Abstract
Mutations in the thick filament associated protein cardiac myosin binding protein-C (cMyBP-C) are a major cause of familial hypertrophic cardiomyopathy. Although cMyBP-C is thought to play both a structural and a regulatory role in the contraction of cardiac muscle, detailed information about the role of this protein in stability of the thick filament and maintenance of the ordered helical arrangement of the myosin cross-bridges is limited. To address these questions, the structure of myosin thick filaments isolated from the hearts of wild-type mice containing cMyBP-C (cMyBP-C+/+) were compared to those of cMyBP-C knockout mice lacking this protein (cMyBp-C−/−). The filaments from the knockout mice hearts lacking cMyBP-C are stable and similar in length and appearance to filaments from the wild-type mice hearts containing cMyBP-C. Both wild-type and many of the cMyBP-C−/− filaments display a distinct 43 nm periodicity. Fourier transforms of electron microscope images typically show helical layer lines to the sixth layer line, confirming the well-ordered arrangement of the cross-bridges in both sets of filaments. However, the “forbidden” meridional reflections, thought to derive from a perturbation from helical symmetry in the wild-type filament, are weaker or absent in the transforms of the cMyBP-C−/− myocardial thick filaments. In addition, the cross-bridge array in the absence of cMyBP-C appears more easily disordered.
Introduction
Cardiac myosin binding protein-C (cMyBP-C) has generated recent interest because mutations in this protein are a major cause of familial hypertrophic cardiomyopathy (see review by Marian and Roberts (1)). The pathological effects of mutations in cMyBP-C are consistent with increasing evidence that it plays both a structural and a regulatory role in myocardial contraction (reviewed by Flashman et al. (2) and Winegrad (3)). Myosin binding protein-C is one of several accessory myosin binding proteins (4) that are present in the vertebrate striated muscle thick filament. These proteins are responsible for a series of 11 transverse stripes with a spacing of 43–43.5 nm across each half of the A-band (4–9). Of these proteins myosin binding protein-C is present in the largest amount, and in skeletal muscle it is present in seven to nine of the stripes (5,9–11).
Although myosin binding protein-C was originally thought to have primarily a structural role in the filament (13), a regulatory role, especially in myocardium, is more compelling (14–21). The cardiac isoform of MyBP-C is specific to cardiac muscle (22,23) and has four sites which can be phosphorylated instead of the one site found in the skeletal muscle isoform (24). Phosphorylation of the cardiac isoform is under adrenergic control (15,24–28), suggesting a regulatory role for cMyBP-C. In addition, extraction or genetic ablation of cMyBP-C has been correlated with changes in calcium sensitivity and force development at submaximal levels of activation (29,30)—again consistent with a regulatory role for cMyBP-C.
Several recent studies (18–20) have suggested that the regulatory role of cMyBP-C may be mediated through its binding to the S2 region of myosin and an effect on cross-bridge extension. Consistent with this, a recent model (31) has suggested that phosphorylation of cMyBP-C may release myosin heads from the thick filament backbone. Other evidence, however, has demonstrated that some of the regulatory and physiological effects of cMyBP-C may be mediated independent of a tethering mechanism (32). Thus, although the evidence for a regulatory role of cMyBP-C is strong, the mechanism by which cMyBP-C exerts its effect is less clear.
The structural role of cMyBP-C is also unclear. Several studies suggest that cMyBP-C is necessary for correct assembly of the thick filament (13,33–35), regulation of its length (34), and stabilization of the filament after assembly (36). However, Harris et al. (37) found that cMyBP-C knockout mice, although displaying hypertrophy and significant contractile defects in the heart, are viable and display well-developed sarcomeres. Thus cMyBP-C does not appear to be an absolute requirement for thick filament formation. From the Harris study, however, it was not possible to determine what, if any, effect the absence of cMyBP-C may have had on the molecular structure of the thick filament or the arrangement of the myosin heads compared to the wild-type filaments.
In this study, we investigated the structure of thick filaments isolated from the hearts of the cMyBP-C knockout mice (cMyBP-C−/−) and compared their structure to that of filaments isolated from wild-type mice (cMyBP-C+/+) using electron microscopy and image analysis. The objective of these studies was to determine the effects of loss of cMyBP-C at the ultrastructural level on the filament length, diameter, and arrangement of the myosin heads. We demonstrate that filaments from the cMyBP-C−/− myocardium are stable in length, diameter, and appearance. The filaments also retain significant amounts of the relaxed periodic cross-bridge arrangement. Compared to wild-type filaments, however, the relaxed periodic cross-bridge array on the cMyBP-C−/− myocardial thick filaments is more easily disrupted and regions of disorder are more often observed.
Materials and methods
Transgenic mice
Cardiac MyBP-C knockout mice (37) were maintained on an SV/129 background. Wild-type Balb/C mice (7–8 weeks of age) were obtained from Harlan (Indianapolis, IN). Care and handling of all mice were performed according to institutional guidelines approved by the Association for Assessment and Accreditation of Laboratory Care International.
Filament isolation
Thick filaments were isolated from mouse ventricular muscle by a modification of the method previously described (38) for isolation of thick filaments from the rabbit heart. In brief, fresh mouse hearts were excised from mice anesthetized with isoflurane, and hearts were placed into an EGTA-mincing solution containing 100 mM NaCl, 2 mM EGTA, 5 mM MgCl2, 1 mM dithiothreitol, and 7 mM phosphate buffer (pH 7). Ventricular muscles were then rapidly dissected and minced into cubes (∼1–2 mm3). The muscles were then teased into fine bundles 0.5 mm or less in diameter with forceps tips, and bundles were placed into fresh EGTA mincing solution 3 or 4 times (1 h each time). Bundles were stored in EGTA mincing solution until filament isolation.
For filament isolation, small amounts of the muscle were incubated with slow stirring for 30 min at room temperature in relaxing solution (EGTA mincing solution plus 2.5 mM ATP) containing 10 mM creatine phosphate. Separation of the filaments was accomplished using the elastase treatment described by Levine et al. (39) as a modification of the original procedure of Magid et al. (40). In this procedure, a small piece of the muscle was incubated in an elastase solution containing the proteolytic inhibitor cocktail of Sellers (41) (1.1 mg/ml elastase in relaxing solution containing 0.44 mg/ml trypsin inhibitor, 0.0004 mg/ml pepstatin A, 0.0004 mg/ml leupeptin, 0.0004 mg/ml aprotinin, and 0.04 mM phenylmethanesulfonyl fluoride) for 3 min and then transferred to an Eppendorf tube containing fresh relaxing solution (with proteolytic inhibitors at 2 mg/liter). This was vigorously shaken by hand for 3 min and centrifuged at 3,000 rpm in an Eppendorf 5415C centrifuge (Eppendorf, Hamburg, Germany) to pellet unbroken cells and debris. The suspension of separated thick and thin filaments was then examined by electron microscopy.
Negative staining and electron microscopy
Negative staining with 1% uranyl acetate was performed as previously described (42). In this procedure, thick filaments were adsorbed onto thin carbon films (5–7 nm thickness) supported on perforated Formvar-coated grids. The grids were rinsed sequentially with eight drops of half-strength relaxing solution (50 mM NaAc, 1 mM-EGTA, 1 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP, and 2 mM imidazole buffer at pH 7.0) and negatively stained with 1% uranyl acetate. All of the rinse and stain solutions were maintained at 25°C on a Thermolyne Dri-bath (Thermolyne, Dubuque, IA) heater to avoid the loss of helical ordering, which has been demonstrated to occur for mammalian and avian thick filaments at temperatures below ∼15°C. Electron microscopy of the negatively stained preparations was performed at 80 kV with a JEOL-1200EXII electron microscope (JEOL, Tokyo, Japan) equipped with an AMT HR60 High Resolution Digital Camera (2 K × 2 K; AMT, Danvers, MA) at 150,000×. At this magnification there were ∼10 axial repeats of the filaments per 1024 pixels.
