The C-terminal catalytic domain of P. multocida toxin, which is the virulence factor of the organism in P. multocida, has been expressed, purified and subsequently crystallized using the sitting-drop vapour-diffusion technique.
Keywords: Pasteurella multocida toxin, catalytic domain, progressive atrophic rhinitis
Abstract
The C-terminal catalytic domain of Pasteurella multocida toxin, which is the virulence factor of the organism in P. multocida, has been expressed, purified and subsequently crystallized using the sitting-drop vapour-diffusion technique. Native diffraction data to 1.9 Å resolution were obtained at the BL44XU beamline of SPring-8 from a flash-frozen crystal at 100 K. The crystals belong to space group C2, with unit-cell parameters a = 111.0, b = 150.4, c = 77.1 Å, β = 105.5°, and are likely to contain one C-PMT (726 residues) per asymmetric unit.
1. Introduction
Pasteurella multocida, a causative agent of progressive atrophic rhinitis in pigs, produces a protein toxin (P. multocida toxin; PMT) with a molecular weight of 146 kDa (Buys et al., 1990 ▶; Lax et al., 1990 ▶; Nakai et al., 1984 ▶; Petersen, 1990 ▶). Several lines of evidence have shown that PMT is one of the major virulence factors causing turbinate atrophy in atrophic rhinitis (Dominick & Rimler, 1986 ▶, 1988 ▶; Elias et al., 1990 ▶; Felix et al., 1992 ▶; Kimman et al., 1987 ▶; Sterner-Kock et al., 1995 ▶). PMT is also known to be a potent mitogen in various types of cells (Higgins et al., 1992 ▶; Ohnishi et al., 1998 ▶; Rozengurt et al., 1990 ▶; Seo et al., 2000 ▶; Zywietz et al., 2001 ▶). Previous studies have shown that the cellular effects of PMT are mediated by at least two different types of GTPases, Gq and G12/13 (Lax & Grigoriadis, 2001 ▶; Orth et al., 2003 ▶, 2004 ▶, 2005 ▶; Seo et al., 2000 ▶). PMT-treated cells have shown increases in inositol 1,4,5-triphosphate and diacylglycerol levels, Ca2+ mobilization and activation of protein kinase C (Higgins et al., 1992 ▶; Murphy & Rozengurt, 1992 ▶; Rozengurt et al., 1990 ▶; Staddon et al., 1990 ▶, 1991 ▶). A PMT-induced Ca2+-dependent Cl− current in Xenopus oocytes could be inhibited by antibodies against PLCβ1 and α-subunits of heterotrimeric GTPase Gq and G11 (Wilson et al., 1997 ▶). These findings clearly indicate the involvement of PLCβ activation in the actions of PMT. However, PMT is also known to cause the formation of stress fibres and focal adhesions and tyrosine phosphorylations of focal adhesion kinase and paxillin, both of which are localized at the focal adhesions (Essler et al., 1998 ▶; Lacerda et al., 1996 ▶; Zywietz et al., 2001 ▶). These effects of the toxin could be blocked by C3 exoenzyme, an inhibitor of RhoA function, indicating that RhoA is involved in these toxic actions. RhoA was activated and stress-fibre formation was induced by PMT in Gq/11 double-deficient fibroblasts (Zywietz et al., 2001 ▶; Orth et al., 2005 ▶). PMT-induced RhoA activation was inhibited by not only a regulator of G protein signalling 2 (RGS2) and RGS16, but also by lscRGS and dominant negative G13GA, indicating the involvement of G12/13 as well as Gq in the effect of PMT on RhoA (Orth et al., 2005 ▶). Thus, PMT stimulates the two different signalling pathways through the heterotrimeric GTPase Gq and G12/13. However, the real target molecules of the toxin and the nature of its molecular action remain unknown.
PMT consists of 1285 amino-acid residues. It is generally accepted that the toxin is structured as a typical AB toxin, composed of a catalytic active A subunit and a B subunit responsible for binding to the cell-surface receptor. Initial studies demonstrated that the N-terminus of PMT is involved in the binding and translocation of the toxin into target cells (Pullinger et al., 2001 ▶), whereas the C-terminal region of PMT has been found to carry the putative toxic effect because it retains the ability to stimulate DNA synthesis and PLC activation (Busch et al., 2001 ▶; Orth et al., 2003 ▶; Pullinger et al., 2001 ▶; Ward et al., 1998 ▶). At its N-terminus PMT shows a significant sequence similarity to the N-terminal part of the cytotoxic necrotizing factor (CNF) of Escherichia coli, which is involved in binding and translocation. Moreover, three amino acids, Cys1165, His1205 and His1223, which are essential for the toxic activity of PMT were identified at the C-terminus (Ward et al., 1998 ▶; Orth et al., 2003 ▶). These findings indicate that the C-terminus of PMT is the pivotal catalytic domain.
