The structure of an uncomplexed form of α-amylase from B. halmapalus is compared with a form in which maltose, glucose and a nonasaccharide derived from acarbose and maltose are bound.
Keywords: α-amylase, maltose-binding site, Bacillus halmapalus
Abstract
Recombinant Bacillus halmapalus α-amylase (BHA) was studied in two different crystal forms. The first crystal form was obtained by crystallization of BHA at room temperature in the presence of acarbose and maltose; data were collected at cryogenic temperature to a resolution of 1.9 Å. It was found that the crystal belonged to space group P212121, with unit-cell parameters a = 47.0, b = 73.5, c = 151.1 Å. A maltose molecule was observed and found to bind to BHA and previous reports of the binding of a nonasaccharide were confirmed. The second crystal form was obtained by pH-induced crystallization of BHA in a MES–HEPES–boric acid buffer (MHB buffer) at 303 K; the solubility of BHA in MHB has a retrograde temperature dependency and crystallization of BHA was only possible by raising the temperature to at least 298 K. Data were collected at cryogenic temperature to a resolution of 2.0 Å. The crystal belonged to space group P212121, with unit-cell parameters a = 38.6, b = 59.0, c = 209.8 Å. The structure was solved using molecular replacement. The maltose-binding site is described and the two structures are compared. No significant changes were seen in the structure upon binding of the substrates.
1. Introduction
α-Amylases (α-1,4-glucan-4-glucanohydrolases; EC 3.2.1.1) are monomeric enzymes that catalyse the hydrolysis of the internal α-1,4-glycosidic bond in starch and related oligosaccharides and polysaccharides. These enzymes are used primarily in the food, detergent and pharmaceutical industries. Most α-amylases belong to the sequence-related glycoside hydrolase family 13 (GH13) in the CAZY database (Coutinho & Henrissat, 1999 ▶; http://www.cazy.org/CAZY/index.html). To date, GH13 contains around 2593 members, for which there are 215 PDB entries (Berman et al., 2000 ▶). These 215 PDB entries cover the structures of 45 members of GH13, which all share a central (β/α)8 TIM-barrel domain with a loop domain inserted between the third β-strand and the third α-helix. The structure of recombinant Bacillus halmapalus α-amylase crystallized in the presence of acarbose and maltose has previously been solved (PDB code 1w9x; Davies et al., 2005 ▶). In this structure, it was found that acarbose had been involved in a transglycosylation reaction as well as an elongation mechanism, in which maltose or other maltooligosaccharides were acceptors, thereby forming a nonasaccharide which was bound to BHA.
B. halmapalus α-amylase has a catalytic core domain (residues 5–110 and 212–398) consisting of a (β/α)8-barrel. Between the third β-strand and the third α-helix, an extended loop domain is present (residues 111–211). The C-terminal domain (residues 399–485) is an eight-stranded β-sheet domain. The active site is situated at the bottom of the substrate-binding cleft, which is positioned at the interface between the (β/α)8-barrel and the extended loop.
In this present study, we have determined the crystal structure of BHA without any substrate analogues bound at cryogenic temperature. Furthermore, we reproduced the previously reported substrate-bound crystal form of BHA and collected data at cryogenic temperature in order to permit direct comparisons of the structures with and without substrates bound. An earlier study on the structure of a maltohexaose-producing amylase (G6-amylase) from the alkalophilic Bacillus sp. 707, which shares 85% sequence identity with BHA (Kanai et al., 2004 ▶), has established that Asp236 is the nucleophile catalyst and Glu266 is a proton donor/acceptor. Kanai et al. (2004 ▶) reported that a change in hydrogen-bonding pattern was observed for the catalytically important Glu266 residue upon substrate binding. In contrast, we did not observe any significant structural differences upon binding the substrate. Our findings indicate for the first time, however, that in addition to the nonasaccharide, a maltose molecule and two glucose molecules are also bound to the protein.
2. Experimental
2.1. Protein expression and purification
Recombinant B. halmapalus α-amylase (EC 3.2.1.1) was provided by Novozymes A/S, Bagsvaerd, Denmark. The amylase was expressed in B. subtilis and purified by Novozymes A/S.