Computer image analysis
Computation of Fourier transforms and filtered images of the filaments was performed as previously described (43). For computation of Fourier transforms, selected areas of filament images at a pixel size corresponding to ∼0.42 nm were floated in 1024 × 1024 arrays. The images were straightened, rescaled, and rotated as necessary to ensure that the helical layer lines fell on the sampled lines of the transform by sizing the filament images to exactly 10 axial repeats per 1024 pixels.
Filtered images of the filaments were computed by Fourier inversion of the layer line data for the first six layer lines. Information about the periodic structure of the filaments is present in the layer line data, whereas information about nonperiodic structures is scattered over the entire Fourier transform. Masking the transform to include only the layer line data excludes much of the noise coming from the nonperiodic structures and background. Performing an inverse Fourier transform of the masked transform gives a filtered image displaying the periodic structure more clearly. Plots of the axial density profile of the filtered images were done with National Institutes of Health ImageJ software.
Measurements of filament diameter and the radius of the center of mass of the cross-bridges
The diameter of the filaments was measured on filtered images of the filaments obtained by inverse Fourier transformation of the data along the first six layer lines in the transforms. Since the images contained exactly 10 repeats of the 43 nm axial repeat in 1024 pixels, the repeat spacing of 43 nm equals 102.4 pixels in the filtered images, allowing conversion of the measured diameters of the filaments in pixels to values in nm by simple proportion using 43 nm = 102.4 pixels.
We also measured the positions of the primary maxima on the first layer line in computed Fourier transforms of each filament to calculate an estimate of the average radius at which the center of mass of the myosin heads lies. If the filaments are three stranded, as our data show, then by helical diffraction theory (44) the radial position (R) of the primary peaks along the first layer line is related to the radius at which the cross-bridges lie (r) by the equation 4.2 = 2πrR, where 4.2 is the value expected for a J3 Bessel function. By carefully setting the scaling of the filament images so that exactly 10 helical repeats (10 × 43 nm) occurred in the 1024 pixels of the 1024 × 1024 transform, it was possible to accurately measure the value of R in nm and use this to calculate an estimate of the average radius of the center of mass of the cross-bridge. It must be noted that the values obtained are only estimates used for comparative purposes and may not be the precise average radius at which the cross-bridge mass is centered. As we describe in the Discussion, the filament is not strictly helical and has cylindrical symmetry. Because of this, both J3 and J6 Bessel functions are likely to contribute to the primary reflection on the first layer line. This may result in an inaccuracy in the values calculated for the average radius at which the cross-bridge mass is centered (r). For comparative purposes between the filaments, however, the calculated values are useful as an alternative measure of the size of the filaments. Although smaller than the total diameter (or radius to which the cross-bridges project), the measurement provides an additional assessment of the filament size that should correlate with the direct measurements of filament diameter.
Results
Appearance of isolated filaments
Thick filaments were isolated from the hearts of both cMyBP-C+/+ mice and cMyBP-C−/− mice. Fig. 1, A–C, shows micrographs of the isolated thick filaments from the hearts of the cMyBP-C+/+ mice. The filaments are bipolar, with a central bare zone and a length of ∼1.6 microns. They display the distinct 43 nm axial periodicity of the myosin heads in the cross-bridge regions (indicated by tick marks) previously shown for isolated rabbit cardiac thick filaments (38,43). This periodicity can best be seen by tilting the micrographs and looking along the long axis of a filament. In addition, in Fig. 1 B, the boxed inset shows two repeats of the 43 nm axial periodicity from the Fourier filtered image of the filament.
Figure 1.

A gallery of electron micrograph images of isolated and negatively stained thick filaments from the wild-type mouse heart (MyBP-C+/+). A shows a slightly lower magnification field of the isolated filaments, and B and C show higher magnification examples of the fields. Note that the filaments show the distinct 43 nm quasihelical periodicity of the myosin heads previously shown for rabbit and rat cardiac filaments. This periodicity is indicated by the tick marks along the filaments. The boxed inset in B shows two repeats of the 43 nm axial periodicity from a Fourier filtered image of the filament calculated using the data along the first six layer lines. The two repeats from the filtered image more clearly show the repeating structure along the filament, which can then be seen to continue along the filament arm. A similar pattern can also be recognized in the filaments in A and C as well. The asterisks indicate the position of the filament bare zone. The magnification bars indicate 50 nm.
The filtered image was calculated by masking the Fourier transform of the filament to include only the data along the first six layer lines and performing a Fourier inversion of this data. Since the information about the periodic structure is localized to the layer lines of the transform, the inverse Fourier transform produces an image excluding the nonperiodic noise and showing the periodic structure more clearly. In Fig. 1 B, the 43 nm axial repeat present in the filtered image inset can be seen to extend along the rest of this filament. A similar periodic structure is evident in the other filaments as well. Close inspection of both the filtered image segments and the rest of the filaments demonstrates that the staining pattern at each cross-bridge crown of the mouse myocardial filaments lacks the bilateral symmetry expected for an even-stranded arrangement of the myosin heads (45). The filaments display the “sawtooth” pattern that we previously described in isolated rabbit cardiac thick filaments (38,43) and that we showed to be consistent with the three-stranded arrangement of the myosin heads.
Fig. 2, A–C, shows micrographs of thick filaments isolated from the cMyBP-C−/− myocardium. The filaments appear to be of normal length and diameter and to be relatively stable in structure. The normal length of the filaments is suggested by the presence of “end filaments” (46) normally seen at the ends of the wild-type filaments and other vertebrate striated thick filaments. The thick filaments of the cMyBP-C−/− myocardium, like the filaments isolated from the cMyBP-C+/+ myocardium, have a bipolar structure, and most display evidence of a distinct 43 nm axial periodicity in the cross-bridge regions. This axial periodicity is indicated by the tick marks along most of the filaments in Fig. 2, A–C, and, again, is best seen by viewing each filament along its long axis. In Fig. 2 B the boxed inset shows two repeats of the 43 nm axial periodicity from a filtered image of the filament. As in the case of the wild-type filaments, the staining pattern in the cross-bridge region of the cMyBP-C−/− filaments is the “sawtooth” pattern characteristic of a three-stranded arrangement of myosin heads (38,43).
Figure 2.
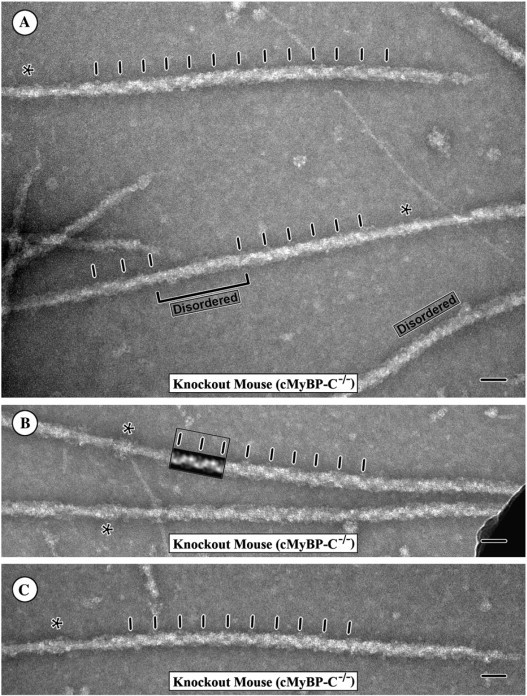
A gallery of electron micrograph images of isolated and negatively stained thick filaments from the cMyBP-C mouse heart (MyBP-C−/−). A shows a field of the isolated filaments, and B and C show examples of the individual filaments. Note that despite the absence of MyBP-C, the filaments appear to be of normal length and width and many of the filaments retain the 43 nm quasihelical arrangement of the myosin heads. The 43 nm periodicity is indicated by the tick marks along the filaments. The boxed inset in B shows two repeats of the 43 nm axial periodicity from a Fourier filtered image of the filament calculated using the data along the first six layer lines. The two repeats from the filtered image more clearly show the repeating structure along the filament, which can then be seen to continue along the filament arm. A similar pattern can also be recognized in the filaments in A and C as well. As shown in A, however, in the preparations of the cMyBP-C−/− filaments an increased number of filaments are either disordered in the periodic arrangement of the cross-bridges (labeled) or have alternating regions of order and disorder of the cross-bridge array. The bracket indicates one such region of disorder along a filament. The asterisks indicate the position of the filament bare zone. The magnification bars indicate 50 nm.
