Abstract
Loss of kidney graft function with tubular atrophy (TA) and interstitial fibrosis (IF) causes most kidney allograft losses. We aimed to identify the molecular pathways involved in IF/TA progression. Kidney biopsies from normal kidneys (n = 24), normal allografts (n = 6), and allografts with IF/TA (n = 17) were analyzed using high-density oligonucleotide microarray. Probe set level tests of hypotheses tests were conducted to identify genes with a significant trend in gene expression across the three groups using Jonckheere-Terpstra test for trend. Interaction networks and functional analysis were used. An unsupervised hierarchical clustering analysis showed that all the IF/TA samples were associated with high correlation. Gene ontology classified the differentially expressed genes as related to immune response, inflammation, and matrix deposition. Chemokines (CX), CX receptor (for example, CCL5 and CXCR4), interleukin, and interleukin receptor (for example, IL-8 and IL10RA) genes were overexpressed in IF/TA samples compared with normal allografts and normal kidneys. Genes involved in apoptosis (for example, CASP4 and CASP5) were importantly overexpressed in IF/TA. Genes related to angiogenesis (for example, ANGPTL3, ANGPT2, and VEGF) were downregulated in IF/TA. Genes related to matrix production-deposition were upregulated in IF/TA. A distinctive gene expression pattern was observed in IF/TA samples compared with normal allografts and normal kidneys. We were able to establish a trend in gene expression for genes involved in different pathways among the studied groups. The top-scored networks were related to immune response, inflammation, and cell-to-cell interaction, showing the importance of chronic inflammation in progressive graft deterioration.
INTRODUCTION
Despite progress in decreasing acute rejection and improving short-term survival (1–4), much of it through improved immunosuppression and anti-infective agents, late kidney allograft loss remains a problem. Progressive graft function deterioration causes most kidney allograft losses and remains the central clinical challenge for improving long-term graft survival rate. With close to 5000 kidney transplants failing per year and returning the recipient to dialysis in the U.S., kidney transplant failure is a leading cause of end-stage renal disease (4).
Interstitial fibrosis and tubular atrophy (IF/TA) in renal allografts are the major hallmarks of progressive graft dysfunction, with the result of a continuous decrease in graft survival after one year posttransplant (3). The mechanisms that lead to IF/TA likely are of multifactorial nature, including immunological and hypertensive injury and immunosuppressant-related toxicity (3,4).
The complex, multistage processes that result in chronic IF/TA are poorly understood. Whereas the determination of organ failure often relies on measurable physiological parameters, the early stages of chronic IF/TA are difficult to diagnose. Detection requires invasive procedures to obtain graft biopsies for histological evaluation. Moreover, current methods for diagnosing allograft dysfunction are inadequate for detecting the significant organ damage that occurs before the establishment of clinical manifestations. The development of assays or novel technologies that will able to detect allograft dysfunction/rejection and predict long-term outcomes is vital for the long-term success of transplantation (1–6).
The original nephron mass present in the donor kidney at transplantation depends on donor quality (e.g., age, hypertension). After kidney transplantation and as a consequence of the surgery stress, preservation methods, ischemia, acute rejection, and other stressors, the number of the initial nephrons in the donor kidney is reduced. Ordinarily, after six months posttransplantation, the number of nephrons is stable, resulting in an allograft with normal function (4). For an important number of grafts, however, the loss of nephrons continues, reflecting continued graft injury. This condition ends in fibrosis and atrophy, with the final event loss of graft function (4).
Although different genes have been associated with the biology of IF/TA (7–9), it is essential to advance the understanding of the immune response and the responses of the transplanted organ to both immune and nonimmune injury mechanisms. Microarray is a powerful technology that detects thousands of genes simultaneously and might be an important tool in elucidating patterns for mechanism, diagnosis, prognosis, and treatment of complex, multifactorial diseases such as IF/TA (10).
In this study, we aimed to characterize the genes involved in the progression to IF/TA. We evaluated gene expression profiles in normal kidneys, normal allografts, and kidney tissues with diagnosis of IF/TA.
MATERIALS AND METHODS
Kidney Samples and Patients
The study included 23 kidney transplant recipients. Written informed consent was obtained from all the patients. The Western Institutional Review Board (WIRB) approved the study protocol. Renal allograft tissue was obtained using an 18-gauge biopsy needle. All the biopsies were histologically evaluated. The biopsies were instantly submerged in RNAlater (Ambion, Austin, TX, USA) and homogenized. All biopsies were evaluated according to the Banff criteria (11). Twenty-four kidney biopsies with normal histopathology were also included in the study.
RNA Isolation from Kidney Tissues, cDNA Synthesis, and In Vitro Transcription for Labeled cRNA Probe
With minor modifications, the sample preparation protocol follows the Affymetrix GeneChip Expression Analysis manual (Santa Clara, CA, USA). Total RNA was extracted from kidney tissue samples using TRIzol (Life Technologies, Rockville, MD, USA). After extraction, integrity of RNA was checked using Agilent 2100 Bioanalyzer. The purity of the RNA preparations was tested by the 260/280 nm ratio (range 1.8–2.0). Briefly, total RNA was reverse-transcribed using T7-polydT primer, and converted into double-stranded cDNA using Superscript Choice System (One-Cycle Target Labeling and Control Reagents, Affymetrix), with templates used for an in vitro transcription reaction at 37°C for 8 h to yield biotin-labeled antisense cRNA. The labeled cRNA was chemically fragmented and made into a hybridization cocktail according to the Affymetrix GeneChip protocol, and was then hybridized to U133A 2.0 GeneChip probe arrays (Affymetrix). The array image was generated by the high-resolution GeneChip Scanner 3000 (Affymetrix). All CEL files were processed using Affymetrix GeneChip Operating software, version 5.0, and appropriate Bio-conductor packages (12) available in the R programming environment (13).
