Abstract
Protein S is an anticoagulant protein containing a Gla (enclosing γ-carboxyglutamic acids) module, a TSR (Thrombin Sensitive Region) module, four EGF (Epidermal Growth Factor)-like modules and an SHBG (Sex Hormone Binding Globulin)-like region. Protein S is a cofactor to activated protein C (APC) in the degradation of coagulation factors Va and VIIIa, but also has APC-independent activities. The function of the fourth EGF module (EGF4) in protein S has so far not been clear. We have now investigated this module through studies of recombinant wild-type protein S and a naturally occurring mutant with Asn217Ser. The mutant has essentially normal APC anticoagulant activity and a previously reported secretion defect. In the wild-type protein, Asn217 is normally β-hydroxylated. The binding of calcium to wild-type protein S is characterised by four high affinity binding sites with KD values ranging from 10-7 to 10-9 M. Three of these binding sites are located in EGF modules. Using surface plasmon resonance, competition with a calcium chelator and antibody-based methods, we found that one high affinity binding site for calcium was lost in protein S Asn217Ser, but that the mutation also affected the calcium dependent conformation of EGF1. We conclude that binding of calcium to EGF4 of protein S, involving Asn217, is important for the maintenance of the structure of protein S. Also, the abolition of calcium binding to EGF4, related to Asn217, impairs both the structure and function of EGF1.
Protein S is a vitamin K-dependent anticoagulant protein with a molecular weight of approximately 70 kDa. Its physiological importance is demonstrated by an increased risk of thrombosis in individuals who are heterozygous for protein S deficiency, a risk which is further enhanced if the deficiency occurs in combination with other prothrombotic genetic defects (1). Homozygosity for protein S deficiency is a serious condition associated with neonatal purpura fulminans. Protein S consists of an N-terminal vitamin K-dependent γ-carboxyglutamic acid (Gla)-containing module, a module sensitive to cleavage by thrombin and factor Xa (TSR), four epidermal growth factor (EGF)-like modules and a sex hormone-binding globulin (SHBG)-like region, that contains 2 laminin G-type repeats in the C-terminal part of the molecule.
One of the major functions of protein S is to enhance active protein C (APC)- dependent proteolytic inactivation of coagulation factors Va and VIIIa, which are cofactors in the prothrombinase and tenase complexes of the coagulation cascade. However, protein S also has APC-independent anticoagulant functions, probably through direct inhibition of both the prothrombinase and the tenase complexes, and it is also involved in apoptosis (2). Protein S reverses the protective effect that factor Xa exhibits in factor Va inactivation, and competes for protein and/or phospholipid binding sites (3-8). Other proposed functions of protein S are the inhibition of the activation of thrombin-activatable fibrinolysis inhibitor, and the acceleration of APC-mediated neutralization of PAI-1 (9,10).
The domains of protein S that have been implicated in the interaction with APC include the TSR, EGF1 and EGF2 modules (2). The Gla module contributes to this interaction by having a high affinity for phospholipid membranes. The SHBG region accommodates the binding site for C4b-binding protein (C4BP), a regulatory protein in the complement system to which 70% of protein S in plasma is bound. When protein S is bound to C4BP, the function of protein S as a cofactor to APC in the degradation of factor Va is abolished (11). Measurements of free protein S, as opposed to total levels, have been shown to be superior for prediction of protein S deficiency (12). In patients with thrombosis, several mutations in protein S have been found that cause low levels of protein S in plasma, but only a few patients with qualitative defects have been described (2, 13).
EGF modules have been found in many different proteins, such as blood clotting factors, complement proteins and membrane proteins. In many of the blood clotting factors, a set of EGF modules functions as a spacer between the Gla module and the serine protease region to position the active site at a distance from the phospholipid membrane that is commensurate with biological activity (14). EGF modules are also directly involved in protein-protein interactions, for example between tissue factor and coagulation factor VIIa (15). In fibrillin, binding of calcium to multiple EGF modules has been shown to stabilize the structure of the protein. Without calcium, fibrillin is highly flexible, but with calcium bound the protein adopts a rod-like conformation (16).
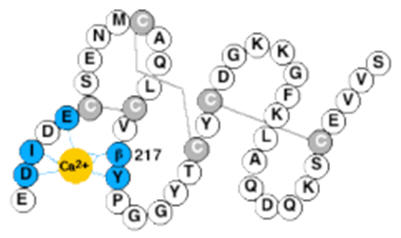
The functions of the third and fourth EGF modules in human protein S remain to be clarified, but very high affinity calcium binding sites have been identified (17, 18). In this study, we have determined the calcium affinity for intact protein S, both the wild-type protein and a variant protein S, harbouring a mutation in EGF4 (Asn217Ser; the Asn residue is normally β-hydroxylated) that has been found in patients with venous thrombosis (Fig. 1) (19, 20). We show that this mutation results in loss of one calcium binding site, and that it also affects the calcium-dependent conformation of EGF1.
FIG. 1. Schematic drawing of protein S.
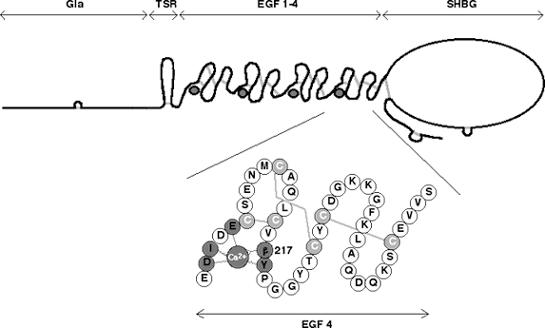
Gla, module with γ-carboxy glutamic acids; TSR, Thrombin Sensitive Region; EGF, Epidermal Growth Factor-like module; SHBG, Sex hormone-binding globulin-like region. Inset: EGF 4 is shown enlarged. The residue 217, which is normally a β-hydroxylated Asn, was mutated to a Ser. The suggested location of the calcium ion is shown in dark grey and the disulfide bridges are indicated in light grey. The dark grey residues are those assumed to be responsible for calcium binding; straight lines represent side-chain ligands and zigzag lines represent backbone ligands.
MATERIAL AND METHODS
Proteins
Recombinant wild-type protein S and protein S with the mutation Asn217Ser were produced in HEK 293 cells as described (20). Site-directed mutagenesis was performed using the Transformer™ Site-Directed Mutagenesis Kit (Clontech, USA). Vitamin K1 (Konakion, Roche, UK) was added to the medium to allow γ-carboxylation of the proteins. Protein S purified from human plasma was purchased from Enzyme Research Laboratories, South Bend, IN, USA.