Although the filaments from the hearts of the cMyBP-C−/− mice typically display a distinct 43 nm axial periodicity of the myosin heads, the helical or quasihelical periodicity of these filaments appears more easily disordered by factors such as changes in the surface properties of the carbon films used for staining than do the wild-type filaments. The decreased stability of the periodic arrangement of the cross-bridges in the cMyBP-C−/− myocardial filaments was evident both in an increased number of filaments with disordered cross-bridges (Fig. 2 A, labeled) and an increased number of filaments in which both regions of order and disorder were observed along the same filament arm (bracket Fig. 2 A). Although we did not quantify numbers, the decreased stability of the helical or quasihelical arrangement of the cross-bridges under relaxing conditions in the cMyBP-C−/− thick filaments was quite clear. It was particularly evident in regions of poor negative staining due to hydrophobicity of the carbon surface. In these regions in which the negative stain was not present to help stabilize the structure, the filaments from the cMyBP-C−/− myocardium typically appeared more disordered than the filaments from the cMyBP-C+/+ myocardium. As illustrated in Fig. 2 A, however, the increased disorder of the filaments was also seen in regions of good negative staining and was not limited to areas of poor negative staining. Thus the disordering cannot be ascribed simply to poor negative staining or the properties of the carbon but most likely reflects a decreased stability of the periodic arrangement of the cross-bridges in the absence of cMyBP-C. These results suggest that cMyBP-C may play a role in stabilizing the relaxed cross-bridge arrangement.
Fourier transforms of the filaments
Fourier transforms of electron micrographs of cross-bridge regions displaying a distinct periodicity were computed and compared for both cMyBP-C+/+ cardiac filaments and filaments from the cMyBP-C−/− myocardium. Fig. 3, A–F, illustrates Fourier transforms obtained from the wild-type thick filaments. The computed transforms show a strong set of layer lines corresponding to a 43 nm quasihelical arrangement of the myosin heads. Typically, the patterns are strong to the 6th layer line with a rapid fall-off in intensity beyond this point, although layer lines out to the 11th or 12th layer line are frequently present. The periodic detail in the filaments therefore extends to ∼4 nm. The first and second off-meridional layer lines are usually strong, whereas the fourth and fifth layer lines are much weaker. The meridional reflections which correspond to the average axial rise of 14.3 nm per cross-bridge crown are present on the third and sixth layer lines but of variable intensity from transform to transform.
Figure 3.
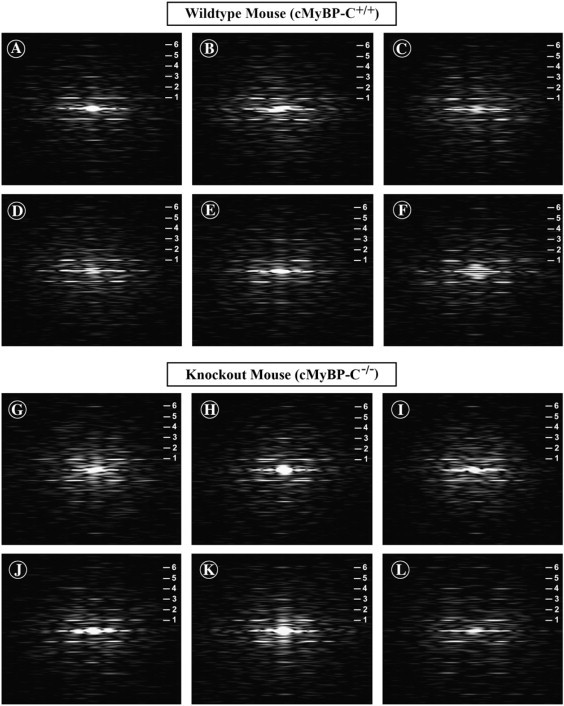
Computed Fourier transforms of electron micrograph images of the negatively stained thick filaments from the wild-type mouse heart (A–F) and the cMyBP-C knockout mouse heart (G–L). The transforms of the wild-type filaments show a strong series of layer lines (indicated by the numerals) corresponding to the 43 nm quasihelical axial repeat of the filament. Note the frequent presence of the meridional reflections on the first, second, fourth, and fifth layer lines, which are not expected for ideal helical symmetry (“forbidden” meridional reflections). The transforms from the filaments of the cMyBP-C knockout mouse heart (G–L) also show a well-developed series of layer lines corresponding to the 43 nm axial repeat. Note that the “forbidden” meridional reflections are very weak or absent in the transforms of the filaments from the hearts of the MyBP-C knockout mice, however, compared to the transforms from the wild-type filaments.
In addition to the meridional reflections expected from ideal helical symmetry on layer lines that are multiples of three, such as 3, 6, 9…, the transforms from the wild-type filaments typically show additional meridional reflections not expected from ideal helical symmetry on the 1st, 2nd, 4th, 5th, 8th, 10th, and 11th layer lines (Fig. 3, A–F). These reflections, on layer lines which are not multiples of three, correspond to the “forbidden” meridional reflections described in the x-ray diffraction pattern of vertebrate skeletal muscle by Huxley and Brown (47). They are thought to arise either from a perturbation from ideal helical symmetry in the cross-bridge array or from the presence of accessory proteins associated with the filament backbone. Overall, these transforms from the wild-type mouse myocardial thick filaments appear very similar to the transforms previously published for isolated rabbit cardiac thick filaments (38,43).
The Fourier transforms computed from the isolated filaments from the cMyBP-C−/− myocardium (Fig. 3, G–L) are similar in showing a strong series of layer lines corresponding to a 43 nm axial periodicity which often extends to the 11th or 12th layer lines. As for the cMyBP-C+/+ filaments, this suggests that periodic detail in the filament images extends to ∼4 nm. Also similar to the transforms from the wild-type filaments, meridional reflections corresponding to the average 14.3 nm axial rise between the cross-bridge crowns are typically present on the third and sixth layer lines. However, one difference from the wild-type filaments is the much stronger relative intensity of the 14.3 nm meridional reflection on the third layer line in the transforms from many of the cMyBP-C−/− filaments (Fig. 3, H–K).