Quality Control
RNA integrity from kidney tissues was checked using the Agilent 2100 Bioanalyzer (Agilent Technologies). To be considered for microarray analysis, the RNA samples needed to pass quality control criteria, 28S/18S ratios >1.5 and A260 nm/A280 nm >2.0. We established a cutoff value of 30% rRNA contribution to the total area under the electropherogram for RNA samples to be considered as intact or undegraded (14). Products of cDNA synthesis and IVT were tested using the Agilent 2100 Bionalyzer (cDNA synthesis 1.5 kb < cDNA < 5.0 kb; IVT 1.0 kb < cRNA < 4.5 kb) (14).
Data Analysis
After hybridization of the cRNA samples to Affymetrix HG-U133A 2.0 Array, probe set expression summaries were estimated using the robust multiarray average method (15). Quality assessment of each GeneChip was performed by assessing the average background, scaling factor, percent present calls, the 3′:5′ ratio for GAPDH, β-actin and ISGF, and the overall 3′:5′ ratio with associated 95% confidence interval (16) using the publicly available application (17). Classic multidimensional scaling (MDS) was applied to the standardized quality control (QC) variables and the resulting two-dimensional MDS plot to determine whether any chips had outlying quality. Before conducting additional statistical analyses, all control probe sets were removed, leaving 22,215 probe sets for statistical analysis.
Probe set level tests of hypothesis tests were conducted to identify genes with a significant trend (increase or decrease) in gene expression across the three groups using Jonckheere-Terpstra test for trend. That is, we were interested in identifying genes with expression levels increasing normal donor > normal allograft > IF/TA, or decreasing normal donor < normal allograft < IF/TA across the three groups.
Interaction Networks and Functional Analysis
Gene ontology and gene interaction analyses were executed using Ingenuity Pathways Analysis tools 3.0 (http://www.ingenuity.com). The gene lists containing Entrez GeneIDs as clone identifiers, as well as fold-change values from corresponding supervised analyses, were mapped to their corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These so-called focus genes were then used in the network generation algorithm, based on the curated list of molecular interactions in IPKB. Significance for the enrichment of the genes in a network with particular biologic functions was determined by the right-tailed Fisher exact test, using a list of all the genes on the array as a reference set.
Validation of Microarray Results
Real-time PCR (QPCR) reactions were used for quantifying the expression of transforming growth factor (TGF)-β, thrombospondin-1 (TSP-1), and epidermal growth factor receptor (EGFR) mRNA in the stock RNA samples that were subjected to microarray study. Each assay consisted of two unlabeled PCR primers and a FAM dye-labeled TaqMan MGB probe. The endogenous control, β-2-microglobulin (B2M), was detected with a VIC dye-labeled TaqMan MGB probe (human B2M endogenous control, VIC/TAMRA Probe; Primer Limited, Applied Biosystems). Total RNA from each sample was subjected to reverse transcription using TaqMan Reverse Transcription Reagents (Applied Biosystems) according to the manufacturer’s protocol. Real-time PCR reactions were carried out in a 25-μL reaction mixture using an ABI Prism 7700 sequence detection system (Applied Biosystems). All amplifications were carried out in duplicate, and threshold cycle (Ct) scores were averaged for calculations of relative expression values. The Ct scores for genes of interest were normalized against Ct scores for the corresponding B2M control. Relative expression was determined by the following calculation where the amount of target is normalized to an endogenous reference (B2M RNA) and relative to an arbitrary calibrator (the reference class of patients used in the comparison) (18): relative expression = 2−ΔΔCt, where ΔΔCt = (ΔCt of experimental group) − (ΔCt of calibrator group).
RESULTS
Patients
Seventeen kidney transplant patients with biopsy-proven diagnosis of IF/TA and six transplant kidneys with normal histopathology were included in the study. Among the patients with IF/TA were eight women and nine men, ranging in age from 29 to 60 years (mean 46.0 ± 9.3), all deceased donor recipients. Among the kidney transplant patients with normal allograft function (all deceased donor recipients), biopsies were performed at 12 ± 3 months posttransplantation. All the patients had creatinine <2 mg/dL and proteinuria <500 mg/24 h. Immunosuppression consisted of either a cyclosporine-based (seven patients) or tacrolimus-based (16 patients) regimen. For patients with IF/TA, biopsies were performed between 15 and 58 months after kidney transplantation (mean 36 ± 12 months). Patient serum creatinine levels ranged from 2.4 to 4.0 mg/dL (mean 2.9 ± 0.5 mg/dL), with proteinuria >500 mg/24 h in all the patients (Table 1). Biopsies were graded 2 or 3 CAN per Banff classification (11) (Table 2).
Table 1.
Characteristics of the studied patients.
| Patient | Sex | Age, years | Transplant type | Creatinine, mg/dLa | Proteinuria, g/24 ha | ACR episodes |
|---|---|---|---|---|---|---|
| 1-IF/TA | M | 29 | DD | 3.2 | 0.8 | 0 |
| 2-IF/TA | F | 48 | DD | 2.6 | 1.2 | 1 |
| 3-IF/TA | M | 43 | DD | 4 | 1.4 | 0 |
| 4-IF/TA | F | 55 | DD | 2.9 | 1.8 | 0 |
| 5-IF/TA | F | 47 | DD | 2.5 | 1.9 | 0 |
| 6-IF/TA | M | 60 | DD | 2.4 | 0.95 | 0 |
| 7-IF/TA | F | 48 | DD | 2.8 | 1.2 | 0 |
| 8-IF/TA | M | 54 | DD | 2.9 | 1.2 | 0 |
| 9-IF/TA | M | 38 | DD | 3.2 | 0.9 | 1 |
| 10-IF/TA | F | 43 | DD | 3.5 | 1.5 | 0 |
| 11-IF/TA | M | 30 | DD | 3.5 | 1.4 | 0 |
| 12-IF/TA | F | 32 | DD | 2.5 | 1.5 | 0 |
| 13-IF/TA | M | 46 | DD | 3.1 | 1.1 | 0 |
| 14-IF/TA | M | 50 | DD | 2.4 | 0.9 | 0 |
| 15-IF/TA | F | 58 | DD | 3.5 | 1.3 | 0 |
| 16-IF/TA | M | 52 | DD | 2.5 | 1 | 0 |
| 17-IF/TA | F | 50 | DD | 2.6 | 1.2 | 1 |
| 1-NKA | M | 49 | DD | 1.1 | Negative | 0 |
| 2-NKA | M | 52 | DD | 1.2 | Negative | 1 |
| 3-NKA | F | 42 | DD | 0.9 | Negative | 0 |
| 4-NKA | M | 48 | DD | 1.2 | Trace | 0 |
| 5-NKA | F | 39 | DD | 1.3 | Negative | 0 |
| 6-NKA | M | 52 | DD | 0.8 | Negative | 0 |
NKA, normal kidney allograft; DD, deceased donor; ACR, acute cellular rejection.