SDS-PAGE and Immunoblotting
Protein samples were analysed by SDS-PAGE, using 12% (w/v) gels stained with Coomassie brilliant blue R-250 (21). For immunoblotting, the proteins were transferred to an Immobilon membrane (Millipore, Bedford, MA, USA), which was blocked with 5% dry milk dissolved in quenching buffer (0.05% Tween 20, 150 mM NaCl, and 10 mM Tris pH 8.0). The membrane was subsequently incubated with polyclonal anti-human protein S (in house) 10μg/ml, then washed and incubated with alkaline phosphatase-conjugated goat anti-rabbit IgG (Dako, Glostrup, Denmark), and thereafter washed again and developed with 0.15 mg/ml 5-bromo-4-chloro-3-indolyl phosphate and 0.3 mg/ml nitro blue tetrazolium.
Preparation of Calcium Free Buffers
Water (18 Ω resistance) and buffers were rendered calcium free by passing them over a column packed with Chelex 100 (Bio-Rad, CA, USA) and they were stored in plastic containers with dialysis bags containing Chelex 100. The pH meter was left for 30 minutes in 0.1 M EDTA pH 8.0, and then washed with calcium free water before use. The concentrations of calcium in both buffers and protein solutions were measured by atomic absorption. The cuvettes were made calcium free by leaving them in 0.1 mM EDTA pH 8.0 for 30 minutes, and then washed with calcium free water, after which they were kept in 1: 1: 3 HF: HNO3: H2O for 3 minutes. Finally, they were washed again with calcium free water.
Protein Purification
Recombinant protein S was purified by affinity chromatography on a HiTrap column (5 ml, Amersham Pharmacia Biotech, Buckinghamshire, UK) with immobilized HPS 21 (a calcium dependent monoclonal antibody directed against the Gla module of human protein S) (22). Benzamidine (final concentration 2 mM), CaCl2 (2 mM) and Triton X100 (0.1%) were added to the medium, after which the column was washed with 50 mM Tris, 150 mM NaCl, 2 mM CaCl2 pH 7.5. To eliminate non-specific binding, a solution containing 1 M NaCl, 50 mM Tris, 2 mM CaCl2 pH 7.5 was applied to the column. The protein was eluted with 0.1 M Glycine pH 2.7, and pH was immediately adjusted by Tris/NaCl to give a final concentration of 50 mM Tris, 150 mM NaCl, pH 7.5. Thereafter, the protein was concentrated (Macrosep 10 K Omega, Pall Life Sciences, MI, USA) to approximately 1 g/l and dialyzed in calcium free 2 mM Tris-HCl pH 7.5. To further lower the concentration of calcium, one volume of protein was mixed with one volume of Chelex 100 and the mixture was gently rocked for six days at +8°C, and then centrifuged at 2000 rpm for 5 min to dispose of the Chelex 100.
Enzyme-linked Immunosorbent Assay (ELISA)
HPS 56 (10 μg/ml in 0.05 M sodium carbonate pH 9.6), a non-calcium dependent monoclonal antibody directed against EGF3 in human protein S (22, 23), was coated on 96-well microtiter plates (Costar, Corning, NY, USA) overnight at +4°C. The plates were washed three times with 50 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.1% Tween, pH 7.5 and then blocked with 1% ovalbumin (Sigma, Germany), 50 mM Tris, 1 mM EDTA pH 7.5 for 1 hour at room temperature. After this, the plates were washed three times with wash buffer containing different concentrations of calcium (from 0-0.9 mM), and from this time and forward in the assay, the same concentrations of calcium were used for each well. Calcium free protein S (wild-type or with the Asn217Ser mutant, 0.5 μg/ml in the same buffers) was added to the plates and incubated for 1 hour at room temperature. Thereafter, the plates were washed four times with the same buffers, followed by the addition of horse-radish peroxidase-conjugated HPS54 (0.2 μg/ml), a calcium dependent monoclonal antibody directed against EGF1 in human protein S (22). After 30 minutes incubation at room temperature, the plates were washed four times with the different buffers. For development, 100 μl/well of TMB peroxidase substrate (Moss, Inc., Pasadena, MA, USA) was added. The reaction was stopped by adding 150 μl of H2SO4/well and absorbance was measured at 450 nM. As a control experiment, HPS61 (a monoclonal, non-calcium dependent antibody directed against the SHBG region of human protein S; 22) was used following biotinylation according to the manufacturers instructions (Pierce, Rockford, IL, USA) with detection using StreptABComplex HRP (Dako).
Factor Va Inactivation Assay
The assay for concentration dependence of protein S was performed as described (24). To determine factor Va degradation by APC as a function of time (25), 0.16nM APC was incubated with 75μM of the phospholipid vesicles (PC:PE:PS 60:20:20%, Avanti Polar Lipids Inc), 4nM factor Va and 600nM protein S in 40mM Tris-HCl, 140mM NaCl, 3mM CaCl2 and 0.3% w/v BSA (0.04nM APC, 19μM phospholipids, 150nM protein S and 1nM FVa; final concentration). The mixture was incubated at 37°C, and 2μl aliquots were removed and added to a prothrombinase mixture, consisting of 75μM phospholipids (PC:PE:PS 60:20:20%), 3nM factor Xa and 1.5μM prothrombin (25μM phospholipids, 1nM factor Xa and 0.5μM prothrombin; final concentration) at defined time points between 0-20 minutes. Each reaction was stopped after three minutes using 5μl ice-cold 0.5M EDTA. 100μl of the reaction mixture was then removed and incubated with 50μl of chromogenic substrate S-2238 (Chromogenix) to assess thrombin generation. The coagulation factors used in these assays were purchased from Haematologic Technologies Inc, Vermont, USA. All experiments were performed in triplicate and kinetic rate constants for cleavage of factor Va were determined by curve fitting (25).
Macroscopic Ca2+ Binding Constants and Number of Binding Sites—Calcium free protein S was titrated with CaCl2 (0.5 mM) in portions of 0.5 μl in the presence of Quin2 (tetrapotassium salt, Fluka Chemie, Switzerland). The concentration of calcium was determined by atomic absorption spectroscopy. Titrations were performed at 25°C in 2 mM Tris pH 7.5, with or without 150 mM NaCl using a total volume of 70-90 μl, and all experiments were made in duplicate. The protein concentration (3-11 μM) was determined by amino acid analysis after acid hydrolysis. The concentration of chelator (15-30μM) was quantitated in a calcium-saturated solution in the absence of protein by measuring the absorbance at 239 nM (ε239 4.2 × 104 litres mol-1 cm-1; 26). The titrations were performed at 263 nM and the absorbance was recorded on a Cary 400E spectrophotometer (Varian, Australia). The number of binding sites and macroscopic binding constants were obtained from a least squares fit to the measured absorbance as a function of the total calcium concentration, using the program CaLigator (27). The KD used for Quin2 was 5.2 × 10-9 M (low salt) and 1.2 × 10-7 M (150 mM NaCl) (26). The KD of the chelator-calcium complex was a fixed parameter, as was also the initial protein, chelator and calcium concentrations. The program corrected for the effect of dilution when calcium was added.