The cMyBP-C−/− transforms also differ from those of the cMyBP-C+/+ filaments in that the “forbidden” meridional reflections on the first, second, fourth, fifth, and eighth layer lines are relatively weak or absent. This difference is particularly evident in the comparison of transforms obtained by averaging the transforms from 20 different cMyBP-C+/+ filaments (Fig. 4 A) with transforms from averaging 20 different cMyBP-C−/− myocardial thick filaments (Fig. 4 B). The averaged transform of the wild-type mouse cardiac filaments (Fig. 4 A) clearly shows the “forbidden” meridional reflections on the first, second, fourth, and fifth layer lines similar to those previously shown for isolated rabbit cardiac thick filaments (38,43). In contrast, the “forbidden” meridional reflections on the first, second, fourth, and fifth layer lines in the averaged transforms of the cMyBP-C−/− myocardial thick filaments (Fig. 4 B) are either very weak or absent. The one exception is the meridional reflection on the 11th layer line (Fig. 4 B inset) that appears to persist in the averaged transforms of the cMyBP-C−/− filaments. The averaged transforms (Fig. 4, A and B) also confirm the relatively stronger intensity of the 14.3 nm meridional reflection on the third layer line in the transforms of the cMyBP-C−/− filaments compared to the transforms of the cMyBP-C+/+ filaments. Transforms from the cMyBP-C−/− myocardial thick filaments thus appear more similar to transforms expected from an ideal helical arrangement of the cross-bridges. These results strongly suggest that although both the cMyBP-C+/+ and cMyBP-C−/− myocardial thick filaments have a 43 nm axial periodicity of the cross-bridge array, the structure is not identical for the two types of filaments.
Figure 4.

A and B represent averages of 20 Fourier transforms from the filaments of the wild-type heart (A) and the cMyBP-C knockout mouse heart (B) showing the strong set of layer lines (indicated by the numerals) corresponding to the 43 nm axial repeat. The boxed areas in both A and B were lightened to show the meridional region from the 7th to the 11th layer line. Although the “Forbidden” meridional reflections are clearly present on first, second, fourth, and fifth layer lines of the transforms from the wild-type mouse heart filaments (A), these reflections are weak or absent in the averaged transform from the filaments of the MyBP-C knockout mouse heart (B). A meridional reflection is present on the 11th layer line in both A and B and indicates that periodic information may extend to ∼4 nm for both the cMyBP-C+/+ and cMyBP-C−/− filaments.
Fourier filtered images of the filaments
Fourier filtered images of the cMyBP-C+/+ (Fig. 5, A–D) and cMyBP-C−/− myocardial thick filaments (Fig. 5, E–H) were computed by inverse Fourier transform of the data along the first six layer lines in the transforms from the filaments. The filtered images from both sets of filaments appear similar and consistent with the staining pattern which we previously shown to correspond to a three-stranded arrangement of the myosin heads (43). The most obvious difference that we observed is that the filtered images of the wild-type filaments typically show a distinct doublet of density at 43 nm axial spacing along one side of the filament (indicated by the brackets in Fig. 5, A–D), which often appears weaker or absent in the filtered images of the cMyBP-C−/− myocardial filaments (indicated by the asterisks in Fig. 5, E–H). In the cMyBP-C+/+ filaments this appears to arise from an additional density adjacent to one of the three cross-bridge crowns in the repeating 43 nm axial repeat of the filament. Plots of the axial density profile of the filtered images confirm the increase in density at this location (arrow in Fig. 5 I) in the cMyBP-C+/+ filaments compared to the cMyBP-C−/− filaments (arrow in Fig. 5 J). This doublet density appearance is also characteristic of the filtered images previously computed for the isolated rabbit cardiac filaments (38,43).
Figure 5.
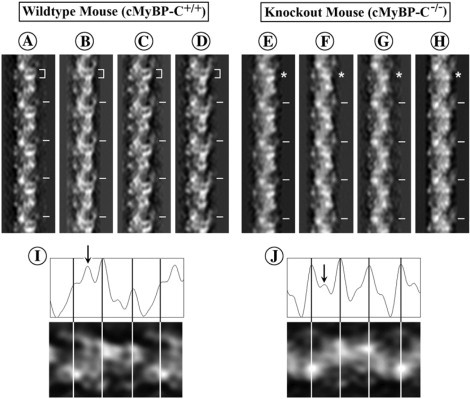
Filtered images obtained by inverse Fourier transform of the data along the first six layer lines of transforms from the filaments of the wild-type heart (A–D) and the cMyBP-C knockout mouse heart (E–H). Both sets of filtered images show the distinct “sawtooth” periodicity and the lack of bilateral symmetry at each cross-bridge crown typical of a three-stranded arrangement of the myosin heads as previously shown for other vertebrate striated muscle thick filaments. Although the filtered images are similar, the filtered images from the filaments of the wild-type hearts (A–D) tend to show an additional strong density between two of the three cross-bridge crowns in the 43 nm axial repeat that gives rise to a distinct density doublet (bracket) which is weaker (asterisks) in the filtered images of the filaments from the cMyBP-C knockout mouse hearts (E and F). The tick marks indicate the 43 nm axial repeat of the filaments. I and J show an axial plot of the density for the three cross-bridge levels in the 43 nm axial repeat in the filtered images of the cMyBP-C+/+ (I) and cMyBP-C−/− filaments (J), respectively. The lines indicate the positions of the cross-bridge crowns. The arrows point to the extra density between cross-bridge levels that is relatively much stronger in the cMyBP-C+/+ filaments than the cMyBP-C−/− filaments. The increased strength of this extra density together with the adjacent strong peak of density for the cross-bridge crown is responsible for the appearance of the doublet of density present in the cMyBP-C+/+ filaments.
Measurements of filament diameter
Since it has been reported that phosphorylation of cMyBP-C in rat cardiac thick filaments causes a change in diameter and an increase in periodicity of the filaments (21), it was important to determine whether the absence of cMyBP-C in the mouse cMyBP-C−/− myocardial thick filaments produced any change in diameter. Two alternative complementary approaches were used to assess differences in diameter between the cMyBP-C+/+ and the cMyBP-C−/− myocardial thick filaments. In both cases, measurements were limited to filaments which retained some degree of periodicity since the extent of projection into the stain of the myosin heads in the disordered filaments is not easily ascertained and therefore measurements are very subjective. For this reason, the comparison of diameters was limited to filaments which displayed at least the first three layer lines in transforms. Since, as shown in Figs. 1 and 2, many of the filaments retained this level of periodicity, this did not appear to impose a limitation on the number of filaments analyzed. In the first procedure, measurements of the maximum diameter of the filament at each 43 nm axial repeat in the C-zone (the region of most constant filament diameter) were made for filtered images of the filaments from the cMyBP-C+/+ and the cMyBP-C−/− myocardium. Because the actual magnification of an electron micrograph can vary up to 10% from that indicated by the instrument, the filament diameter measurements were calculated for each filament using the 43 nm helical periodicity of the filament as a ruler. The measured values of diameter using this procedure corresponded to the average maximum diameter of the filament.
As an alternative procedure, we also measured the positions (R) of the primary maxima on the first layer line in computed Fourier transforms of the filaments to calculate an estimate of the average radius (r) at which the center of mass of the myosin heads lies for each filament. According to helical diffraction theory (44), the radius (r) can be calculated using the equation 4.2 = 2πrR, where 4.2 is the expected value for a J3 Bessel function corresponding to the three-stranded structure of the filament. As described in Materials and Methods, although the values calculated for the average radius of the cross-bridge mass may not be strictly accurate in this case because the thick filament has a perturbed cross-bridge array and is not an ideal helix, the calculated values provide an additional parameter for comparison with the direct measurements of the diameter of the isolated thick filaments. As in the case of the direct measurements of the diameter of the filament, the axial helical periodicity of 43 nm was used in these measurements as an internal yardstick to ensure the accuracy of the calculated values.