A† biopsy time.
Table 2.
Histological evaluation of IF/TA biopsies using Banff 97 (11)
| Patient | Interstitial fibrosis | Hyaline/vascular | Cellular infiltrate | Vascular chronic rejection | Banff score(total) |
|---|---|---|---|---|---|
| 1-IF/TA | ci2 | ah2 | i2 | cv1 | CAN IIb |
| 2-IF/TA | ci3 | ah2 | i3 | cv3 | CAN IIIb |
| 3-IF/TA | ci2 | ah0 | i1 | cv2 | CAN IIb |
| 4-IF/TA | ci2 | ah1 | i3 | cv2 | CAN IIb |
| 5-IF/TA | ci2 | ah1 | i3 | cv2 | CAN IIb |
| 6-IF/TA | ci2 | ah1 | i3 | cv2 | CAN IIb |
| 7-IF/TA | ci2 | ah1 | i3 | cv2 | CAN IIb |
| 8-IF/TA | ci2 | ah2 | i2 | cv1 | CAN IIb |
| 9-IF/TA | ci3 | ah2 | i3 | cv3 | CAN IIIb |
| 10-IF/TA | ci2 | ah1 | i3 | cv3 | CAN IIb |
| 11-IF/TA | ci2 | ah2 | i2 | cv1 | CAN IIb |
| 12-IF/TA | ci2 | ah2 | i2 | cv1 | CAN IIb |
| 13-IF/TA | ci2 | ah2 | i2 | cv1 | CAN IIb |
| 14-IF/TA | ci2 | ah2 | i2 | cv1 | CAN IIb |
| 15-IF/TA | ci3 | ah2 | i2 | cv1 | CAN IIb |
| 16-IF/TA | ci2 | ah2 | i2 | cv1 | CAN IIb |
| 17-IF/TA | ci3 | ah2 | i2 | cv3 | CAN IIIb |
IF, interstitial fibrosis; TA, tubular atrophy; CAN, chronic allograft nephropathy.
Quality Control
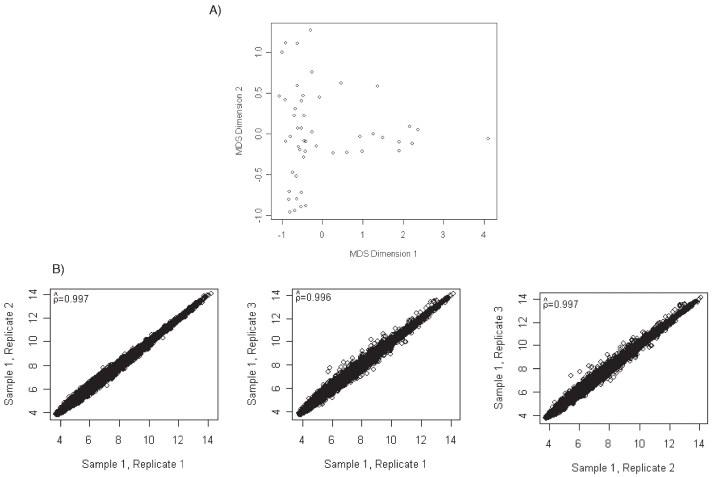
All RNA samples met all of the quality control criteria for sample preparation. Moreover, RNA amount isolated from all the kidney biopsies was sufficient for the microarray studies using the One-Cycle Target Labeling and Control Reagents (Affymetrix). IVT and cDNA synthesis were checked in all the samples, showing satisfactory results in concordance with the pre-established quality control criteria. Results from the quality assessments revealed that eight GeneChips had a lower confidence limit exceeding 1-however, these lower limits only ranged from 1.01 to 1.76. Due to the other quality assessments indicating similarity across GeneChips, these samples were ascertained to meet quality standards. A scatterplot of the MDS plot revealed no further quality concerns (Figure 1A).
Figure 1.
(A) Plot of two-dimensional multidimensional scaling coordinates derived from quality assessment data for the included microarrays. The plot revealed similar distributions, as desired. (B) Pairwise scatterplots and Pearson correlation for the three independent isolations from one kidney sample.
Interassay Reproducibility Analysis for Kidney Tissue Samples
To further assess reproducibility using kidney samples, three different isolations were performed on two kidney tissue samples. Pairwise scatterplots and Pearson correlation again revealed high correspondence between probe set expression summaries obtained from the three independent extractions (Figure 1B).
Gene Expression Profiles among the Kidney Tissue Study Biopsies
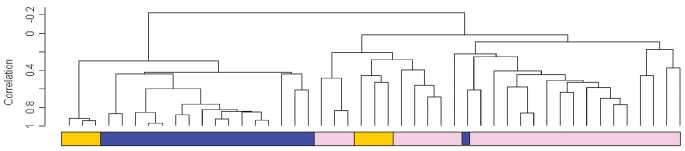
Relationships between the expression profiles of all kidney tissues were further explored by unsupervised hierarchical clustering. The resulting dendrogram showed two major branches (Figure 2). All but one of the kidney tissue samples with IF/TA clustered together in one branch with high correlation. Similar behavior was shown for the normal donor kidney tissues that clustered together in the second branch. The normal allograft kidney samples, however, clustered in closer association with normal kidney (n = 3) and in an independent subbranch closer to the IF/TA group (n = 3). From this analysis, we observed a high homogeneity in the IF/TA kidney tissue group.
Figure 2.
Unsupervised hierarchical clustering analysis including all the study kidney sample tissues. The dendrogram shows that the IF/TA tissue samples clustered with high correlation (black boxes, IF/TA samples).