2.2. Crystallization of BHA in the presence of acarbose and maltose
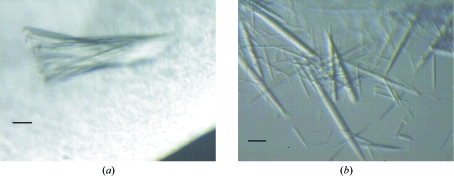
Crystals were grown at room temperature by the sitting-drop vapour-diffusion method (McPherson, 1982 ▶) using BHA solutions containing 10 mg ml−1 BHA, 20 mM Tris pH 7.5 and 1 mM CaCl2 (Davies et al., 2005 ▶). The reservoir solution contained 25 mM acarbose, 18%(w/v) PEG 5000, 100 mM maltose and 6.4 mM Tris pH 7.5. The drop consisted of 2 µl BHA solution and 4 µl reservoir solution and was equilibrated over a 1000 µl reservoir. Crystals suitable for X-ray diffraction formed within two weeks (see Fig. 1 ▶ a).
Figure 1.
Light microscopy showing (a) the assembly of crystals when BHA was crystallized in the presence of acarbose and (b) crystals of BHA crystallized in the absence of substrate analogues. The scale in the pictures corresponds to 50 µm.
2.3. Crystallization of BHA in the absence of substrate analogues
From previous experiments, it is known that pH-induced crystallization of BHA without substrate analogues is possible. A MES–HEPES–boric acid buffer was chosen to solubilize the protein owing to its high buffering capacity over a broad pH range. The pH of the MHB buffer was adjusted using NaOH and crystallization was induced by lowering the pH with acetic acid. Trial experiments showed that crystallization was only possible at a temperature of 298 K or more, since the solubility of BHA in MHB increased with decreasing temperature.
Crystals were grown at 303 K by the sitting-drop vapour-diffusion method using BHA solutions consisting of 42 mg ml−1 BHA, 10 mM MHB pH 9.5 and 5 mM CaCl2. The reservoir solution contained 10 mM MHB pH 7.5. The drop consisted of 2 µl BHA solution, 0.6 µl 87 mM acetic acid and 4 µl reservoir solution and was equilibrated over a 1000 µl reservoir. Crystals suitable for X-ray diffraction formed within 2 d (see Fig. 1 ▶ b).
2.4. Data collection and processing
Diffraction data from the two crystal forms were collected at I911-3, MAX-lab (Lund, Sweden) at cryogenic temperature. A piece of the crystal assembly shown in Fig. 1 ▶(a) was mounted with 20% glycerol added to the reservoir. The uncomplexed crystal form was cryocooled in a solution consisting of the reservoir buffer with 35% glycerol added. Integration, scaling and merging of the intensities were carried out using XDS and XSCALE (Kabsch, 1993 ▶). Data points around ice rings were removed for the uncomplexed crystal form. This is the reason for the low completeness of these data. Data-collection statistics are presented in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the outermost resolution shell.
| Complexed crystal form | Uncomplexed crystal form | |
|---|---|---|
| Beamline | I911-3, MAX-lab | I911-3, MAX-lab |
| Wavelength (Å) | 1.2 | 1.0 |
| Data-collection temperature (K) | 120 | 120 |
| Resolution (Å) | 15.0–1.9 (2.0–1.9) | 20.0–2.1 (2.2–2.1) |
| No. of reflections | 254373 (34311) | 177770 (23604) |
| No. of unique reflections | 41708(6792) | 26081 (3664) |
| Completeness (%) | 98.9 (98.0) | 89.9 (98.6) |
| Rsym | 0.148 (0.618) | 0.155 (0.374) |
| Crystal mosaicity (°) | 0.45 | 0.15–0.4 |
| I/σ(I) | 7.99 (2.43) | 10.24 (5.15) |
| Crystal system | Orthorhombic | Orthorhombic |
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å) | a = 47.0, b = 73.5, c = 151.1 | a = 38.6, b = 59.0, c = 209.8 |
| VM (Å3 Da−1) | 2.36 | 2.16 |
| Solvent content (%) | 48 | 43 |
| Redundancy | 6.1 (5.1) | 6.7 (6.4) |
2.5. Structure determination and refinement
Both structures were refined using REFMAC5 (Murshudov et al., 1997 ▶). In between refinement cycles, the model and the σA-weighted 2F obs − F calc and F obs − F calc electron-density maps were inspected using the program O v.9.0.7 (Jones et al., 1991 ▶) in order to help identify which alterations were required. To evaluate the refinement progress using the R free/R factor, 5% of the reflections were set aside. Water molecules were added at the end of the refinement process using the automatic ARP/wARP (Lamzin & Wilson, 1993 ▶) procedure in REFMAC5. Afterwards, they were checked manually in O and water molecules without significant electron density were removed. The structures were validated using PROCHECK (Laskowski et al., 1993 ▶). The first four residues did not show up in the electron-density maps and have therefore not been included in the two structures. Coordinates and structure factors for both structures have been deposited with the PDB. The refinement statistics are given in Table 2 ▶.