Surface plasmon resonance studies
The interaction between protein S and HPS54 antibody was studied by surface plasmon resonance technology, using the Biacore3000 apparatus (Biacore AB, Neuchâtel, Switzerland) at 25 °C. The flow buffer was 10 mM Tris/HCl, pH 7.4, with 0.15 M NaCl, 0.005 % Tween 20, 0.02 % NaN3, with different free Ca2+ concentrations ranging from 32 nM to 10 mM. Ca2+ concentrations in the range of 32-430 nM were achieved by mixing calcium free buffer with 1 mM EDTA and 0.1 – 0.95 mM CaCl2 (total concentration), and the free Ca2+ concentration was quantified using quin 2 fluorescence. All buffers were filtrated through sterile 0.22 μm filters before use and degassed for at least 30 min.
Immobilization of HPS54 to the sensor chip was performed through amine coupling. 10mM HEPES/NaOH at pH 7.4, 0.15 M NaCl, 0.005% Tween20, 0.02% NaN3 and 3.4 mM EDTA was used as flow buffer during coupling. Amine coupling was performed as described earlier at a constant flow rate of 5 μl/min (28). Equal volumes of 0.1 M NHS and 0.4 M EDC were mixed, and 25 μl of the mixture was allowed to flow over the sensor chip surface to activate the carboxymethylated dextran (5 min). Three solutions (70 μl of 50, 60 or 75 μg/ml HPS54 in sodium acetate buffer at pH 5.0) was then injected over three different lanes of the sensor chip. Typically three levels of immobilization ranging from 4000 to 11000 RU were achieved on each chip. For a second set of experiments, coupling levels between 800 and 4000 RU were achieved using lower concentrations of HPS54 in the coupling step (70 μl of 20-50 μg/ml HPS54 in sodium acetate buffer at pH 5.0). Injection of ethanolamine for 4 minutes deactivated excess reactive groups on the chip. One of the four flow cells on each sensorchip was reserved for a blank immobilization with no protein in the coupling step, and was used as reference. The signal from the reference cell was subtracted from the signal in cells with HPS54. Each chip with immobilized HPS54 was used for one week.
The kinetics in the reconstitution reactions was studied at constant flow rates of 10 - 30 μl/min with no significant change in the rate constants obtained. Therefore a flow rate of 10 μl/min was used in subsequent experiments. The association of protein S (wild-type or mutant) to the immobilized HPS54 and the following dissociation was studied at several different Ca2+ concentrations ranging from 25 nM to 10 mM. Protein stock solutions were diluted using the flow buffer and 300 μl was injected during the association phase, which was followed for 30 minutes. The dissociation process was followed for 30-45 hours. After each experiment, the surfaces were regenerated by injecting 10 mM glycine/HCl pH 2.9 for 5 min.
The data were evaluated using Levenberg-Marqardt non-linear least-square method. The data analysis was made using the software Kaleidagraph (Synergy Software, Reading, PA, USA). The dissociation of the complex was modeled as a single exponential decay, and attempts were made to fit the signal using a single exponential decay plus a constant baseline
| (1) |
or a sum of two exponential decays plus a constant baseline
| (2) |
where koff, koff1 and koff2 are dissociation rate constants, and C, C1 and C2, are the amplitudes of the corresponding processes.
Other Methods
N-terminal sequences were determined on an Applied Biosystems 494 Procise Sequencer (Foster City, CA, USA). Protein concentrations were determined by amino acid analysis of acid hydrolysates as previously described (11). The amount of Gla was measured following alkaline hydrolysis as described (29).
RESULTS
Characterization of Recombinant Proteins
Wild-type protein S and protein S with the mutation Asn217Ser were produced in HEK293 cells. The affinity-purified proteins were more than 95% homogenous as judged by N-terminal sequencing and SDS-page, and there were no signs of internal cleavages. Amino acid analysis of alkaline hydrolysates established that the protein S preparations were almost fully carboxylated (10.1 mol of Gla/mol of protein S for the wild-type protein and 9.8 for the Asn217Ser protein). Western blot of the proteins was performed after extensive decalcification (Fig. 2).
FIG. 2. Immunoblotting of protein S.
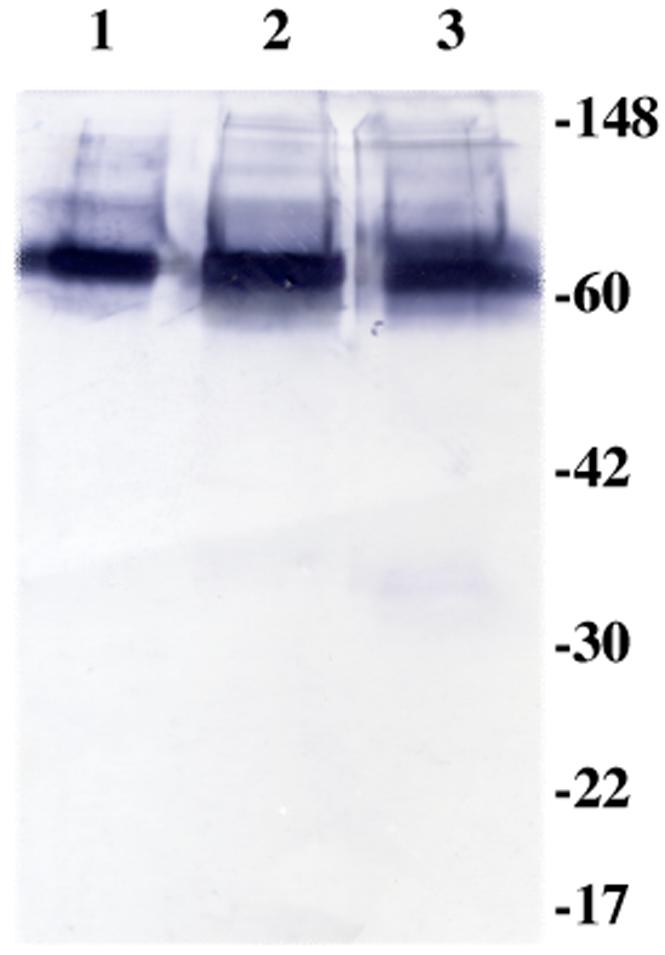
The recombinant proteins had been rocked for 6 days with Chelex 100, and thereafter centrifuged to dispose of the Chelex. Lane 1 shows purified protein S from human plasma, lane 2 wild-type recombinant protein S and lane 3 recombinant protein S with mutation Asn217Ser. Polyclonal anti-human protein S was used as primary antibody and goat anti-rabbit as secondary antibody. The gel was performed under non-reducing conditions: there is no evidence of increased slow mobility bands, suggesting that decalcification does not cause appreciable aggregation of the protein S preparations.