Identifying Trends in Gene Expression among Normal Kidneys, Normal Allografts, and IF/TA Samples
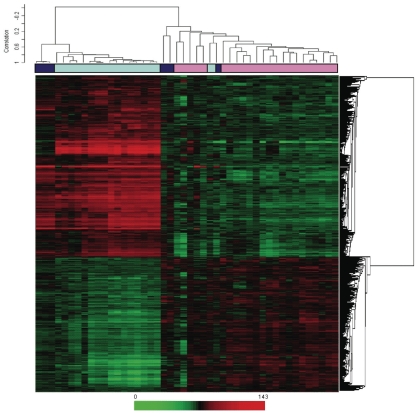
Probe set level tests of hypotheses tests were conducted to identify genes with a significant trend (increase or decrease) in gene expression across the three groups using Jonckheere-Terpstra test for trend. Using the Bonferroni adjustment for multiple hypothesis testing, 1794 probe sets were identified as having a significant trend across the three groups (Supplemental Table 1). Probe sets are listed in ascending order by P value, so those at the top of the list have the smallest P values. Included in the list is the gene annotation information as well as the median by each group (normal kidney, normal allograft, IF/TA) and the Spearman rank correlation (trend). Of the 1794 significant probe sets, 764 have a negative trend while 1030 have a positive trend. Figure 3 shows two-dimensional hierarchical clustering results for the differentially expressed genes among kidney samples.
Figure 3.
Two-dimensional hierarchical clustering results for differentially expressed genes among kidney samples. This set was created using Pearson (centered) correlation cluster analysis, using BRB-ArrayTools version 3.0.2. Each vertical column represents an independent experiment. The fold changes in mRNA levels in the samples are represented by green and red squares, showing decreased and increased levels between the RNA samples, respectively. The color scale indicates the magnitude of fold changes. Dark blue boxes, normal kidney allograft samples; light blue boxes, IF/TA kidney samples; pink boxes, normal kidney samples.
We evaluated the more important pathways where the significant probe sets were classified. Specifically, a positive trend in gene expression levels was observed with expression levels increasing normal kidney > normal allograft > IF/TA for apoptosis. The more important differentially expressed genes associated with apoptosis included TNFRSF7 (5.2, 5.6, 7.2; Spearman rank correlation trend 0.83), TNFAIP8 (6.4, 7.9, 8.4; trend 0.81), FAS (6.9, 7.8, 8.2; trend 0.77), FASL6 (4.2, 4.4, 4.6; trend 0.75), CASP5 (4.9, 5.1, 5.2; trend 0.73), CASP1 (4.5, 5.6, 7.3; trend 0.85), and CASP3 (5.4, 6.4, 6.7; trend = 0.8). Genes involved in homing and chemotaxis also showed a positive trend with increasing expression levels in IF/TA samples (for example, CD44 (5.5, 6.6, 8.1; trend 0.83), CCL5 (4.3, 6.9, 8.7; trend 0.86), CCL4 (5.4, 7.4, 8.2; trend 0.85), among others).
Interestingly, a negative trend of gene expression was observed for genes related to angiogenesis, with gene expression levels decreasing normal kidney < normal allograft < IF/TA across the three groups, for example, ANG (7.9, 6.9, 4.6; trend −0.82), ANGPTL3 (7.9, 6.3, 4.6, trend −0.72), and EGF (8.2, 5.6, 4.7; trend −0.84).
From the analysis of the cell types present in the IF/TA tissues, activation of the humoral branch of the immunological response was observed (immunoglobulins were clearly overexpressed in IF/TA tissues, including IGHG3, IGHA1, IGHM, among others). However, genes related to both cell types (B and T) were involved in IF/TA profiles, and a predominantly immunologic pattern and/or response was not clearly identified (for example, CD4, CD40, CD8B, CD3D, among others).
As previously published (14), genes related to matrix production and deposition (for example, LAMA4, COL1A2, MMP9) were overexpressed in samples with IF/TA compared with normal kidneys and normal allografts. However, TGF-β1 was overexpressed in IF/TA samples compared with normal kidneys, but not compared with normal allografts.
Functional Analysis of the Differential Gene Expression between Normal Kidneys, Normal Allografts, and IF/TA
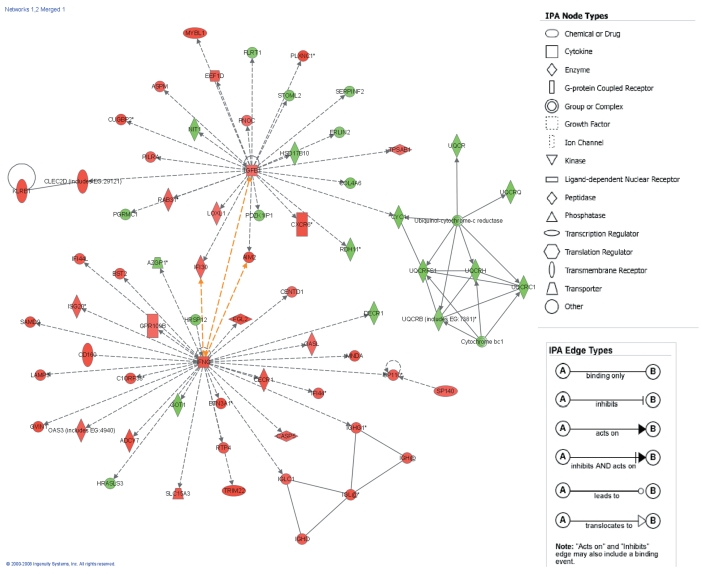
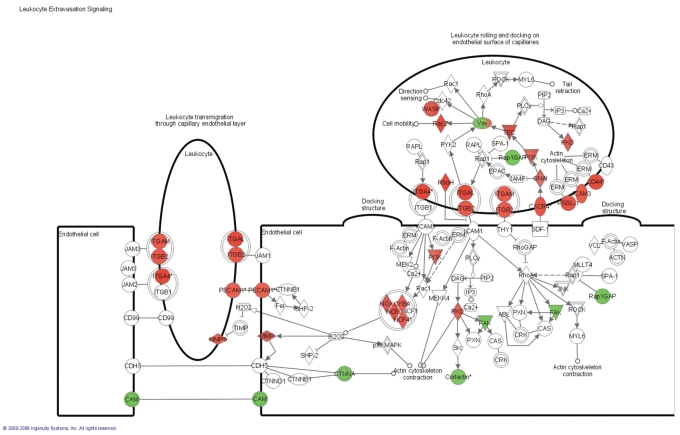
The 1794 differentially expressed probe sets were overlaid onto a global molecular network developed from information contained in the IPKB. Networks of these genes were then algorithmically generated based on their connectivity, or interactions between one another. Biological networks were ranked (Table 3), where the score corresponds to the likelihood of a set of genes being found in the networks due to chance; i.e., a score of 3 indicates that there is a 1/1000 chance that the focus genes are in a network due to chance. Therefore, scores of 3 or higher have a 99.9% confidence of not being generated by chance alone. This score was used as the cutoff for identifying gene networks significantly affected in IF/TA samples. Interconnections of significant functional networks were generated, principally for two of the most significant functional networks (Figure 4). These networks included molecules involved in the immune response and cell-to-cell signaling and interaction, showing the important role that chronic inflammation and chronic inflammatory cell infiltrates might play in IF/TA progression. Moreover, from the analysis of the canonical pathways, we observed that antigen presentation pathway, leukocyte extravasation signaling, and natural killer cell signaling were among the more important. Figure 5 shows the molecules that were found to be differentially expressed in the IF/TA samples in the leukocyte extravasation signaling pathway.