Table 2. Refinement statistics for the two BHA crystal forms studied.
| Complexed crystal form | Uncomplexed crystal form | |
|---|---|---|
| Resolution range (Å) | 15.0–1.9 | 20.0–2.1 |
| Overall R | 0.172 | 0.199 |
| Rfree | 0.214 | 0.242 |
| No. of reflections | 39523 | 26413 |
| Free reflections (%) | 5 | 5 |
| No. of atoms | ||
| Peptide chain | 3940 (485 residues) | 3915 (485 residues) |
| Na+ ions | 1 | 1 |
| Ca2+ ions | 3 | 3 |
| Nonasaccharide | 100 | |
| Maltose | 23 | |
| Glucose | 22 | |
| Water O atoms | 215 | 271 |
| Acetate ions | 4 | |
| Average B factors (Å2) | ||
| Peptide main chain | 17.0 | 13.0 |
| Peptide side chain | 19.4 | 13.3 |
| Metal ions | 12.4 | 12.2 |
| Nonasaccharide | 33.7 | |
| Maltose | 24.8 | |
| Glucose | 53.3 | |
| Acetate | 25.0 | |
| Solvent | 26.7 | 19.9 |
2.5.1. The complexed crystal form
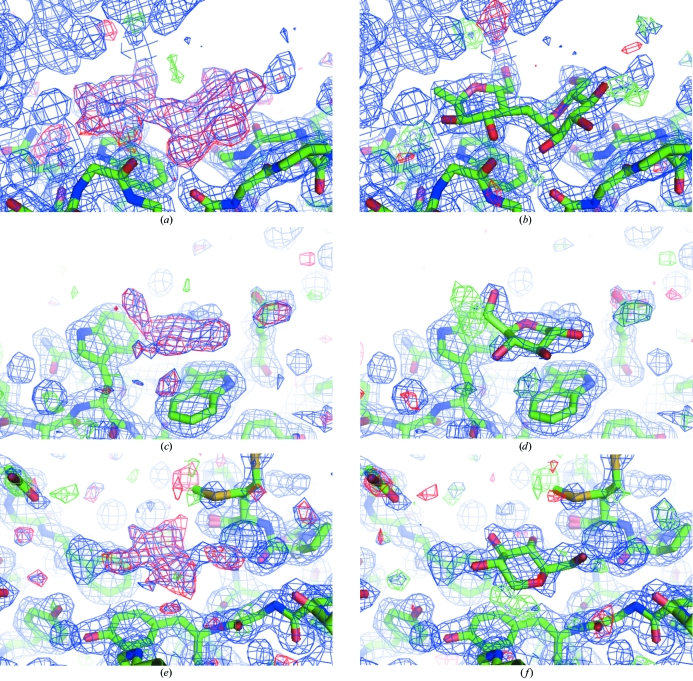
The structure of the complexed crystal form had already been solved by Davies et al. (2005 ▶), which gave a good starting point for further refinement of the structure using our low-temperature data. The refinement was started using PDB code 1w9x with all water molecules removed. 1w9x contains an alanine at position 485. However, the F obs − F calc map contoured at 3σ showed clear density for the side chain of Arg485, so it could be incorporated completely. Furthermore, the residues Gln22, Met323 and Asn283 were modelled in alternate conformations. Clear positive electron density was identified in the difference density maps at several places on the surface of the molecule. At the interface between the (β/α)8-barrel domain and the β-sheet domain this electron density was in accordance with a maltose molecule and could be modelled as such (see Figs. 2 ▶ a and 2 ▶ b) and at the surface of the β-sheet domain and at the bottom of the (β/α)8-barrel domain two glucose molecules were modelled (see Figs. 2 ▶ c, 2 ▶ d, 2 ▶ e and 2 ▶ f). In the Ramachandran plot Tyr152 was found outside the allowed regions, as also observed by Davies et al. (2005 ▶). Furthermore, Tyr203 was in the generously allowed region. Both residues were associated with clear electron density.