Factor Va Inactivation Assay
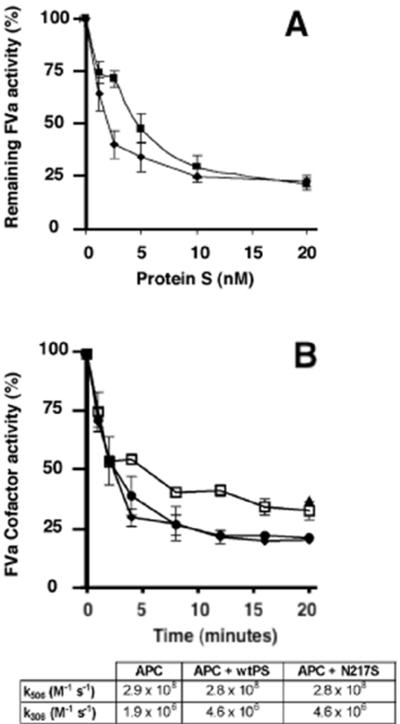
Preliminary results demonstrated high dilutions of wild-type protein S dialysed into assay buffer from culture medium and purified plasma-derived protein S exhibited the same APC cofactor activities in a factor Va inactivation assay. Different concentrations of recombinant protein S (1.25, 2.5, 5, 10 and 20nM) were therefore employed to asses the ability of the mutated protein to function as a cofactor to APC. The Asn217Ser mutant showed minimal reduction in cofactor function compared to wild-type protein S, but only at low protein S concentration (Fig. 3A). Dialyzed conditioned medium from non transfected cells, diluted to the same extent as the wild-type protein S, had no activity in the assay. Neither the wild-type nor the mutated protein S (Asn217Ser) had any APC-independent effects in this assay. In order to confirm that the mutant had essentially normal activity, a higher (∼plasma) concentration (150nM) of protein S was used and factor Va degradation measured as a function of time. As can be seen in Fig 3B, the Asn217Ser mutant was as effective as wild-type protein S in enhancing factor Va inactivation, compared to the absence of protein S. The time course curves were fitted using an equation describing biphasic factor Va proteolysis. Identical kinetic constants were obtained for mutant and wild-type protein S, see inset table in Fig. 3B.
FIG. 3.
A. Concentration dependence of protein S on enhancement of APC-dependent factor Va inactivation. Protein S in different concentrations was incubated with 0.1 nM APC, 3 nM factor Va, and phospholipid vesicles (PC:PE:PS, 40:40:20%; 25μM) at 37°C. Remaining factor Va activity was quantified in a prothrombinase assay using a chromogenic substrate. Results are expressed relative to the remaining activity, with no protein S present (100%), as mean of 3 experiments ± SD. ◆ wild-type protein S and ■ protein S with mutation Asn217Ser. B. Time course of inactivation of factor Va by APC in the presence of protein S. Factor Va (1nM) was incubated with APC (0.04nM) and phospholipids (PC:PE:PS, 60:20:20%; 25μM) in the presence or absence of recombinant protein S (150nM). At specified time-points, an aliquot was removed and the factor Va cofactor activity determined using a prothrombinase assay (phospholipids (25μM), FXa (1nM) and prothrombin (1.5 μM), as described in Materials and Methods. APC in the absence of protein S, ( ); APC and wild-type protein S (◆) and APC with protein S variant N217S (●) were assessed, and the kinetic rate constants for factor Va proteolysis determined (see table, inset).
Affinity of Ca2+ Binding and Number of Binding Sites
To determine the number of calcium binding sites and their binding constants, the Chelex-treated protein was used for calcium titrations in the presence of the chelator Quin2. All protein concentrations were determined by acid hydrolysis. The buffers and solutions with Quin2 were found to contain between 0.25-0.4 μM calcium before addition of the protein. As a control, titrations were made with only Quin2 and the different buffers, and the data could easily be fitted to a straight line with the correct concentration of Quin2 in accordance with one strong binding site (not shown). After introducing the protein into the cuvette, approximately 1 mol of calcium was bound per mol of protein. Binding constants and stoichiometry were obtained from least square fitting to the data points using the program CaLigator (Fig. 4). The macroscopic binding constants obtained are shown in Table I, and they reveal four high affinity calcium-binding sites in wild-type protein S and three sites in protein S Asn217Ser. The values represent the average of two determinations. To evaluate the accuracy of the stoichiometry, the program was forced to use one less or more high affinity sites, compared to the best fit. This resulted in an increase in the error square sum by a factor of 5-10 or more. Hence, it was not possible to fit the data to a model with more or less than four sites for the wild-type or three sites for the mutant. In the evaluation of the accuracy of the fits, a change in log KA for an individual site of 0.5 resulted in an increase in the error square sum by a factor of at least 5-10. When the individual macroscopic binding constants K1, K2 and K3 for the wild-type protein were compared with the values for the mutated protein, the changes were not significant, while for K4, representing the fourth calcium ion bound, the change was clearly significant. Also, when the product K1K2K3 was considered, the change was significant. Therefore, it can be concluded that the Asn217Ser mutation results in loss of one binding site for calcium, and also that it results in slightly reduced affinity of calcium in one or more of the other three sites. It cannot be concluded, however, from these measurements whether the effect is located to one site or if it is due to smaller changes in all the other three sites.
FIG. 4. Ca2+titration of Quin 2 in the presence of protein S.
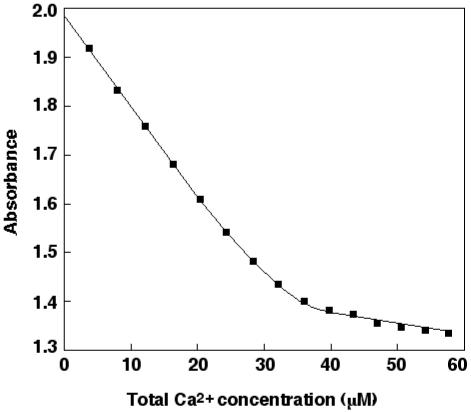
The experiment shown was performed with the mutated protein S Asn217Ser (3.2 μM) in 2 mM Tris pH 7.5. The absorbance at 263 nM is plotted versus the total concentration of calcium. ■ indicates experimental points and the line represent the curve obtained by least square fitting to the data.