Table 3.
Biological networks ranked by score in kidney tissues from patients with IF/TA.
| ID | Molecules in network | Score | Focus molecules | Top functions |
|---|---|---|---|---|
| 1 | ADCY7, AZGP1, BST2, BTN3A1, C1ORF38, CASP5, CD160, CECR1, CENTD1, DECR1, FGL2, GOT1, GPR109B, GVIN1, HRASLS3, HRSP12, IFI44, IFI44L, IFNG, IGH@, IGHD, IGHG1, IGL2, IGLC1, ISG20, LAMP3, MNDA, OAS3 (includes EG:4940), OASL, RTP4, SAMD9, SLC15A3, SP110, SP140, TRIM22 | 45 | 35 | Immune response, cell-to-cell signaling and interaction, hematologic system development and function |
| 2 | AIM2, ASPM, CLEC2D (includes EG:29121), COL4A6, CUGBP2, CXCR6, CYC1, cytochrome bc1, EEF1D, ERLIN2, FLRT1, HSD17B10, IFI30, KLRB1, LOXL1, MYBL1, NIT1, PDZK1IP1, PGRMC1, PILRA, PLXNC1, PNOC, RAB31, RDH11, SERPINF2, STOML2, TGFB1, TPSAB1, ubiquinol-cytochrome-c reductase, UQCR, UQCRB (includes EG:7381), UQCRC1, UQCRFS1, UQCRH, UQCRQ | 40 | 33 | Carbohydrate metabolism, cell signaling, energy production |
| 3 | ARHGEF6, CALCR, CD47, CD69, CDKN1B, CLEC2B, COPS8, CTNNA1, CTSH, CYFIP2, DGKA, Dynamin, EPB41L2, EVL, Foxo, FYB, IGHA1, ITGA4, LAMP2, LGALS1, LGALS3, LPXN, MCM5, MCM6, NCK1 (includes EG:4690), NCKAP1, NPAT, PARD3, POU2AF1, PRSS1 (includes EG:5644), SIRPG, SSTR2, STAG2, TFRC, YWHAH | 40 | 33 | Cell cycle, cellular development, cellular movement |
| 4 | AIFM1, APIP, ARHGDIB, BCL2A1, CASP1, CASP3, Caspase, CCL5, CIDEB, CYBA, Cyba-Ncf1c-Ncf2-Nox-Ncf4, CYBB, CYCS, ENDOG, GNLY, GZMB, GZMK, HSPE1, IFNγ, IL16, MTCH1, NAIP, NCF4, NGFRAP1, PECAM1, PMAIP1, Pmca, PSAP, PYCARD, RAC2, SERPINB9, SRGN, STK3, TPD52L1, TXN2 | 35 | 31 | Cell death, cancer, free radical scavenging |
| 5 | ALDH7A1, CD180, Ciap, CST7, CYLD, FABP5, FAM46A, IGKC, IL-1R/TLR, IL10RA, IL1RL1, Itgal-Itgb2, LSP1, LY86, LY96, MYD88, Myd88-Tlr1-Tlr2, NFkB, PPIF, PRKD2, RHOH, RSAD2, ST18, STK10, TACSTD1, TLR1, TLR2, TLR4, TLR5, TLR7, TLR8, TNFAIP8, TPMT, TRAFD1, UMOD | 33 | 30 | Cell signaling, immune response, cell-to-cell signaling and interaction |
| 6 | Adenosine-tetraphosphatase, AGT, ATP1B1, ATP5A1, ATP5B, ATP5C1, ATP5D, ATP5G3, ATP5O, COX17, COX4I1, COX5A, COX5B, COX6A1, COX6B1, COX6C (includes EG:1345), COX7A1, COX7A2, COX7B, COX7C, COX8A, cytochrome c oxidase, ESRRG, IDH2, IDH3A, NADP isocitrate dehydrogenase, PPARGC1A, PRDX3, SDHA, SDHAL1, SDHB, SDHD, succinate dehydrogenase, succinate dehydrogenase (ubiquinone), TNS1 | 33 | 30 | Carbohydrate metabolism, cell signaling, energy production |
| 7 | CCL8, CCR2, CCR5, CD2, CLC, CSF2RA (includes EG:1438), CSF2RB, CXCR4, DDB2, DNAJA3, GBP1, IFI27, IFNαreceptor, IFNGR1, IL10RB, IL21R, IL2RB, IL2RG, IRF1, JAK, JAK2, LIFR, NMI, OAS1, PTPN2, PTPN6, PTPRC, SOCS, SOCS2, STAT, STAT1, STAT4, STAT5a/b, UBE1L, WARS | 33 | 30 | Cellular growth and proliferation, cellular development, hematologic system development and function |
| 8 | AACS, ACSL, ACSL1, ACSL3, ACSL5, BCL11B, CFD, CKB, CYB5A, CYP51A1, CYP7B1, DBI, ESRRA, ETFDH, EZH2, FDX1, GAD, GAD1, Hdac, HDAC9, histone h3, HMG CoA synthase, IKZF1, LPL, MAOB, MEF2C, MTUS1, RPS9, SC4MOL, SC5DL, SMC4, SP3, SREBF1, SUCLG1, WWC1 | 33 | 30 | Lipid metabolism, small molecule biochemistry, molecular transport |
| 9 | 3α-Hydroxysteroid dehydrogenase (A-specific), ACTR2, AKR1C1, AKR1C2, AKR1C3, ARHGEF2, Arp2/3, ARPC2, ARPC5, BAG3, CDH11, CTTN, DAPP1, EGF, Erm, F actin, FGR, FSTL1, HCLS1, HSD17B, HSD17B1, HSD17B12, MAP4K1, MARCKS (includes EG:4082), PDCD4, PFKM, PPAP2B, RALGDS, RAP2B, REPS2, SH2B3, TOM1L1, trans-1,2-dihydrobenzene-1,2-diol dehydrogenase, TUFM, WAS | 31 | 29 | Cellular assembly and organization, endocrine system development and function, lipid metabolism |
| 10 | ACAT1, ACAT2, Acetyl-CoA C-acetyltransferase, ARG2, B2M, CSRP2, CTH, EBI2, FXYD5, HADHB, HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, HLA-G, HMGCS1, IRF2, KLRC2, KLRD1, KLRK1, LILRB1, MHC class I, MHC Iα, NDUFB8, PDGF BB, PLK2, RPL13, SFRS7, SLC1A1, SLC2A3, Sphk, Tap, TAP1, TYROBP | 31 | 29 | Cell signaling, immune response, cellular growth and proliferation |
Figure 4.
The top-two scoring network of interactions among the differentially expressed genes between normal, normal allograft, and IF/TA kidney tissues. Interconnections of significant functional networks in IF/TA are indicated. The meaning of the node shapes and interaction edges is also indicated.
Figure 5.
Canonical pathway analysis. Leukocyte transvasation signaling: genes differentially expressed in IF/TA samples. Red, overexpressed genes; green, underexpressed genes.
Validation of Microarray Studies
Expression levels of TGF-β, EGFR, and TSP-1 were further confirmed using a quantitative real time RT-PCR. The results from the microarray were reproduced by real time RT-PCR (r = 0.85, P < 0.001; r = 0.6, P = 0.0012; r = 0.67, P < 0.001).
DISCUSSION