Figure 2.
Difference density maps before (left) and after (right) insertion of the maltose and the glucose molecules. The σA-weighted 2F obs − F calc electron-density map is in blue and is contoured at 1σ. The σA-weighted F obs − F calc electron density in red is contoured at 3σ and in green is contoured at −3σ. (a) and (b) maltose, (c) and (d) glucose near Trp439 and Trp469 and (e) and (f) glucose near Tyr363. The figure was produced using PyMOL (DeLano, 2002 ▶).
2.5.2. The uncomplexed crystal form
The structure of the uncomplexed crystal form was solved by molecular replacement using the program MOLREP (Vagin & Teplyakov, 1997 ▶), which belongs to the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The peptide chain of the complexed crystal form was used as a search model and a solution was found in space group P212121 with one molecule in the asymmetric unit (α, β, γ, t x, t y, t z) = (114.07°, 54.56°, 64.55°, 0.207, 0.017, 0.141) (R = 0.44, Corr = 0.51). During refinement an acetate ion was incorporated in the model. As was seen for the complexed crystal form, Tyr152 was outside the allowed regions in the Ramachandran plot. Ser342 was found in the generously allowed region in the Ramachandran plot. These residues were associated with clear electron density.
3. Results and discussion
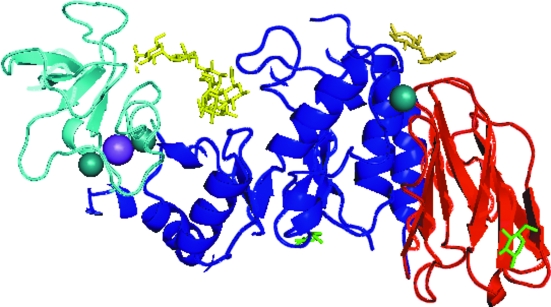
The tertiary structure of BHA in which the nonasaccharide, maltose and glucose are bound is shown in Fig. 3 ▶, with a close-up of the region where the nonasaccharide is positioned in Fig. 4 ▶ and close-ups of the region where maltose is bound in Fig. 5 ▶.
Figure 3.
The tertiary structure of B. halmapalus α-amylase in the complexed crystal form. The N-terminal (β/α)8-barrel domain is shown in blue, the loop domain in turquoise and the C-terminal β-sheet domain in red. The nonasaccharide and maltose are shown as yellow sticks and the two glucose molecules are shown as green sticks. Metal ions are shown as spheres. The figure was produced using PyMOL (DeLano, 2002 ▶).
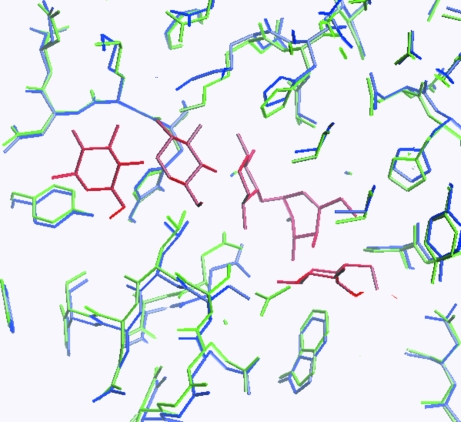
Figure 4.
Part of the nonasaccharide-binding site. The two crystal forms are superimposed. The complexed crystal form is coloured blue, the uncomplexed crystal form is green and the nonasaccharide is red. The figure illustrates that the nonasaccharide is wedged into the structure, forcing apart the structure of the complexed crystal form. The figure was produced using O (Jones et al., 1991 ▶).