To clarify the structural bases for the activity of PMT, which have not yet been identified, we present here the crystallization and preliminary X-ray diffraction analysis of the C-terminal catalytic domain of the P. multocida toxin.
2. Methods and results
2.1. Cloning
2.2 kbp of PMT DNA fragment digested with SnaBI and NotI from pSN131 was cloned into the StuI and NotI residues 569–1285 of the PMT sequence) with an initial methionine and a 28-amino-acid spacer sequence including a six-histidine tag sequence (MGHHHHHHDYDIPTTENLYFQGAHMGIQR) at the N-terminal of the recombinant protein. Finally, His-tagged C-PMT was cloned and expressed in E. coli BL21-CodonPlus (DE3)-RIL (Stratagene).
2.2. Purification of His-C-PMT
50 ml of an overnight culture of E. coli BL21-CodonPlus (DE3)-RIL cells transformed with pPROEX-1-C-PMT was inoculated into 5 l of LB medium with 50 µg ml−1 ampicillin and 50 µg ml−1 chloramphenicol. The culture was grown at 310 K with vigorous shaking until the Abs600nm reached 0.6. Recombinant protein expression was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside (IPTG) to the culture to a final concentration of 1 mM. 6 h after induction, the cells were harvested by centrifugation at 5000 rev min−1 for 20 min at 277 K, washed with ice-cold deionized distilled water, suspended in 50 mM sodium phosphate buffer pH 8.0 including 0.3 M NaCl and 1 mM imidazole and then disrupted by sonication using a S-350 sonifier (Branson) at 273 K. The disrupted cell suspension was centrifuged at 12 000 rev min−1 for 20 min at 277 K in order to pellet insoluble material. The supernatant was purified by nickel–NTA affinity agarose gel (HIS-Select Nickel Affinity Gel, Sigma) column chromatography and the soluble recombinant protein was then eluted with an imidazole concentration gradient (1–500 mM). The C-PMT fraction was collected and diluted tenfold with 10 mM Tris–HCl pH 8.0. Further purification was carried out by DEAE Sephacel (GE Healthcare) column chromatography with a NaCl concentration gradient (0–1.0 M). Finally, C-PMT was purified by gel-filtration chromatography on a Sephacryl S-200 (GE Healthcare) column equilibrated and eluted with 10 mM Tris–HCl pH 8.0 including 0.2 M NaCl. The protein concentration was determined with the BCA protein-assay kit (Pierce) using bovine serum albumin as a standard. The C-PMT fraction was concentrated to 15 mg ml−1 using a Vivaspin 20 (Sartorius) and sterile-filtrated using 0.1 µm Ultrafree-MC (Millipore). The homogeneity of the purified preparation was confirmed by 12.5% SDS–PAGE and native PAGE.
2.3. Crystallization
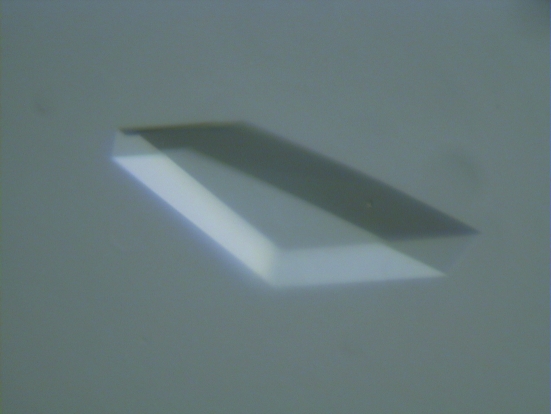
For crystallization experiments, a 15 mg ml−1 solution of the recombinant C-PMT in 0.01 M Tris–HCl pH 7.5 was employed. Crystallization trials were set up at room temperature as sitting-drop vapour-diffusion experiments on Linbro crystallization plates. Initial screening was performed using the sparse-matrix method (Jancarik & Kim, 1991 ▶) with commercial crystal screening kits (Hampton Research). Crystals of the recombinant C-PMT proteins were readily grown from polyethylene glycol solutions, as well as under different physico-chemical conditions (ammonium phosphate, ammonium citrate, potassium/sodium phosphate), yielding isomorphous crystal forms. The best crystals were obtained through equilibration against a solution containing 1.6 M ammonium phosphate and 0.1 M MES buffer pH 6.5 at 293 K in sitting-drop vapour-diffusion setups. The crystallization droplets consisted of 2 µl protein solution and 2 µl reservoir solution and were equilibrated against 500 µl reservoir solution; triangle-shaped crystals appeared within a few days and grew to maximal dimensions of about 0.5 × 0.5 ×0.1 mm (Fig. 1 ▶).