Using these two approaches, we found that the relaxed periodic cMyBP-C+/+ mouse cardiac thick filaments have a mean diameter of 31.0 ± 1.7 nm (n = 123 filtered images) compared to a mean diameter of 31.1 ± 1.4 nm (n = 126 filtered images) for the cMyBP-C−/− myocardial thick filaments. The center of mass of the cross-bridges was calculated at a mean radius of 13.4 ± 1.4 nm (n = 159 transforms) for the cMyBP-C+/+ mouse filaments, compared to a mean radius of 13.5 ± 1.4 nm (n = 182 transforms) for the cMyBP-C−/− myocardial thick filaments. As shown in Fig. 6, A–D, the distributions for the measurements appear unimodal, consistent with a single population of filaments, among those filaments that retain an ordered arrangement of myosin heads. Both sets of measurements suggest that there is no significant difference in diameter between the relaxed ordered thick filaments of the cMyBP-C+/+ and the cMyBP-C−/− myocardia. Thus, no uniform change in the extension of the cross-bridges occurs in the absence of cMyBP-C for those filaments that retain the 43 nm axial periodicity of the cross-bridge array.
Figure 6.
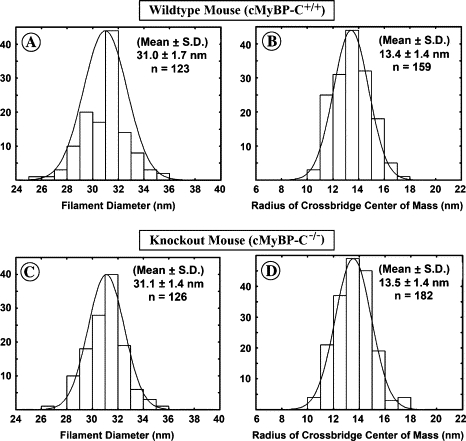
Histograms of the distribution of filament diameters measured from the filtered images from the filaments of the wild-type mouse heart (A) and the MyBP-C knockout mouse heart (C) and histograms of the distribution of the measured values of the average radius at which the cross-bridge mass is centered for the filaments from the wild-type mouse heart (B) and the cMyBP-C knockout mouse heart (D). The histograms demonstrate that the measured diameter of the filaments (A and C) and the mean radius at which the cross-bridge mass is centered (B and D) are very similar for the filaments from the wild-type mouse heart and the cMyBP-C knockout mouse hearts.
Discussion
The recently developed cMyBP-C knockout mouse (37), in combination with the use of electron microscopy and image analysis, provides a powerful tool for studying the functional and structural roles of cMyBP-C in the cardiac thick filament. Although chemical methods exist for the removal of MyBP-C from the thick filament, these generally result in incomplete loss of the MyBP-C and can remove other sarcomeric proteins as well (48). The incomplete removal of MyBP-C prohibits meaningful interpretation of structural changes in individual isolated filaments and complicates interpretation of functional changes. In the case of the cMyBP-C knockout mouse, both sodium dodecylsulfate-polyacrylamide gel electrophoresis and Western blot analyses for cMyBP-C (37,49) confirmed that cMyBP-C is not present in the hearts of the cMyBP-C knockout mice. Harris et al. (37) also demonstrated that skeletal muscle MyBP-C was not upregulated to compensate for the loss of the cMyBP-C. Consistent with the absence of cMyBP-C, physiological studies demonstrated contractile deficits and hypertrophy of the hearts in the mutant mice. Surprisingly, however, gross sarcomeric structure appeared normal in the hearts of the cMyBP-C knockout mice.
Although a number of earlier studies suggested that cMyBP-C is necessary for sarcomere assembly, myofibrillogenesis, and stability of the filament (13,22,35,50,51), in this study, we demonstrated that thick filaments can be isolated from the hearts of the cMyBP-C knockout mice. The presence of the thick filaments confirms the conclusion of Harris et al. (37) that cMyBP-C is not necessary for thick filament formation.
The cMyBP-C−/− myocardial thick filaments appear stable and of normal length. Many of the filaments retain an ordered periodic arrangement of the myosin heads under relaxing conditions. Measurements of filament diameters demonstrate that the filaments from cMyBP-C knockout mouse hearts have diameters similar to thick filaments from normal mouse hearts. This conclusion is further supported by the similarity of the calculations of the average radius at which the cross-bridge mass lies. The mean diameter of 31.0–31.1 nm and the mean radius of 13.4–13.5 nm at which the cross-bridge mass is centered are similar to the values previously reported for relaxed rabbit heart filaments (43). The similarity of the appearance, length, and diameter of the filaments from the wild-type and the cMyBP-C knockout mouse hearts thus suggests that a major macromolecular change in the structure of thick filaments does not occur in the absence of cMyBP-C. As a corollary, the results also suggest that if titin is acting as a molecular ruler for determining filament formation and control of filament length as has been suggested (52–57), binding to cMyBP-C is not required for this function.
Although the filaments from the cMyBP-C+/+ and cMyBP-C−/− myocardium appear similar, several observations suggest that subtle differences exist. The clearest difference is seen in the weakness or absence of the “forbidden” meridional reflections in the Fourier transforms of the cMyBP-C−/− myocardial thick filaments compared to those of the cMyBP-C+/+ filaments. This is particularly evident in averaged transforms from the filaments but is also seen in the transforms from individual filaments. Huxley and Brown (47) first described these reflections, which are not expected from helical symmetry, in the x-ray diffraction patterns of vertebrate skeletal muscle. They proposed that the “forbidden” meridional reflections arise either from a perturbation from ideal helical symmetry in the cross-bridge array or from the presence of accessory proteins associated with the filament backbone. Three-dimensional (3D) reconstructions of both frog and fish skeletal muscle thick filaments (58,59) demonstrated a perturbation from ideal helical symmetry in cross-bridge arrangement. This perturbation results in the cross-bridge arrangement having cylindrical symmetry rather than strict helical symmetry. In the case of strict helical symmetry, the axial translation of 14.3 nm between cross-bridge crowns would be strictly paired with a 40° rotation of each crown relative to the adjacent cross-bridge crown. By contrast, in 3D reconstructions of the fish skeletal muscle thick filament rotational angles between the three cross-bridge crowns in the 43 nm axial repeat are 0°, 60°, and 60° (59). Furthermore, with an ideal three-stranded helix with an axial repeat every third cross-bridge crown, meridional reflections would be expected only on layer lines which are multiples of three, such as the third, sixth and ninth layer lines. With cylindrical symmetry, in contrast, a meridional reflection (described by a J0 Bessel function) is expected on each layer line, including layer lines which are not multiples of three (discussed in Stewart and Kensler (58)).
One possible explanation for the observed weakness of the “forbidden” meridional reflections in the transforms of the cMyBP-C−/− myocardial thick filaments is that in the absence of cMyBP-C the perturbation is not present and the cross-bridges are helically arranged. It has previously been suggested that MyBP-C might be responsible for the perturbation from ideal helical symmetry observed in the cross-bridge arrangement (58,59) and that this might place the myosin heads in a more favorable position to interact with the six surrounding actins (59). If the “forbidden” meridional reflections arise from the perturbation in the cross-bridge array, as the cross-bridge arrangement becomes more helical these reflections will be weakened or absent.
A second possibility is that the periodic density of cMyBP-C along the filament backbone at 43 nm axial spacings directly contributes to the intensity of the “forbidden” meridional reflections. Although Hudson et al. (60) suggested that the mass of MyBP-C may be too small to contribute strongly to the myosin layer line pattern, a strong band of density on the filament backbone that could correspond to MyBP-C was observed in the 3D reconstruction of the frog thick filament (58). This banding of the filament backbone at 43 nm axial spacings could contribute to the intensity of the “forbidden” meridional reflections. If cMyBP-C contributes to a similar banding of the backbone of the cardiac thick filament at axial spacings of 43 nm, the “forbidden” reflections may be weakened in the absence of cMyBP-C.