Progressive graft deterioration with IF/TA remains an important problem in renal transplantation because its pathogenesis is still not understood and thus treatment is not available. IF/TA might be the end result of a series of time-dependent injuries to the allograft resulting in permanent damage and loss of nephrons. Destroyed nephrons cannot be replaced, leading to accumulation of damage in a progressive and cumulative manner, as confirmed in prospective protocol biopsy studies (19,20). The damage occurring to the transplanted kidney might be the result of a combination of pre-existing donor disease and subsequent insults to the transplant leading to cumulative injury. Specific histological lesions that define the etiology have increasingly been identified. These include arteriolar hyalinosis, which is the hallmark of CNI (calcineurin inhibitors) toxicity; cellular interstitial infiltration found with subclinical rejection; specific electron-microscopy or immunofluorescence findings found with recurrent glomerulonephritis; and tubular cell inclusions suggestive of viral infection. Currently, data from large-scale prospective studies suggest that initial immune injury occurs within the first 12 months, often presenting as subclinical rejection, which can progress to chronic immune injury. A second level of injury occurring after 12 months is associated with arteriolar hyalinosis, ischemic glomerulosclerosis, and secondary fibrosis, which is predominantly due to CNI toxicity (1,2). At the molecular level, there is increasing identification of chemokine, growth factor, and cytokine impact on tubulointerstitial damage (21–23). Other pathways include cellular senescence, epithelial-to-mesenchymal tubular cell transition, cytokine excess, and structural damage with loss of tubules leading to the formation of atubular glomeruli (24). To find appropriate therapeutic strategies, more detailed insights into the molecular pathogenesis are essential.
Analysis of gene-expression patterns will provide a window on the biology and pathogenesis of IF/TA. Despite the high number of published references on single gene analysis and chronic allograft injury (25–31), mechanisms and predictors of progressive alloimmune scarring remain unclear.
Because differential degradation of RNA could lead to misleading conclusions, verification of RNA quality is an important aspect of any microarray experiment. In this study, we used pre-established quality control criteria for evaluating sample quality at every reaction step (RNA isolation, cDNA synthesis, IVT reaction, and hybridization). In addition, we were able to obtain sufficient RNA concentration for running the microarrays without the necessity of an amplification method. These methods of quality control greatly strengthen the data and conclusions found in this study.
A distinctive gene expression pattern was observed in IF/TA samples, compared with normal allografts and normal kidneys. It was clearly demonstrated in an unsupervised cluster analysis in which all but one IF/TA kidney biopsy clustered together with high correlation. However, we did not observe associations between specific histopathologic features of progressive graft deterioration with IF/TA and gene expression profiles.
Using Jonckheere-Terpstra test for trend analysis, we were able to establish a trend in gene expression for genes involved in different pathways among the studied groups. A biopsy of a chronically failing kidney transplant usually shows nonspecific or end-stage changes, so the relative contributions of pre-existing disease in the allograft and immunologic and nonimmunologic factors become difficult to distinguish. A major impediment has been the lack of prospective, longitudinal histologic data from studies of humans with chronic graft dysfunction. Thus, we evaluated how the gene expression changed from normal donor kidneys, normal allografts, and IF/TA samples. We understand the limitation related to interindividual variation, but this study design provides an initial step for the analysis of the more important pathways leading to progressive graft deterioration with IF/TA. Also, this study described the gene expression signature at the specific study point related with the patient condition (normal kidney, normal allograft, IF/TA). The biopsy time was different between groups (normal allograft versus IF/TA), but we included as a normal allograft group kidney transplantation patients with >9 months posttransplantation, normal kidney function, and nonhistological evidence of IF/TA. It has been reported that in protocol biopsies performed during the first 6 months after kidney transplantation, 25% to 50% of patients already display variable histological evidence of chronic allograft nephropathy (19,20).