Figure 1.
Crystal of C-PMT. Results from a typical C-PMT crystal-growth droplet. The crystal displayed measures approximately 0.3 × 0.5 × 0.1 mm.
2.4. Data collection and processing
X-ray diffraction data were collected from C-PMT crystals at 100 K in a nitrogen stream, supplementing the mother-liquor solution with 30% trehalose as cryoprotectant. Initially, X-ray diffraction data were collected from the crystals of the C-terminal domain of PMT using a Jupiter 210 CCD detector system on beamline BL38B1 at SPring-8 (Harima, Japan). Subsequently, a high-resolution native data set at 1.90 Å was collected from a single crystal on beamline BL44XU at SPring-8 (Table 1 ▶). The X-ray wavelength was 0.9 Å, the oscillation angle was 1.0° and the crystal-to-detector distance was 300 mm. Analysis of the diffraction pattern and of the systematic absences allowed assignment of C-PMT crystals to the centred monoclinic space group C2, with unit-cell parameters a = 111.0, b = 150.4, c = 77.1 Å, β = 105.5° (α and γ are defined by the Bravais system; see Table 1 ▶). A total of 93 581 unique reflections were obtained using the HKL-2000 program package (Otwinowski & Minor, 1997 ▶). Intensity data in the resolution range 50.0–1.9 Å were processed with an R merge of 9.7%. Assuming a molecular weight of 80.833 kDa for the expressed C-PMT domain, packing-density calculations indicate the most probable value for V M to be 3.80 Å3 Da−1, with one C-PMT chain per asymmetric unit. This corresponds to a solvent fraction of about 67.7%, a typical value for protein crystals (Matthews, 1968 ▶). Amino-acid sequence searches did not highlight any structural homology with proteins of known three-dimensional structure. Therefore, the crystallographic analysis of C-PMT will be based on the multiple isomorphous replacement method or the MAD method; a search for heavy-atom derivatives is presently in progress.
Table 1. Data-collection statistics for C-PMT native data sets.
Values in parentheses refer to the outer resolution shell.
| Native 1 | Native 2 | |
|---|---|---|
| Beamline | BL38B1 | BL44XU |
| Wavelength (Å) | 1.00 | 0.90 |
| Space group | C2 | C2 |
| Unit-cell parameters | ||
| a (Å) | 112.4 | 111.0 |
| b (Å) | 150.5 | 150.4 |
| c (Å) | 78.1 | 77.1 |
| β (°) | 105.0 | 105.5 |
| Resolution (Å) | 50–2.6 (2.69–2.60) | 50–1.9 (1.97–1.90) |
| Total reflections | 136030 | 335746 |
| Unique reflections | 36642 | 93581 |
| I/σ(I) | 4.1 (7.3) | 5.8 (13.0) |
| Rmerge† (%) | 5.7 (21.9) | 9.7 (44.8) |
| Completeness (%) | 94.7 (74.4) | 97.3 (81.3) |
R
merge = 100
 , where 〈I〉h is the mean intensity of all symmetry-related reflections I
h.
, where 〈I〉h is the mean intensity of all symmetry-related reflections I
h.
Acknowledgments
We are grateful to all members of beamlines BL44XU and BL38B1 at SPring-8 and NW-12 and BL-5A at Photon Factory for help in collecting data. We thank Dr Nagai for pSN131 and Ms Hattori for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Science and Technology of Japan.