TABLE I. KD (M) for binding of Ca2+ to protein S.
KD values were obtained from titration of Ca2+ with the chelator Quin2 in the presence or absence of 150 mM NaCl, in 2 mM Tris pH 7.5, ± one standard deviation. N.D., values were too low to be determined with the chelator Quin2.
| K1 | K2 | K3 | K4 | |
|---|---|---|---|---|
| Wild-type, low salt | 2.5 ± 0.5 × 10-9 | 3.9 ± 2.1 × 10-8 | 3.1 ± 0.4 × 10-8 | 2.0± 1.2 × 10-7 |
| Wild-type, 150 mM NaCl | 6.3 ± 0 × 10-9 | 4.0 ± 0.7 × 10-7 | 5.0 ± 0.6 × 10-7 | 8.0 ± 0.5 × 10-7 |
| Asn217Ser, low salt | 1.2 ± 0.2 × 10-8 | 6.3 ± 0.5 × 10-8 | 3.2 ± 2.2 × 10-7 | N.D. |
| Asn 217Ser 150 mM NaCl | 1.0 ± 0 × 10-8 | 5.0 ± 1.2 × 10-7 | 8.0 ± 0.9 × 10-7 | N.D. |
Enzyme-linked Immunosorbent Assay (ELISA)
To further investigate the effect of the mutation Asn217Ser on the other EGF modules, especially EGF1, an ELISA was performed (Fig. 5). Protein S was bound to the micro-titre plate through the calcium-independent antibody HPS56, directed against EGF3. A calcium titration was accomplished using the strongly calcium dependent antibody HPS 54, directed against EGF1, as a reporter. There was a clear difference between the wild-type and mutated proteins, indicating that the mutation Asn217Ser in EGF4 affects binding of calcium to EGF1. As a control experiment, the calcium-independent antibody HPS61, directed against the SHBG region, was used instead of HPS54. With this antibody there was no difference in absorbance for the wild-type protein when different calcium concentrations were used (not shown), indicating that the studied effect of calcium is dependent on HPS54.
FIG. 5. Enzyme-linked immuno sorbent assay.
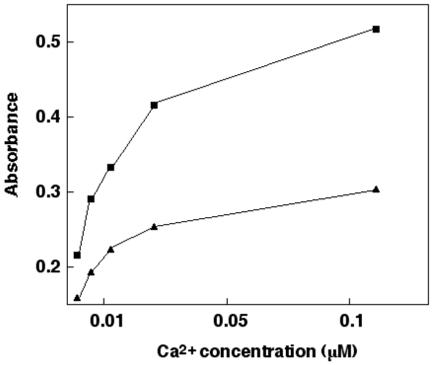
Protein S was bound to the micro-titre plate through the non-calcium dependent monoclonal antibody HPS56. Peroxidase-conjugated HPS 54, a calcium dependent monoclonal antibody directed against EGF1, was used as a marker. ■ indicates wild-type protein S and ▲ indicates protein S with mutation Asn217Ser. All experiments were performed with 1 mM EDTA in the assay, and the shown concentrations of calcium are the concentrations calculated not to be bound to EDTA.
Surface plasmon resonance studies of protein S binding to monoclonal antibody HPS54
The interaction between protein S wild-type and HPS54 occurs with extremely high affinity in the presence of Ca2+, and the dissociation is so slow that a large fraction of protein S remains bound to HPS54 even after 30-60 hours of dissociation (Fig. 6). Both the association and dissociation processes are clearly Ca2+-dependent, although the affinity is still very high at 32 nM Ca2+-concentration. The dissociation data can in most cases only be fitted if two exponential decays are included (equation 2), representing one very slow process and one extremely slow process. The rate of the extremely slow process is so slow that we can only say that koff2 is less than 10-6 s-1. The rate of the slow process decreases with increasing Ca2+ concentration. The extremely slow process represents around 50% of the bound wild-type protein at low nM concentration of free Ca2+, and its fraction increases as the Ca2+ concentration is raised. In contrast, the Asn217Ser mutant displays Ca2+-independent kinetics of dissociation from HPS54. At all Ca2+ concentrations examined, the dissociation phase is dominated by an extremely slow process (koff<10-6 s-1). Dissociation data obtained using different levels of immobilized HPS54 ranging from 800 to 11000 RU display the same behavior with Ca2+-dependent kinetics for the wild-type and Ca2+-independent kinetics for the mutant. The association phase has considerably lower amplitude for the mutant compared to wild-type for all Ca2+ concentrations examined.
FIG. 6. Binding of protein S to HPS54.
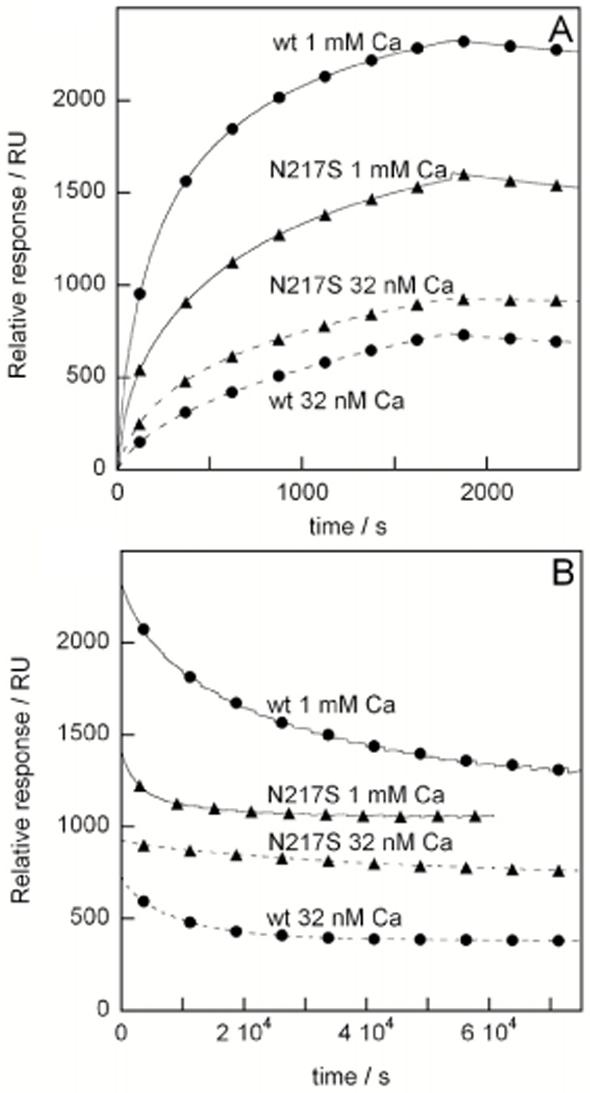
Surface plasmon resonance performed with the monoclonal antibody HPS54 coupled to the chip and human recombinant protein S (wild-type or with the mutation Asn217Ser) in the flow buffer. Several different Ca2+ concentrations were used; the data shown here are 32 nM and 1 mM. A. Association phase data. B. Dissociation phase data.