With regard to the contribution of other accessory proteins to the “forbidden” meridional reflections, the presence on the 11th layer line of a meridional reflection that persists in the averaged transforms of both the cMyBP-C+/+ and the cMyBP-C−/− filaments is of interest. In the transforms of rabbit cardiac thick filaments, we observed a similar reflection on the 11th layer line and presented evidence that it may correspond to a series of 11 stripes or densities every 43 nm on the backbone of the filament (61). Since titin is expected to have 11 domains per 43 nm (57), it is tempting to suggest that the stripes correspond to the domains of titin. However, the possibility that this striping arises from the packing and staining of charged regions on the myosin rod could not be excluded. It is clear that this reflection does not arise solely from the perturbation in the cross-bridge arrangement. This supports the idea that other proteins or features of the filament backbone such as cMyBP-C might also contribute to the intensity of the “forbidden” meridional reflections.
A third possible explanation is that cMyBP-C normally contributes to both a perturbation from helical symmetry in the cross-bridge array and a banding of the filament backbone density. The weakening of the “forbidden” meridional reflections in the cMyBP-C−/− myocardium may therefore reflect both contributions.
It was hoped that comparing the filtered images of the thick filaments from the cMyBP-C+/+ and the cMyBP-C−/− myocardium would explain how the absence of cMyBP-C contributes to the changes in intensity of the “forbidden” meridional reflections in the transforms, but the differences in the filtered images are quite subtle. The main difference noted is that a doublet of density present at one of the three cross-bridge crowns in the filtrations of the cMyBP-C+/+ filaments is weaker or absent in many of the filtrations of the cMyBP-C−/− myocardial filaments. It is interesting that the difference in the filtered images occurs at the cross-bridge crown (crown 1) that Al-Khayat et al. (59) have shown to have additional mass in the 3D reconstruction of the isolated fish skeletal muscle thick filament. They suggested that MyBP-C lying near this cross-bridge crown might be responsible for the strong azimuthial perturbation from helical symmetry observed in the cross-bridge array in the 3D reconstruction of the fish thick filament. Although the change in the filtered images of the mouse cardiac filaments is suggestive, it is not possible from two-dimensional filtered images alone to ascertain whether a change has occurred in the azimuthial perturbation of the cross-bridge array or whether this is the location of cMyBP-C.
The other major difference observed between the cMyBP-C+/+ and the cMyBP-C−/− myocardial filaments was that in filaments lacking cMyBP-C, the helical or quasihelical arrangement of the cross-bridges was more easily disrupted. Although most of the filaments from the cMyBP-C−/− myocardium displayed a 43 nm axial periodicity, disruption of the ordered cross-bridge arrangement was evident both as an increased number of filaments with disrupted cross-bridge order and as an increased number of filaments in which both regions of order and disorder were observed along the same filament arm (Fig. 2 A).
Several studies suggest that cMyBP-C may tether the myosin heads near the backbone (31,48), acting as a restraint on actomyosin interactions. In the absence of cMyBP-C or upon phosphorylation of cMyBP-C (21), this tether or restraint may be released, allowing extension of the cross-bridges from the backbone. Consistent with this idea, Levine et al. (21) reported that phosphorylation of cMyBP-C in isolated rat myocardial thick filaments causes an extension of the myosin heads from the backbone of the filament. This was reported to result in a population of highly ordered thick filaments with a mean diameter of ∼35 nm, compared to the diameter of 30 nm for filaments, which were only partly phosphorylated. Since phosphorylation of cMyBP-C is thought to release the tethering of the myosin head, the absence of cMyBP-C in the cMyBP-C−/− myocardial thick filament might induce analogous effects.
The decreased stability of the helical or quasihelical arrangement of relaxed cross-bridges observed in filaments from cMyBP-C−/− hearts reported here is consistent with the release of a tether, resulting in greater mobility of cross-bridge heads in the absence of cMyBP-C. However, in contrast to observations of phosphorylated rat thick filaments (21), measurements of thick filament diameters from cMyBP-C−/− mice did not show evidence of a population of highly ordered filaments with an increased diameter. Histograms of the distributions of diameter of the cMyBP-C−/− myocardial filaments appeared unimodal, with a mean diameter of ∼31.1 nm compared to a similar distribution with a mean diameter of 31.0 nm for the cMyBP-C+/+ filaments. This diameter is similar to that reported for rabbit cardiac thick filaments (38). Also the cMyBP-C−/− filaments showed increased disorder of the myosin heads, rather than increased order and decreased flexibility of the heads as reported for the isolated rat cardiac thick filaments (21). In the case of the isolated rat myocardial thick filaments, it was suggested (21) that phosphorylation of cMyBP-C resulted in a loosening of a collar formed by cMyBP-C around the filament and that consequent loosening of the packing of myosin in the backbone correlated with an extension of the cross-bridges. By contrast, we saw no evidence of a loosening of the myosin packing in the filament backbone of isolated cMyBP-C−/− thick filaments. Taken together, these observations suggest that phosphorylation of cMyBP-C may not be the structural equivalent to its removal. In this regard there is evidence that cMyBP-C binds actin (62,63) and affects actomyosin interactions through mechanisms not involving myosin S2 (64). Additional studies are therefore necessary to fully describe the molecular mechanisms by which cMyBP-C affects cross-bridge interactions.
It may seem paradoxical that we observed that cross-bridge order appeared more labile in the absence of cMyBP-C but that significant numbers of the filaments still retained relaxed cross-bridge order. This apparent inconsistency can be resolved if cMyBP-C increases the stability of the relaxed myosin head arrangement but is not an absolute requirement for maintenance of this arrangement. Other factors such as electrostatic binding of the myosin heads to the backbone or a nucleotide-dependent decrease in flexibility of the cross-bridge (65) may be sufficient to maintain the periodicity of the cross-bridge arrangement in the absence of cMyBP-C. This may be particularly true, as in our studies, for isolated thick filaments which are no longer surrounded by the lattice of actin filaments. In intact muscle, in contrast, the formation of weak-binding attachments to adjacent actin thin filaments may represent a significant disordering factor not present in the isolated filaments.
Both the observed increased tendency for disorder in the relaxed cross-bridge arrangement and the changes in the Fourier transforms of the cMyBP-C−/− myocardial filaments are significant. Although the similarity of the cMyBP-C+/+ and cMyBP-C−/− myocardial filaments suggests that gross changes in the macromolecular packing of the myosin in the filament do not occur in the absence of cMyBP-C, these differences between the filaments indicate that smaller changes in structure may correlate with changes in the physiological properties of the muscle after ablation or mutation of cMyBP-C (30,37,66). To fully understand these changes in the properties of the muscle that occur upon phosphorylation or ablation of cMyBP-C, it is important to elucidate the precise relationship between cMyBP-C and the myosin heads in the thick filament. The 43 nm axial periodicity shown here to be present in many of the cMyBP-C−/− myocardial thick filaments is significant since it means that calculations of 3D reconstructions of these filaments are possible. Perez-Zoghbi et al. (67,68) have recently reported the calculation of preliminary 3D reconstructions of the cMyBP-C+/+ and cMyBP-C−/− mouse cardiac thick filament. The cMyBP-C+/+ reconstruction shows densities that may correspond to cMyBP-C and titin. Comparisons of 3D reconstructions of the cMyBP-C−/− myocardial filaments with this reconstruction should provide answers to many unresolved questions about the functional and structural roles of cMyBP-C.
Acknowledgments
This work was supported by a National Institutes of Health (NIH) MBRS grant S06 GM08224 and in part by funding from NIH Institutional Research Centers in Minority Institutes grant G12RR-03051 to R.W.K. and NIH HL080367 to S.P.H.
Footnotes
Editor: Leepo C. Yu.