References
- Busch, C., Orth, J., Djouder, N. & Aktories, K. (2001). Infect. Immun.69, 3628–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buys, W. E., Smith, H. E., Kamps, A. M., Kamp, E. M. & Smits, M. A. (1990). Nucleic Acids Res.18, 2815–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominick, M. A. & Rimler, R. B. (1986). Am. J. Vet. Res.47, 1532–1536. [PubMed] [Google Scholar]
- Dominick, M. A. & Rimler, R. B. (1988). Vet. Pathol.25, 17–27. [DOI] [PubMed] [Google Scholar]
- Elias, B., Boros, G., Albert, M., Tuboly, S., Gergely, P., Papp, L., Barna Vetro, I., Rafai, P. & Molnar, E. (1990). Nippon Juigaku Zasshi, 52, 677–688. [DOI] [PubMed] [Google Scholar]
- Essler, M., Hermann, K., Amano, M., Kaibuchi, K., Heesemann, J., Weber, P. C. & Aepfelbacher, M. (1998). J. Immunol.161, 5640–5646. [PubMed] [Google Scholar]
- Felix, R., Fleisch, H. & Frandsen, P. L. (1992). Infect. Immun.60, 4984–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, T. E., Murphy, A. C., Staddon, J. M., Lax, A. J. & Rozengurt, E. (1992). Proc. Natl Acad. Sci. USA, 89, 4240–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Kimman, T. G., Lowick, C. W., van de Wee-Pals, L. J., Thesingh, C. W., Defize, P., Kamp, E. M. & Bijvoet, O. L. (1987). Infect. Immun.55, 2110–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda, H. M., Lax, A. J. & Rozengurt, E. (1996). J. Biol. Chem.271, 439–445. [DOI] [PubMed] [Google Scholar]
- Lax, A. J., Chanter, N., Pullinger, G. D., Higgins, T., Staddon, J. M. & Rozengurt, E. (1990). FEBS Lett.277, 59–64. [DOI] [PubMed] [Google Scholar]
- Lax, A. J. & Grigoriadis, A. E. (2001). Int. J. Med. Microbiol.291, 261–268. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Nakai, T., Sawata, A., Tsuji, M., Samejima, Y. & Kume, K. (1984). Infect. Immun.46, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A. C. & Rozengurt, E. (1992). J. Biol. Chem.267, 25296–25303. [PubMed] [Google Scholar]
- Ohnishi, T., Horiguchi, Y., Masuda, M., Sugimoto, N. & Matsuda, M. (1998). J. Vet. Med. Sci.60, 301–305. [DOI] [PubMed] [Google Scholar]
- Orth, J. H., Blocker, D. & Aktories, K. (2003). Biochemistry, 42, 4971–4977. [DOI] [PubMed] [Google Scholar]
- Orth, J. H., Lang, S. & Aktories, K. (2004). J. Biol. Chem.279, 34150–34155. [DOI] [PubMed] [Google Scholar]
- Orth, J. H., Lang, S., Taniguchi, M. & Aktories, K. (2005). J. Biol. Chem.280, 36701–36707. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Petersen, S. K. (1990). Mol. Microbiol.4, 821–830. [DOI] [PubMed] [Google Scholar]
- Pullinger, G. D., Sowdhamini, R. & Lax, A. J. (2001). Infect. Immun.69, 7839–7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozengurt, E., Higgins, T., Chanter, N., Lax, A. J. & Staddon, J. M. (1990). Proc. Natl Acad. Sci. USA, 87, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, B., Choy, E. W., Maudsley, S., Miller, W. E., Wilson, B. A. & Luttrell, L. M. (2000). J. Biol. Chem.275, 2239–2245. [DOI] [PubMed] [Google Scholar]
- Staddon, J. M., Barker, C. J., Murphy, A. C., Chanter, N., Lax, A. J., Michell, R. H. & Rozengurt, E. (1991). J. Biol. Chem.266, 4840–4847. [PubMed] [Google Scholar]
- Staddon, J. M., Chanter, N., Lax, A. J., Higgins, T. E. & Rozengurt, E. (1990). J. Biol. Chem.265, 11841–11848. [PubMed] [Google Scholar]
- Sterner-Kock, A., Lanske, B., Uberschar, S. & Atkinson, M. J. (1995). Vet. Pathol.32, 274–279. [DOI] [PubMed] [Google Scholar]
- Ward, P. N., Miles, A. J., Sumner, I. G., Thomas, L. H. & Lax, A. J. (1998). Infect. Immun.66, 5636–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, B. A., Zhu, X., Ho, M. & Lu, L. (1997). J. Biol. Chem.272, 1268–1275. [DOI] [PubMed] [Google Scholar]
- Zywietz, A., Gohla, A., Schmelz, M., Schultz, G. & Offermanns, S. (2001). J. Biol. Chem.276, 3840–3845. [DOI] [PubMed] [Google Scholar]