DISCUSSION
In protein S, there are four EGF modules. So far, no structure of protein S has been determined. However, there is a recent structure of a fragment comprising the SHBG domain of growth arrest specific protein 6 (Gas6) that is homologous to protein S (30). The structure of other coagulation proteins with EGF modules, such as factors VIIa and IXa, have been determined as well as the structure of single or double EGF modules. The structure of a pair of EGF modules may be extended as in fibrillin, or bent with the calcium binding area acting as a hinge, as in many of the coagulation proteins. It can also be U-shaped as in Plasmodium Falciparum (31). A recent NMR and SAXS study of the four EGF modules from protein S found that there is exchange between two conformations corresponding to one straight and one bent arrangement (32). The structures of the individual EGF modules are similar with a characteristic 1-3, 2-4 and 5-6 pairing of the six cysteines into three disulfide bonds. More than 25% of the EGF modules described in the literature bind one calcium ion. The calcium ion is bound in the N-terminal part of the module in a pentagonal bipyramidal geometry, coordinated by oxygen atoms that are supplied either from the backbone or from the side chains of the protein (14). The residues that can be assumed to be responsible for calcium binding in protein S EGF4 are Asp202, Ile203, Glu205, Asn217 and Tyr218 (Fig. 1). Residue 217 normally carries a post-translationally modified asparagine (erythro-β-hydroxyasparagine). The function of this modification is not known, but in aspartyl β-hydroxylase knock-out mice there are developmental defects that have been assumed to be caused by defects in EGF-containing Notch gene family members (33). For coagulation factors IX and X, it has been shown that the β-hydroxylation does not affect binding of calcium, probably because the β-hydroxyl group points away from the calcium binding site (34).
In this paper, we wanted to clarify the function and importance of the fourth EGF module in protein S. Patients with the naturally occurring mutation Asn217Ser in EGF4 have thrombosis, indicating that residue 217 is important for maintaining the anticoagulant function of protein S. Since it has been suggested that 217 is a calcium-coordinating residue, we wanted to compare the calcium affinity of the mutated protein with wild-type protein S. For bovine protein S, four high affinity sites have been found (35). We have now shown that also human protein S has four high affinity binding sites for calcium (table I). Stenberg et al have shown (using a variety of fragments) that there are high affinity sites in human EGF 2-4, with the highest affinity (nanomolar range) site in EGF4 (17, 23). Since the affinity for calcium is so high, direct methods cannot be used and we therefore employed chelator methods instead. In our studies, it was not possible to see a calcium binding site in EGF1, but the fragments used did not include the TSR module which might have affected the binding. Studies of the protein S-C4BP interaction using protein S/factor IX chimeras show that the calcium-dependence of this interaction is not due to the EGF-domains of protein S (36), suggesting that one of the high affinity sites is present in the SHBG domain. Indeed, a calcium ion is observed in the crystal structure of SHBG (37), as well as in the SHBG domain of Gas6 (30).
Dahlbäck et al have produced a monoclonal antibody directed against human EGF1 (HPS54), and this antibody requires less than 1 μM of calcium for maximum binding (22). Addition of another module (Gla- or EGF-) N-terminal to an EGF module has been known to increase the affinity of the calcium site (17, 38). The binding of calcium to the Gla module is probably not of enough strength to be seen in our calcium binding studies. This is in line with earlier investigations on other coagulation proteins, which showed calcium affinities to the Gla modules in the milli-molar range, and it is also in line with studies of the monoclonal antibody HPS21 (directed against the Gla module in human protein S) which requires 1 mM calcium for maximal binding (22, 39). In EGF1 in protein S there is no complete consensus sequence for binding of calcium, but it has been shown in the EGF module of the LDL receptor that calcium can bind to an EGF module even though it contains only part of the consensus sequence (40). Therefore, of the four high affinity binding sites for calcium in human protein S, three are most likely located in EGF 2-4, and one in the SHBG domain.
We investigated the calcium affinity for the mutated protein Asn217Ser, and found that in the range accessible for the chelator used, one high affinity binding site was lost. This shows that residue 217 is important for forming the calcium binding site. Moreover, the calcium affinity is affected also for the other sites. Whether this effect is located to only one other EGF module, or whether it is a smaller effect on several other sites, cannot be determined from these investigations. However, to confirm the existence of long range intramolecular effects in protein S, we studied the mutated protein in a sandwich assay including the calcium dependent monoclonal antibody HPS54. There was a clear difference between the wild-type protein S and the mutated protein, indicating that the mutation in EGF4 affects calcium-induced response in the conformation of EGF1. More importantly, in the surface plasmon resonance studies the mutated protein did not show the pronounced calcium dependence in binding to HPS54 as found in the case of wild-type protein S. When the calcium-binding site in EGF4 is destroyed, the structure might be more flexible and this affects other parts of the protein, such as EGF1.
One of the major functions of protein S is to be a cofactor to APC in the degradation of factor Va. We have now shown that the calcium binding region in EGF4 is not critical for this interaction, as the Asn217Ser mutation had minimal, if any, effect upon this function. Our finding of essentially normal function is compatible with prior studies that showed crucial roles for the TSR-EGF1-EGF2 region (2) and with a domain deletion/swap study in which complete removal of EGF4 resulted in a mutant that retained ∼30% activity (41). It is interesting to note that cellular expression of the Asn217Ser mutant following transient transfection of COS-1 cells was reduced to ∼30% of normal, and that pulse chase experiments following metabolic labeling suggested decreased stability of the mutant within the cell (20). It may be that one of the main functions of the high affinity calcium binding site in EGF4 is to keep the protein in the correct conformation for secretion. The abolition of this binding site will lead to intracellular degradation and reduced secretion of protein S.
ACKNOWLEDGEMENTS
We thank Ann-Marie Thämlitz for her skilful work in the laboratory and Dr Cristina Razarri for help in preparing the wild-type and mutant protein S. The work has been supported by grants from the Wellcome Trust, the British Heart Foundation and the Brazilian Government (Agencia CAPES).