References
- 1.Marian A.J., Roberts R. The molecular genetic basis for hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 2001;33:655–670. doi: 10.1006/jmcc.2001.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flashman E., Redwood C., Moolman-Smook J., Watkins H. Cardiac myosin binding protein C: its role in physiology and disease. Circ. Res. 2004;94:1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 3.Winegrad S. Cardiac myosin binding protein C. Circ. Res. 1999;84:1117–1126. doi: 10.1161/01.res.84.10.1117. [DOI] [PubMed] [Google Scholar]
- 4.Starr R., Offer E. Polypeptide chains of intermediate molecular weight in myosin preparations. FEBS Lett. 1971;15:40–44. doi: 10.1016/0014-5793(71)80075-3. [DOI] [PubMed] [Google Scholar]
- 5.Craig R., Offer G. The location of C-protein in rabbit skeletal muscle. Proc. R. Soc. Lond. B. Biol. Sci. 1976;192:451–461. doi: 10.1098/rspb.1976.0023. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto K., Moos C. A comparative study of C-proteins from heart and skeletal muscles. Biophys. J. 1981;33:A237. (Abstr.) [Google Scholar]
- 7.Starr R., Offer G. H-protein and X-protein. Two new components of the thick filaments of vertebrate skeletal muscle. J. Mol. Biol. 1983;170:675–698. doi: 10.1016/s0022-2836(83)80127-2. [DOI] [PubMed] [Google Scholar]
- 8.Bennett P., Starr R., Elliot A., Offer G. The structure of C-protein and X-protein molecules and a polymer of X-protein. J. Mol. Biol. 1985;184:297–309. doi: 10.1016/0022-2836(85)90381-x. [DOI] [PubMed] [Google Scholar]
- 9.Bennett P., Craig R., Starr R., Offer G. The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J. Muscle Res. Cell Motil. 1985;7:550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 10.Pepe F.A., Drucker B. The myosin filament. III. C-protein. J. Mol. Biol. 1975;99:609–617. doi: 10.1016/s0022-2836(75)80175-6. [DOI] [PubMed] [Google Scholar]
- 11.Rome E., Offer G., Pepe F.A. X-ray diffraction of muscle labelled with antibody to C-protein. Nat. New Biol. 1973;244:152–154. doi: 10.1038/newbio244152a0. [DOI] [PubMed] [Google Scholar]
- 12.Reference deleted in proof.
- 13.Seiler S.H., Fischman D.A., Leinwand L.A. Modulation of myosin filament organization by C-protein family members. Mol. Biol. Cell. 1996;7:113–127. doi: 10.1091/mbc.7.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeacocke S., England P. Phosphorylation of a myofibrillar protein of Mr 150,000 in perfused rat heart, and the tentative identification of this as C-protein. FEBS Lett. 1980;122:129–132. doi: 10.1016/0014-5793(80)80418-2. [DOI] [PubMed] [Google Scholar]
- 15.Hartzell H.C., Titus L. Effects of cholinergic and adrenergic agonists on phosphorylation of a 165,000-dalton myofibrillar protein in intact cardiac muscle. J. Biol. Chem. 1982;257:2111–2120. [PubMed] [Google Scholar]
- 16.Garvey J.L., Kranias E.G., Solaro R.J. Phosphorylation of C-protein, troponin I, and phospholamban in isolated rabbit hearts. Biochem. J. 1988;249:709–714. doi: 10.1042/bj2490709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlender K., Bean L.J. Phosphorylation of chick cardiac C-protein by calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 1990;266:2811–2817. [PubMed] [Google Scholar]
- 18.Weisberg A., Winegrad S. Alteration of myosin cross-bridges by phosphorylation of myosin-binding protein C in cardiac muscle. Proc. Natl. Acad. Sci. USA. 1996;93:8999–9003. doi: 10.1073/pnas.93.17.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisberg A., Winegrad S. Relation between crossbridge structure and actomyosin ATPase activity in rat heart. Circ. Res. 1998;83:60–72. doi: 10.1161/01.res.83.1.60. [DOI] [PubMed] [Google Scholar]
- 20.Kunst G., Kress M., Gruen M., Utenweiler D., Gautel M., Fink R.H.A. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ. Res. 2000;86:51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 21.Levine R., Weisberg A., Kulikovskaya I., McClellan G., Winegrad S. Multiple structures of thick filaments in resting cardiac muscle and their influence on cross-bridge interactions. Biophys. J. 2001;81:1070–1082. doi: 10.1016/S0006-3495(01)75764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautel M., Furst D.O., Cocco A., Schiaffino S. Isoform transitions of the myosin binding protein C family in developing human and mouse muscles: lack of isoform transcomplementation in cardiac muscle. Circ. Res. 1998;82:124–129. doi: 10.1161/01.res.82.1.124. [DOI] [PubMed] [Google Scholar]
- 23.Kurasawa M., Sato N., Matsuda A., Koshida S., Totsuka T., Obinata T. Differential expression of C-protein isoforms in developing and degenerating mouse striated muscles. Muscle Nerve. 1999;22:196–207. doi: 10.1002/(sici)1097-4598(199902)22:2<196::aid-mus7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Gautel M., Zuffardi O., Freiburg A., Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 1995;14:1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartzell H.C. Phosphorylation of C-protein in intact amphibian cardiac muscle. Correlation between 32P incorporation and twitch relaxation. J. Gen. Physiol. 1984;83:563–588. doi: 10.1085/jgp.83.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlender K.K., Hegazy M.G., Thysseril T.J. Dephosphorylation of cardiac myofibril C-protein by protein phosphatase 1 and protein phosphatase 2A. Biochim. Biophys. Acta. 1987;928:312–319. doi: 10.1016/0167-4889(87)90191-1. [DOI] [PubMed] [Google Scholar]
- 27.Gruen M., Gautel M. Mutations in β-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J. Mol. Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 28.McClellan G., Kulikovskaya I., Winegrad S. Changes in cardiac contractility related to calcium-mediated changes in phosphorylation of myosin-binding protein C. Biophys. J. 2001;81:1083–1092. doi: 10.1016/S0006-3495(01)75765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korte F.S., McDonald K.S., Harris S.P., Moss R.L. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ. Res. 2003;93:752–758. doi: 10.1161/01.RES.0000096363.85588.9A. [DOI] [PubMed] [Google Scholar]
- 30.Stelzer J.E., Fitzsimons D.P., Moss R.L. Ablation of myosin-binding protein-C accelerates force development in mouse myocardium. Biophys. J. 2006;90:4119–4127. doi: 10.1529/biophysj.105.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calaghan S.C., Trinick J., Knight P.J., White E. A role for C-protein in the regulation of contraction and intracellular Ca2+ in intact rat ventricular myocytes. J. Physiol. 2000;528:151–156. doi: 10.1111/j.1469-7793.2000.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris S.P., Rostkova E., Gautel M., Moss R.L. Binding of myosin binding protein-C to myosin subfragment S2 affects contractility independent of a tether mechanism. Circ. Res. 2004;95:930–936. doi: 10.1161/01.RES.0000147312.02673.56. [DOI] [PubMed] [Google Scholar]
- 33.Moos C., Offer G., Starr R., Bennett P. Interaction of C-protein with myosin, myosin rod and light meromyosin. J. Mol. Biol. 1975;97:1–9. doi: 10.1016/s0022-2836(75)80017-9. [DOI] [PubMed] [Google Scholar]
- 34.Koretz J.F. Effects of C-protein on synthetic myosin filament structure. Biophys. J. 1979;27:433–446. doi: 10.1016/S0006-3495(79)85227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis J.S. Interaction of C-protein with pH 8.0 synthetic thick filaments prepared from the myosin of vertebrate skeletal muscle. J. Muscle Res. Cell Motil. 1988;9:174–183. doi: 10.1007/BF01773739. [DOI] [PubMed] [Google Scholar]