Activation of the humoral branch of the immunological response was observed. However, as we reported recently (14,32), genes related to both cell types (B and T) were involved in IF/TA sample profiles, and a predominantly immunologic pattern and/or response was not clearly identified. Moreover, when samples for different groups were compared for identifying specific cell types and/or immunological branch predominance, a positive trend leading to IF/TA was also observed, reaffirming our initial results.
Moreover, we observed a negative trend in gene expression of genes related to angiogenesis in IF/TA samples. In chronic inflammation, where tissue destruction and mononuclear cell infiltration are dominant, the persistent delivery and local expression of angiogenesis factors can serve to sustain the angiogenesis response (33–38). In its normal guise, angiogenesis is thought to facilitate the repair of injured tissues and to restore oxygenation. In diseases such as glomerulonephritis, ischemic nephropathy, and tubulointerstitial fibrosis, and in the aging process, accelerated attrition of the microvasculature (as a result of inefficient delivery of angiogenesis factors and/or EPC [endothelial progenitor cells]) results in ongoing and persistent hypoxia, which can result in further tissue destruction (39–42). Angiogenesis might play an important role in IF/TA progression and deserves further evaluation and study. Finally, as was previously demonstrated, matrix production and deposition gene expression were increased in kidneys with IF/TA.
Interaction networks and functional analysis were used for evaluating the different pathways involved in the progression to IF/TA. This approach revealed highly connected coexpression networks with significant gene-gene associations in relation to kidney tissue status (normal kidney, normal allograft, IF/TA). The 10 networks with the highest complexity score (see “Methods”) were identified as potentially important in the multifactorial pathology of IF/TA, as shown in Table 3. Moreover, almost all the top-scored networks were related to immune response, inflammation, and cell-to-cell interaction, showing the importance of the chronic inflammation and chronic cell infiltrates in IF/TA progression.
Prospective studies including protocol biopsies (pre-implantation and follow-up during the first 12 months posttransplantation) are needed for establishing potential biomarkers as predictors of IF/TA.
ACKNOWLEDGMENTS
This study has been supported by a grant from ROCHE Laboratories Investigator Initiated Research Program.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors of this manuscript do not have any conflict of interest.
REFERENCES
- 1.Hariharan S, et al. Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med. 2000;342:605–12. doi: 10.1056/NEJM200003023420901. [DOI] [PubMed] [Google Scholar]
- 2.Meier-Kriesche HU, et al. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am J Transplant. 2004;4:378–83. doi: 10.1111/j.1600-6143.2004.00332.x. [DOI] [PubMed] [Google Scholar]
- 3.Solez K, et al. Banff ‘05 Meeting Report: differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy (‘CAN’) Am J Transplant. 2007;7:518–26. doi: 10.1111/j.1600-6143.2006.01688.x. [DOI] [PubMed] [Google Scholar]
- 4.Halloran PF, et al. Assessing long-term nephron loss: is it time to kick the CAN grading system? Am J Transplant. 2004;11:1729–30. doi: 10.1111/j.1600-6143.2004.00662.x. [DOI] [PubMed] [Google Scholar]
- 5.Nankivell BJ, Chapman JR. Chronic allograft nephropathy: current concepts and future directions. Transplantation. 2006;81:643–54. doi: 10.1097/01.tp.0000190423.82154.01. [DOI] [PubMed] [Google Scholar]
- 6.Nankivell BJ, et al. Natural history, risk factors, and impact of subclinical rejection in kidney transplantation. Transplantation. 2004;78:242–9. doi: 10.1097/01.tp.0000128167.60172.cc. [DOI] [PubMed] [Google Scholar]
- 7.Mas V, et al. Intragraft messenger RNA expression of angiotensinogen: relationship with transforming growth factor beta-1 and chronic allograft nephropathy in kidney transplant patients. Transplantation. 2002;4:718–21. doi: 10.1097/00007890-200209150-00022. [DOI] [PubMed] [Google Scholar]
- 8.Szeto CC, et al. Messenger RNA expression of target genes in the urinary sediment of patients with chronic kidney diseases. Nephrol Dial Transplant. 2005;20:105–13. doi: 10.1093/ndt/gfh574. [DOI] [PubMed] [Google Scholar]
- 9.Nicholson ML, Waller JR, Bicknell GR. Renal transplant fibrosis correlates with intra-graft expression of tissue inhibitor of metalloproteinase messenger RNA. Br J Surg. 2002;89:933–7. doi: 10.1046/j.1365-2168.2002.02118.x. [DOI] [PubMed] [Google Scholar]
- 10.Mansfield ES, Sarwal MM. Arraying the orchestration of allograft pathology. Am J Transplant. 2004;4:853–62. doi: 10.1111/j.1600-6143.2004.00458.x. [DOI] [PubMed] [Google Scholar]
- 11.Racusen LC, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55:713–23. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 12.Irizarry RA, Bolstad BM, Collin F, Cope LCM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbell E, Liu W-M, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–92. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 14.Mas V, et al. Establishing the molecular pathways involved in chronic allograft nephropathy for testing new noninvasive diagnostic markers. Transplantation. 2007;83:448–57. doi: 10.1097/01.tp.0000251373.17997.9a. [DOI] [PubMed] [Google Scholar]