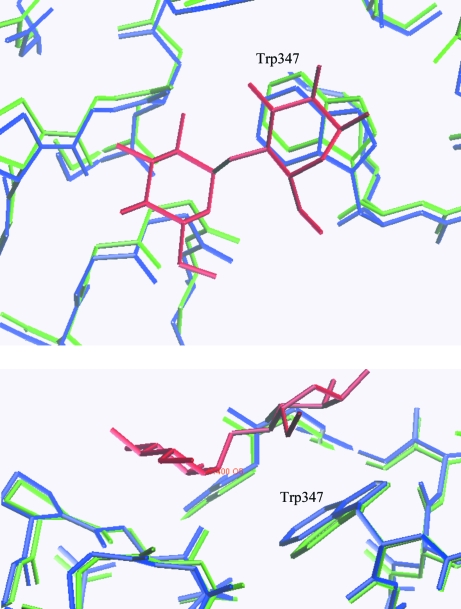
Figure 5.
Maltose (red) shown in the complexed crystal structure (blue). The uncomplexed crystal form is superimposed (green). The illustration was produced using O (Jones et al., 1991 ▶).
As described by Davies et al. (2005 ▶), the nonasaccharide binds at the active site at the interface between the (β/α)8-barrel domain and the loop domain. Also observed by Davies et al. (2005 ▶) were two calcium ions and a sodium ion between the (β/α)8-barrel domain and the loop domain and a third calcium ion positioned at the interface between the (β/α)8-barrel domain and the β-sheet domain.
In addition to the nonasaccharide, we found a maltose bound to the surface at the interface between the (β/α)8-barrel domain and the β-sheet domain. In Table 3 ▶ the hydrogen-bond network of this maltose molecule is listed. Hydrogen bonds are seen between maltose O2 and the backbone O of Asp432 and between maltose O3 and Pro434 N. The maltose molecule mediates intermolecular contacts through a hydrogen bond between the maltose O1′ and the backbone O atom of Asn128. In Fig. 5 ▶ a close-up of the maltose-binding site is shown and it is observed that the ring systems of Trp347 and Pro434 create a platform where the sugar rings of the maltose molecule may stack. The distance between the stacking maltose ring and the tryptophan is between 3.5 and 3.9 Å, indicating a π-stack.
Table 3. The intermolecular interactions between maltose and BHA.
| Source atom | Target atom | Distance (Å) |
|---|---|---|
| Maltose O2′ | Water O46 | 2.8 |
| Maltose O2 | Asp432 O | 2.5 |
| Maltose O4 | Water O43 | 2.4 |
| Maltose O1′ | Asn128 O† | 2.5 |
| Maltose O3 | Pro434 N | 3.4 |
Interaction with Asn128 O through symmetry operation x′ = x + 1/2, y′ = −y + 1/2, z′ = −z + 1.
Two glucose molecules were incorporated in the model. One stacks on top of Trp439 and Trp469 at the outer surface of the β-sheet domain (as seen in Figs. 2 ▶ d and 3 ▶) and the other stacks on Tyr363 situated at the bottom of the (β/α)8-barrel domain (as seen in Figs. 2 ▶ f and 3 ▶). The distances between the glucose molecules and the underlying aromatic systems are between 3.6 and 4.0 Å. The thermal displacement parameters for both are rather high (∼50 Å2), which leads to some uncertainty as to the precise orientation of these molecules.
The peptide chains of the two crystal forms were superimposed using the program LSQKAB (Kabsch, 1976 ▶), giving a mean r.m.s.d. of 0.45 Å, whilst superimposing only the two (β/α)8-barrel domains gave a mean r.m.s.d. of 0.33 Å. Although the structures can be almost perfectly aligned, it can be seen in Fig. 4 ▶ that the nonasaccaride is wedged into the enzyme, forcing the complexed structure apart. While the side of the enzyme containing the loop domain (coloured turquoise in Fig. 3 ▶) is fixed, the loops containing residues 339 and 379 move approximately 1 Å upon substrate binding. A search for domain movements was therefore performed using the program HINGEFIND (Wriggers & Schulten, 1997 ▶), but no significant overall domain movements could be detected.