- 36.Sebillon P., Bonne G., Flavigny J., Venin S., Rouche A., Fiszman M., Vikstrom K., Leinwand L., Carrier L., Schwartz K. COOH-terminal truncated human cardiac MyBP-C alters myosin filament organization. C. R. Acad. Sci. III. 2001;324:251–260. doi: 10.1016/s0764-4469(00)01292-0. [DOI] [PubMed] [Google Scholar]
- 37.Harris S.P., Bartley C.R., Hacker T.A., McDonald K.S., Douglas P.S., Greaser M.L., Powers P.A., Moss R.L. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ. Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 38.Kensler R.W. Mammalian cardiac muscle thick filaments: their periodicity and interactions with actin. Biophys. J. 2002;82:1497–1508. doi: 10.1016/S0006-3495(02)75503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine R., Kensler R., Yang Z., Stull J. Myosin light chain phosphorylation affects the structure of rabbit skeletal muscle thick filaments. Biophys. J. 1996;71:898–907. doi: 10.1016/S0006-3495(96)79293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magid A., Ting-Beal H., Carwell M., Kontis T., Lucaveche C. Connecting filaments, core filaments, and side struts: a proposal to add three new load-bearing structures to the sliding filament model. Adv. Exp. Med. Biol. 1984;170:307–328. doi: 10.1007/978-1-4684-4703-3_26. [DOI] [PubMed] [Google Scholar]
- 41.Sellers J. Phosphorylation-dependent regulation of Limulus muscle. J. Biol. Chem. 1981;256:9274–9278. [PubMed] [Google Scholar]
- 42.Kensler R.W., Levine R.J.C., Stewart M. Electron microscope and optical diffraction analysis of the structure of scorpion muscle thick filaments. J. Cell Biol. 1985;101:395–401. doi: 10.1083/jcb.101.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kensler R.W. The mammalian cardiac muscle thick filament: crossbridge arrangement. J. Struct. Biol. 2005;149:303–312. doi: 10.1016/j.jsb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Klug A., Crick F.H.C., Wykoff W.W. Diffraction by helical structures. Acta Crystallogr. 1958;11:199–213. [Google Scholar]
- 45.Moody M.F. Structure of the sheath of bacteriophage T4. I. Structure of the contracted sheath and polysheath. J. Mol. Biol. 1967;25:167–200. doi: 10.1016/0022-2836(67)90136-2. [DOI] [PubMed] [Google Scholar]
- 46.Trinick J. End-filaments, a new structural element of vertebrate skeletal muscle thick filaments. J. Mol. Biol. 1981;151:309–314. doi: 10.1016/0022-2836(81)90517-9. [DOI] [PubMed] [Google Scholar]
- 47.Huxley H.E., Brown W. The low angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and vigor. J. Mol. Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 48.Hofmann P.A., Hartzell H.C., Moss R.L. Alterations in Ca2+ sensitive tension due to partial extraction of C-protein from rat skinned cardiac myocytes and rabbit skeletal muscle fibers. J. Gen. Physiol. 1991;97:1141–1163. doi: 10.1085/jgp.97.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colson B.A., Bekyarova T., Fitzsimons D.P., Irving T.C., Moss R.L. Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. J. Mol. Biol. 2007;367:36–41. doi: 10.1016/j.jmb.2006.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultheiss T., Lin Z.X., Lu M.H., Murray J., Fischman D.A., Weber K., Masaki T., Imamura M., Holtzer H. Differential distribution of subsets of myofibrillar proteins in cardiac nonstriated and striated myofibrils. J. Cell Biol. 1990;110:1159–1172. doi: 10.1083/jcb.110.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClellan G., Kulikovskaya I., Flavigny J., Carrier L., Winegrad S. Effect of cardiac myosin-binding protein C on stability of the thick filament. J. Mol. Cell. Cardiol. 2004;37:823–835. doi: 10.1016/j.yjmcc.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 52.Furst D.O., Vinkemeyer U., Weber K. Mammalian skeletal muscle C-protein: purification from bovine muscle, binding to titin and the characterization of a full length cDNA. J. Cell Sci. 1992;102:769–778. doi: 10.1242/jcs.102.4.769. [DOI] [PubMed] [Google Scholar]
- 53.Labeit S., Gautel M., Lakey A., Trinick J. Towards a molecular understanding of titin. EMBO J. 1992;11:1711–1716. doi: 10.1002/j.1460-2075.1992.tb05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koretz J.F., Irving T.C., Wang K. Filamentous aggregates of native titin and binding of C-protein and AMP-deaminase. Arch. Biochem. Biophys. 1993;304:305–309. doi: 10.1006/abbi.1993.1354. [DOI] [PubMed] [Google Scholar]
- 55.Soteriou A., Gamage M., Trinick J. A survey of interactions made by the giant protein titin. J. Cell Sci. 1993;104:119–123. doi: 10.1242/jcs.104.1.119. [DOI] [PubMed] [Google Scholar]
- 56.Freiburg A., Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur. J. Biochem. 1996;235:317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 57.Labeit S., Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- 58.Stewart M., Kensler R.W. Arrangement of heads in relaxed thick filaments from frog skeletal muscle. J. Mol. Biol. 1986;192:831–851. doi: 10.1016/0022-2836(86)90032-x. [DOI] [PubMed] [Google Scholar]
- 59.Al-Khayat H.A., Morris E.P., Kensler R.W., Squire J.M. 3D structure of relaxed fish muscle myosin filaments by single particle analysis. J. Struct. Biol. 2006;155:202–217. doi: 10.1016/j.jsb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 60.Hudson L., Harford J., Denny R., Squire J. Myosin head configuration in relaxed fish muscle: resting state myosin heads must swing axially by up to 150 Å or turn upside down to reach rigor. J. Mol. Biol. 1997;273:440–455. doi: 10.1006/jmbi.1997.1321. [DOI] [PubMed] [Google Scholar]
- 61.Kensler R.W. The mammalian cardiac muscle thick filament: backbone contributions to meridional reflections. J. Struct. Biol. 2005;149:313–324. doi: 10.1016/j.jsb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 62.Razumova M.V., Shaffer J.F., Tu A.Y., Flint G.V., Regnier M., Harris S.P. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: evidence for long-lived crossbridges. J. Biol. Chem. 2006;281:35846–35854. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 63.Kulikovskaya I., McClellan G., Flavigny J., Carrier L., Winegrad S. Effect of MyBP-C binding to actin on contractility in heart muscle. J. Gen. Physiol. 2003;122:761–774. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaffer J.F., Razumova M.V., Tu A.Y., Regnier M., Harris S.P. Myosin S2 is not required for effects of myosin binding protein-C on motility. FEBS Lett. 2007;581:1501–1504. doi: 10.1016/j.febslet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Xu S., Gu J., Rodes T., Belknap B., Rosenbaum G., Offer G., White H., Yu L.C. The M.ADP.P(i) state is required for helical order in the thick filaments of skeletal muscle. Biophys. J. 1999;77:2665–2676. doi: 10.1016/s0006-3495(99)77101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palmer B.M., McConnell B.K., Li G.H., Seidman C.E., Seidman J.G., Irving T.C., Alpert N.R., Maughan D.W. Reduced cross-bridge dependent stiffness of skinned myocardium from mice lacking cardiac myosin binding protein-C. Mol. Cell. Biochem. 2004;263:73–80. doi: 10.1023/B:MCBI.0000041849.60591.45. [DOI] [PubMed] [Google Scholar]
- 67.Perez-Zoghbi M.E., Woodhead J.L., Craig R. Structure of vertebrate cardiac myosin thick filaments by single particle analysis. Biophys. J. 2007;92:373a. [Google Scholar]
- 68.Perez-Zoghbi, M., J. L. Woodhead, R. L. Moss, and R. Craig. 2008. Three-Dimensional structure of vertebrate cardiac muscle myosin filaments. PNAS. In Press. [DOI] [PMC free article] [PubMed]


