysis of DNA bending by transient electric birefringence. Biopolymers. 2003;70:270–288. doi: 10.1002/bip.10458. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y.J., Weers B.D., Stellwagen N.C. Intrinsic curvature in the VP1 gene of SV40: comparison of solution and gel results. Biophys. J. 2005;88:1191–1206. doi: 10.1529/biophysj.104.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stellwagen N.C., Lu Y.J. The use of transient electric birefringence to analyze curvature in naturally occurring, mixed-sequence DNA molecules. In: Stoylov S.P., Stoimenova M.V., editors. Molecular and Colloidal Electro-Optics. CRC Press; Boca Raton, FL: 2007. pp. 285–299. [Google Scholar]
- 29.Lu Y.J., Weers B.D., Stellwagen N.C. Analysis of the intrinsic bend in the M13 origin of replication by atomic force microscopy. Biophys. J. 2003;85:409–415. doi: 10.1016/S0006-3495(03)74485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagaich A.K., Bhattacharyya D., Brahmachari S.K., Bansal M. CA/TG sequence at the 5′ end of oligo(A)-tracts strongly modulates DNA curvature. J. Biol. Chem. 1994;269:7824–7833. [PubMed] [Google Scholar]
- 31.Milton D.L., Casper M.L., Wills N.M., Gesteland R.F. Guanine tracts enhance sequence directed DNA bends. Nucleic Acids Res. 1990;18:817–820. doi: 10.1093/nar/18.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abagyan R.A., Mironov V.N., Chernov B.K., Chuprina V.P., Ulyanov A.V. Electrophoretic behavior of d(GGAAAAAAGG)n, d(CCAAAAAACC)n, and (CCAAAAAAGG)n and implications for a DNA bending model. Nucleic Acids Res. 1990;18:989–992. doi: 10.1093/nar/18.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Møllegaard N.E., Bailly M., Waring J., Nielsen P.E. Effects of diaminopurine and inosine substitutions on A-tract induced DNA curvature. Importance of the 3′-A-tract junction. Nucleic Acids Res. 1997;25:3497–3502. doi: 10.1093/nar/25.17.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dlakic M., Harrington R.E. Unconventional helical phasing of repetitive DNA motifs reveals their relative bending contributions. Nucleic Acids Res. 1998;26:4274–4279. doi: 10.1093/nar/26.18.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merling A., Sagaydakova N., Haran T.E. A-tract polarity dominate the curvature in flanking sequences. Biochemistry. 2003;42:4978–4984. doi: 10.1021/bi020662w. [DOI] [PubMed] [Google Scholar]
- 36.Faiger H., Ivanchenko M., Cohen I., Haran T.E. TBP flanking sequences: asymmetry of binding, long-range effects and consensus sequences. Nucleic Acids Res. 2006;34:104–119. doi: 10.1093/nar/gkj414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes D.L., Stellwagen N.C. Estimation of polyacrylamide gel pore size from Ferguson plots of normal and anomalously migrating DNA fragments. I. Gels containing 3% N,N′-methylenebisacrylamide. Electrophoresis. 1991;12:253–263. doi: 10.1002/elps.1150120405. [DOI] [PubMed] [Google Scholar]
- 38.Lu Y.J., Weers B., Stellwagen N.C. DNA persistence length revisited. Biopolymers. 2002;61:261–275. doi: 10.1002/bip.10151. [DOI] [PubMed] [Google Scholar]
- 39.Stellwagen N.C. Transient electric birefringence of two small DNA restriction fragments of the same molecular weight. Biopolymers. 1991;31:1651–1667. doi: 10.1002/bip.360311318. [DOI] [PubMed] [Google Scholar]
- 40.Bevington P.R. McGraw-Hill; New York: 1969. Data Reduction and Error Analyses for the Physical Sciences. [Google Scholar]
- 41.Stellwagen N.C., Gelfi C., Righetti P.G. The free solution mobility of DNA. Biopolymers. 1997;42:687–703. doi: 10.1002/(SICI)1097-0282(199711)42:6<687::AID-BIP7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 42.Fredericq E., Houssier C. Clarendon Press; Oxford: 1973. Electric Dichroism and Electric Birefringence. [Google Scholar]
- 43.Stellwagen N.C. Electric birefringence of restriction enzyme fragments of DNA: optical factor and electric polarizability as a function of molecular weight. Biopolymers. 1981;20:399–434. doi: 10.1002/bip.1981.360200302. [DOI] [PubMed] [Google Scholar]
- 44.Williams S.L., Parkhurst L.K., Parkhurst L.J. Changes in DNA bending and flexing due to tethered cations detected by fluorescence resonance energy transfer. Nucleic Acids Res. 2006;34:1028–1035. doi: 10.1093/nar/gkj498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McFail-Isom L., Sines C.C., Williams L.D. DNA structure: cations in charge? Curr. Opin. Struct. Biol. 1999;9:298–304. doi: 10.1016/S0959-440X(99)80040-2. [DOI] [PubMed] [Google Scholar]
- 46.Williams L.D., Maher L.J., III Electrostatic mechanisms of DNA deformation. Annu. Rev. Biophys. Biomol. Struct. 2000;29:497–521. doi: 10.1146/annurev.biophys.29.1.497. [DOI] [PubMed] [Google Scholar]
- 47.Mendieta J., Pérez-Lago L., Salas M., Camacho A. DNA sequence-specific recognition by a transcriptional regulator requires indirect readout of A-tracts. Nucleic Acids Res. 2007;35:3252–3261. doi: 10.1093/nar/gkm180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stellwagen N.C. Curved DNA molecules migrate anomalously slowly in polyacrylamide gels even at zero gel concentration. Electrophoresis. 2006;27:1163–1168. doi: 10.1002/elps.200500612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Supplement 1