REFERENCES
- 1.Zöller B, Garcia de Frutos P, Hillarp A, Dahlbäck B. Thrombophilia as a multigenic disease. Haematologia. 1999;84:59–70. [PubMed] [Google Scholar]
- 2.Rezende SM, Simmonds RE, Lane DA. Coagulation, inflammation, and apoptosis: different roles for protein S and the protein S-C4b binding protein complex. Blood. 2004;103:1192–1201. doi: 10.1182/blood-2003-05-1551. [DOI] [PubMed] [Google Scholar]
- 3.Heeb MJ, Mesters RM, Tans G, Rosing J, Griffin JH. Binding of protein S to factor Va associated with inhibition of prothrombinase that is independent of activated protein C. J. Biol. Chem. 1993;268:2872–2877. [PubMed] [Google Scholar]
- 4.Hackeng TM, van't Veer C, Meijers JC, Bouma BN. Human protein S inhibits prothrombinase complex activity on endothelial cells and platelets via direct interactions with factors Va and Xa. J. Biol. Chem. 1994;269:21051–21058. [PubMed] [Google Scholar]
- 5.Heeb MJ, Rosing J, Bakker HM, Fernandez JA, Tans G, Griffin JH. Protein S binds to and inhibits factor Xa. Proc. Natl. Acad. Sci. USA. 1994;91:2728–2732. doi: 10.1073/pnas.91.7.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koppelman SJ, Hackeng TM, Sixma JJ, Bouma BN. Inhibition of the intrinsic factor X activating complex by protein S: evidence for a specific binding of protein S to factor VIII. Blood. 1995;86:1062–1071. [PubMed] [Google Scholar]
- 7.van Wijnen M, Stam JG, van't Veer C,, Meijers JC, Reitsma PH, Bertina RM, Bouma BN. The interaction of protein S with the phospholipid surface is essential for the activated protein C-independent activity of protein S. Thromb. Haemost. 1996;76:397–403. [PubMed] [Google Scholar]
- 8.Solymoss S, Tucker MM, Tracy PB. Kinetics of inactivation of membrane-bound factor Va by activated protein C. Protein S modulates factor Xa protection. J. Biol. Chem. 1988;263:14884–14890. [PubMed] [Google Scholar]
- 9.Mosnier LO, Meijers JC, Bouma BN. The role of protein S in the activation of thrombin activatable fibrinolysis inhibitor (TAFI) and regulation of fibrinolysis. Thromb. Haemost. 2001;86:1040–1046. [PubMed] [Google Scholar]
- 10.de Fouw NJ, Haverkate F, Bertina RM, Koopman J, van Wijngaarden A, van Hinsberg VW. The cofactor role of protein S in the acceleration of whole blood clot lysis by activated protein C in vitro. Blood. 1986;67:1189–1192. [PubMed] [Google Scholar]
- 11.Dahlbäck B. Inhibition of protein Ca cofactor function of human and bovine protein S by C4b-binding protein. J. Biol. Chem. 1986;261:12022–12027. [PubMed] [Google Scholar]
- 12.Zöller B, Garcia de Frutos P, Dahlbäck B. Evaluation of the relationship between protein S and C4b-binding protein isoforms in hereditary protein S deficiency demonstrating type I and type III deficiencies to be phenotypic variants of the same genetic disease. Blood. 1995;85:3524–3531. [PubMed] [Google Scholar]
- 13.Persson KEM, Dahlbäck B, Hillarp A. Diagnosing protein S deficiency: analytical considerations. Clin. Lab. 2003;49:103–110. [PubMed] [Google Scholar]
- 14.Stenflo J, Stenberg Y, Muranyi A. Calcium-binding EGF-like modules in coagulation proteinases: function of the calcium ion in module interactions. Biochim. Biophys. Acta. 2000;1477:51–63. doi: 10.1016/s0167-4838(99)00262-9. [DOI] [PubMed] [Google Scholar]
- 15.Banner DW, D’Arcy A, Chène C, Winkler FK, Guha A, Koningsberg WH, Nemerson Y, Kirchhofer D. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 16.Cardy CM, Handford PA. Metal ion dependency of microfibrils supports a rod-like conformation for fibrillin-1 calcium-binding epidermal growth factor-like domains. J. Mol. Biol. 1998;276:855–860. doi: 10.1006/jmbi.1997.1593. [DOI] [PubMed] [Google Scholar]
- 17.Stenberg Y, Linse S, Drakenberg T, Stenflo J. The high affinity calcium-binding sites in the epidermal growth factor module region of vitamin K-dependent protein S. J. Biol. Chem. 1997;272:23255–23260. doi: 10.1074/jbc.272.37.23255. [DOI] [PubMed] [Google Scholar]
- 18.Stenberg Y, Drakenberg T, Dahlbäck B, Stenflo J. Characterization of recombinant epidermal growth factor (EGF)-like modules from vitamin-K-dependent protein S expressed in Spodoptera cells--the cofactor activity depends on the N-terminal EGF module in human protein S. Eur. J. Biochem. 1998;251:558–564. doi: 10.1046/j.1432-1327.1998.2510558.x. [DOI] [PubMed] [Google Scholar]
- 19.Formstone CJ, Wacey AI, Berg L, Rahman S, Bevan D, Rowley M, Voke J, Bernardi F, Legnani C, Simioni P, Girolami A, Tuddenham EGD, Kakkar VV, Cooper DN. Detection and characterization of seven novel protein S (PROS) gene lesions: evaluation of reverse transcript-polymerase chain reaction as a mutation screening strategy. Blood. 1995;86:2632–2641. [PubMed] [Google Scholar]
- 20.Rezende SM, Lane DA, Zöller B, Mille-Baker B, Laffan M, Dahlbäck B, Simmonds RE. Genetic and phenotypic variability between families with hereditary protein S deficiency. Thromb. Haemost. 2002;87:258–265. doi: 10.1055/s-0037-1612982. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–684. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Dahlbäck B, Hildebrand B, Malm J. Characterization of functionally important domains in human vitamin K-dependent protein S using monoclonal antibodies. J. Biol. Chem. 1990;265:8127–8135. [PubMed] [Google Scholar]
- 23.Stenberg Y, Julenius K, Dahlqvist I, Drakenberg T, Stenflo J. Calcium-binding properties of the third and fourth epidermal-growth-factor-like modules in vitamin-K-dependent protein S. Eur. J. Biochem. 1997;248:163–170. doi: 10.1111/j.1432-1033.1997.t01-1-00163.x. [DOI] [PubMed] [Google Scholar]