- 15.Gentleman R, et al. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer: New York; 2005. [Google Scholar]
- 16.Gentelman R. Using GO for statistical analysis. In: Antioch R, editor. COMPSTAT: Proceedings in Computational Statistics. Physica-Verlag/Springer; Heidelberg: 2004. p. 171. [Google Scholar]
- 17.Ali S, et al. Renal transplantation: examination of the regulation of chemokine binding during acute rejection. Transplantation. 2005;79:672–9. doi: 10.1097/01.tp.0000155961.57664.db. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR the 2(-Delta Delta C (T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Solez K, Vincenti F, Filo RS. Histopathologic findings from 2-year protocol biopsies from a U.S. multicenter kidney transplant trial comparing tacrolimus versus cyclosporine: a report of the FK506 Kidney Transplant Study Group. Transplantation. 1998;66:1736–40. doi: 10.1097/00007890-199812270-00029. [DOI] [PubMed] [Google Scholar]
- 20.Nankivell BJ, et al. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349:2326–33. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 21.Mas V, et al. Intragraft expression of transforming growth factor-beta 1 by a novel quantitative reverse transcription polymerase chain reaction ELISA in long lasting kidney recipients. Transplantation. 2000;70:612–6. doi: 10.1097/00007890-200008270-00014. [DOI] [PubMed] [Google Scholar]
- 22.Szeto CC, et al. mRNA expression of target genes in the urinary sediment as a noninvasive prognostic indicator of CKD. Am J Kidney Dis. 2006;47:578–86. doi: 10.1053/j.ajkd.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 23.Bicknell GR, et al. Differential effects of cyclosporin and tacrolimus on the expression of fibrosis-associated genes in isolated glomeruli from renal transplants. Br J Surg. 2000;87:1569–89. doi: 10.1046/j.1365-2168.2000.01577.x. [DOI] [PubMed] [Google Scholar]
- 24.Chapman JR. Longitudinal analysis of chronic allograft nephropathy: clinicopathologic correlations. Kidney Int. 2005;99(Suppl):S108. doi: 10.1111/j.1523-1755.2005.09920.x. [DOI] [PubMed] [Google Scholar]
- 25.Pribylova-Hribova P, et al. TGF-beta1 mRNA upregulation influences chronic renal allograft dysfunction. Kidney Int. 2006;69:1872–9. doi: 10.1038/sj.ki.5000328. [DOI] [PubMed] [Google Scholar]
- 26.Muthukumar T, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005;353:2342–51. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- 27.Eikmans M, et al. Expression of surfactant protein-C, S100A8, S100A9, and B cell markers in renal allografts: investigation of the prognostic value. J Am Soc Nephrol. 2005;16:3771–86. doi: 10.1681/ASN.2005040412. [DOI] [PubMed] [Google Scholar]
- 28.Djamali A, et al. Heat shock protein 27 in chronic allograft nephropathy: a local stress response. Transplantation. 2005;79:1645–57. doi: 10.1097/01.tp.0000164319.83159.a7. [DOI] [PubMed] [Google Scholar]
- 29.Koop K, et al. Differentiation between chronic rejection and chronic cyclosporine toxicity by analysis of renal cortical mRNA. Kidney Int. 2004;66:2038–46. doi: 10.1111/j.1523-1755.2004.00976.x. [DOI] [PubMed] [Google Scholar]
- 30.Eikmans M, et al. Renal mRNA levels as prognostic tools in kidney diseases. J Am Soc Nephrol. 2003;14:899–907. doi: 10.1097/01.asn.0000056611.92730.7b. [DOI] [PubMed] [Google Scholar]
- 31.Yehia M, et al. Predictors of chronic allograft nephropathy from protocol biopsies using histological and immunohistochemical techniques. Nephrology (Carlton) 2006;11:261–6. doi: 10.1111/j.1440-1797.2006.00561.x. [DOI] [PubMed] [Google Scholar]
- 32.Hotchkiss H, et al. Differential expression of profibrotic and growth factors in chronic allograft nephropathy. Transplantation. 2006;81:342–9. doi: 10.1097/01.tp.0000195773.24217.95. [DOI] [PubMed] [Google Scholar]
- 33.Polverini PJ. Role of the macrophage in angiogenesis-dependent diseases. EXS. 1997;79:11–28. doi: 10.1007/978-3-0348-9006-9_2. [DOI] [PubMed] [Google Scholar]
- 34.Libby P, Zhao DX. Allograft arteriosclerosis and immune-driven angiogenesis. Circulation. 2003;107:1237–9. doi: 10.1161/01.cir.0000059744.64373.08. [DOI] [PubMed] [Google Scholar]
- 35.Libby P, Pober JS. Chronic rejection. Immunity. 2001;14:387–97. doi: 10.1016/s1074-7613(01)00119-4. [DOI] [PubMed] [Google Scholar]
- 36.Reinders ME, Briscoe DM. Angiogenesis and allograft rejection. Graft. 2002;5:96–8. [Google Scholar]
- 37.Leibovich SJ, et al. Macrophage-induced angiogenesis is mediated by tumor necrosis factor-alpha. Nature. 1987;329:630–2. doi: 10.1038/329630a0. [DOI] [PubMed] [Google Scholar]
- 38.Leibovich SJ, Wiseman DM. Macrophages, wound repair and angiogenesis. Prog Clin Biol Res. 1988;266:131–45. [PubMed] [Google Scholar]
- 39.Kang DH, et al. Role of the microvascular endothelium in progressive renal disease. J Am Soc Nephrol. 2002;13:806–16. doi: 10.1681/ASN.V133806. [DOI] [PubMed] [Google Scholar]
- 40.Kang DH, et al. Impaired angiogenesis in the aging kidney: vascular endothelial growth factor and thrombospondin-1 in renal disease. Am J Kidney Dis. 2001;37:601–11. doi: 10.1053/ajkd.2001.22087. [DOI] [PubMed] [Google Scholar]
- 41.Masuda Y, et al. Vascular endothelial growth factor enhances glomerular capillary repair and accelerates resolution of experimentally induced glomerulonephritis. Am J Pathol. 2001;159:599–608. doi: 10.1016/S0002-9440(10)61731-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iruela-Arispe L, et al. Participation of glomerular endothelial cells in the capillary repair of glomerulonephritis. Am J Pathol. 1995;147:1715–27. [PMC free article] [PubMed] [Google Scholar]