Interestingly, the structure of a maltohexaose-producing G6-amylase from the alkalophilic Bacillus sp. 707 (which shares 85% sequence identity with BHA) has also been solved (Kanai et al., 2004 ▶) after crystallization from 2-methylpentane-2,4-diol and phosphate at pH 8.5. The active binding sites are almost identical. The structure of G6-amylase was solved with (PDB code 1wpc) and without (PDB code 1wp6) the same nonasaccharide as found in BHA. The procedure was different, however, as in this case the complex was obtained by soaking the apo crystals in acarbose and maltotriose (Kanai et al., 2004 ▶). The authors, however, do find conformational differences in G6-amylase between the two crystal forms. Most noteworthy is the observation of a conformational change of Glu266, which is believed to be important for catalysis. In the complexed form of G6-amylase, the side-chain OE1/2 of Glu266 is hydrogen bonded to Ala237 N and to N4A and O3A in the AC1 unit at subsite +1 of the nonasaccharide. In the uncomplexed form Glu266 turns and hydrogen bonds to Asp333 OD2 and Arg234 NH2.
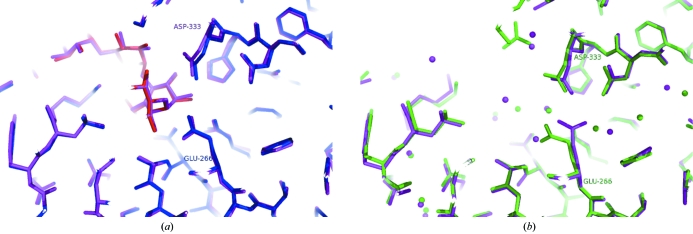
A superposition of the BHA complex structure with the complexed structure of the G6-amylase (PDB code 1wpc) is shown in Fig. 6 ▶(a). These two structures are seen to be almost identical. However, if we superimpose the two uncomplexed structures of BHA and G6-amylase differences appear, as seen in Fig. 6 ▶(b). In the uncomplexed crystal form of BHA in which a water molecule replaces the substrate, Glu266 retains the hydrogen bond to Ala237 N. For BHA we do not observe any conformational change of Glu266: it has the same conformation in both forms. According to our data it seems that substrate binding to BHA is not necessarily connected with a conformational change of Glu266. It may be in this conformation already and it cannot be ruled out that the crystallization conditions are important for these details in the structure.
Figure 6.
(a) Superposition of the alkalophilic G6-amylase (PDB code 1wcs, violet) and BHA (blue and red) with nonasaccharide. (b) The same area in G6-amylase (PDB code 1wc6, violet) and BHA (green) without substrate analogues. The figure was produced using PyMOL (DeLano, 2002 ▶).
Supplementary Material
PDB reference: α-amylase complexed with acarbose and maltose, 2gjp, r2gjpsf
PDB reference: uncomplexed, 2gjr, r2gjrsf
Acknowledgments
The authors are grateful to MAX-lab, Lund, Sweden for providing beam time for the project and to DANSYNC and the EU for contribution to travel expenses under the ‘Access to Research Infrastructure’ program.
References
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res.28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Coutinho, P. M. & Henrissat, B. (1999). Recent Advances in Carbohydrate Bioengineering, edited by H. J. Gilbert, G. Davies, B. Henrissat & B. Svensson, pp. 3–12. Cambridge: The Royal Society of Chemistry.
- Davies, G. J., Brzozowski, A. M., Dauter, Z., Rasmussen, M. D., Borchert, T. V. & Wilson, K. S. (2005). Acta Cryst. D61, 190–193. [DOI] [PubMed] [Google Scholar]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1976). Acta Cryst. A32, 922–923. [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Kanai, R., Haga, K., Akiba, T., Yamane, K. & Harata, K. (2004). Biochemistry, 43, 14047–14056. [DOI] [PubMed] [Google Scholar]
- Lamzin, V. S. & Wilson, K. S. (1993). Acta Cryst. D49, 129–147. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., Moss, D. S. & Thornton, J. M. (1993). J. Mol. Biol.231, 1049–1067. [DOI] [PubMed] [Google Scholar]
- McPherson, A. J. (1982). Preparation and Analysis of Protein Crystals. New York: Wiley.
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]
- Wriggers, W. & Schulten, K. (1997). Proteins, 29, 1–14. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: α-amylase complexed with acarbose and maltose, 2gjp, r2gjpsf
PDB reference: uncomplexed, 2gjr, r2gjrsf