- 24.Rezende SM, Lane DA, Mille-Baker B, Samama MM, Conard J, Simmonds RE. Protein S Gla-domain mutations causing impaired Ca(2+)-induced phospholipid binding and severe functional protein S deficiency. Blood. 2002;100:2812–2819. doi: 10.1182/blood-2002-03-0909. [DOI] [PubMed] [Google Scholar]
- 25.Nicolaes GA, Tans G, Thomassen MC, Hemker HC, Pabinger I, Varadi K, Schwarz HP, Rosing J. Peptide bond cleavages and loss of functional activity during inactivation of factor Va and factor VaR506Q by activated protein C. J. Biol. Chem. 1995;270:21158–21166. doi: 10.1074/jbc.270.36.21158. [DOI] [PubMed] [Google Scholar]
- 26.Linse S, Brodin P, Drakenberg T, Thulin E, Sellers P, Elmdén K, Grundström T, Forsén S. Structure-function relationships in EF-hand Ca2+-binding proteins. Protein engineering and biophysical studies of calbindin D9k. Biochemistry. 1987;26:6723–6735. doi: 10.1021/bi00395a023. [DOI] [PubMed] [Google Scholar]
- 27.André I, Linse S. Measurement of Ca2+-binding constants of proteins and presentation of the CaLigator software. Analyt. Biochem. 2002;305:195–205. doi: 10.1006/abio.2002.5661. [DOI] [PubMed] [Google Scholar]
- 28.Berggård T, Julenius K, Ogard A, Drakenberg T, Linse S. Fragment complementation studies of protein stabilization by hydrophobic core residues. Biochemistry. 2001;40:1257–64. doi: 10.1021/bi0014812. [DOI] [PubMed] [Google Scholar]
- 29.Persson KEM, Knobe KE, Stenflo J. An anti-EGF monoclonal antibody that detects intramolecular communication in factor IX. BBRC. 2001;286:1039–1044. doi: 10.1006/bbrc.2001.5398. [DOI] [PubMed] [Google Scholar]
- 30.Sasaki T, Knyazev PG, Cheburkin Y, Gohring W, Tisi D, Ullrich A, Timpl R, Hohenester E. Crystal structure of a C-terminal fragment of growth arrest-specific protein Gas6. Receptor tyrosine kinase activation by laminin G-like domains. J. Biol. Chem. 2002;277:44164–44170. doi: 10.1074/jbc.M207340200. [DOI] [PubMed] [Google Scholar]
- 31.Morgan WD, Birdsall B, Frenkiel TA, Gradwell MG, Burghaus PA, Syed SEH, Uthaipibull C, Holder AA, Feeney J. Solution structure of an EGF module pair from the Plasmodium falciparum merozoite surface protein 1. J. Mol. Biol. 1999;289:113–122. doi: 10.1006/jmbi.1999.2753. [DOI] [PubMed] [Google Scholar]
- 32.Drakenberg T, Ghasriani H, Thulin E, Thamlitz A-M, Muranyi A, Annila A, Stenflo J. Solution structure of the Ca2+-binding EGF 3-4 pair from vitamin K-dependent protein S: identification of an unusual fold in EGF3. Biochemistry. 2005;44:8782–8789. doi: 10.1021/bi050101f. [DOI] [PubMed] [Google Scholar]
- 33.Dinchuk JE, Focht RJ, Kelley JA, Henderson NL, Zolotarjova NI, Wynn R, Neff NT, Link J, Huber RM, Burn TC, Rupar MJ, Cunningham MR, Selling BH, Ma J, Stern AA, Hollis GF, Stein RB, Friedman PA. Absence of post-translational aspartyl beta-hydroxylation of epidermal growth factor domains in mice leads to developmental defects and an increased incidence of intestinal neoplasia. J. Biol. Chem. 2002;277:12970–12977. doi: 10.1074/jbc.M110389200. [DOI] [PubMed] [Google Scholar]
- 34.Selander Sunnerhagen M, Persson E, Dahlqvist I, Drakenberg T, Stenflo J, Mayhew M, Robin M, Handford PA, Tilley JW, Campbell ID, Brownlee GG. The effect of aspartate hydroxylation on calcium binding to epidermal growth factor-like modules in coagulation factors IX and X. J. Biol. Chem. 1993;268:23339–23344. [PubMed] [Google Scholar]
- 35.Dahlbäck B, Hildebrand B, Linse S. Novel type of very high affinity calcium-binding sites in beta-hydroxyasparagine-containing epidermal growth factor-like domains in vitamin K-dependent protein S. J. Biol. Chem. 1990;265:18481–18489. [PubMed] [Google Scholar]
- 36.He X, Shen L, Malmborg AC, Smith KJ, Dahlbäck B, Linse S. Binding site for C4b-binding protein in vitamin K-dependent protein S fully contained in carboxy-terminal laminin-G-type repeats. A study using recombinant factor IX-protein S chimeras and surface plasmon resonance. Biochemistry. 1997;36:3745–3754. doi: 10.1021/bi962315q. [DOI] [PubMed] [Google Scholar]
- 37.Grishkovskaya I, Avvakumov GV, Sklenar G, Dales D, Hammond GL, Muller YA. Crystal structure of human sex hormone-binding globulin: steroid transport by a laminin G-like domain. EMBO J. 2000;19:504–512. doi: 10.1093/emboj/19.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persson KEM, Astermark J, Björk I, Stenflo J. Calcium binding to the first EGF-like module of human factor IX in a recombinant fragment containing residues 1-85. Mutations V46E and Q50E each manifest a negligible increase in calcium affinity. FEBS. 1998;421:100–104. doi: 10.1016/s0014-5793(97)01546-9. [DOI] [PubMed] [Google Scholar]
- 39.Stenflo J. Contributions of Gla and EGF-like domains to the function of vitamin K-dependent coagulation factors. Crit. Rev. Eucar. Gene. Expr. 1999;9:59–88. [PubMed] [Google Scholar]
- 40.Malby S, Pickering R, Saha S, Smallridge R, Linse S, Downing AK. The first epidermal growth factor-like domain of the low-density lipoprotein receptor contains a noncanonical calcium binding site. Biochemistry. 2001;27:2555–2563. doi: 10.1021/bi002322l. [DOI] [PubMed] [Google Scholar]
- 41.Mille-Baker B, Rezende SM, Simmonds RE, Mason PJ, Lane DA, Laffan MA. Deletion or replacement of the second EGF-like domain of protein S results in loss of APC cofactor activity. Blood. 2003;101:1416–1418. doi: 10.1182/blood-2002-08-2353. [DOI] [PubMed] [Google Scholar]