The crystallization of B. cereus chitinase is reported.
Keywords: chitinase, Bacillus cereus NCTU2
Abstract
Chitinases (EC 3.2.1.14) are found in a broad range of organisms, including bacteria, fungi and higher plants, and play different roles depending on their origin. A chitinase from Bacillus cereus NCTU2 (ChiNCTU2) capable of hydrolyzing chitin as a carbon and nitrogen nutrient has been identified as a member of the family 18 glycoside hydrolases. ChiNCTU2 of molecular weight 36 kDa has been crystallized using the hanging-drop vapour-diffusion method. According to the diffraction of chitinase crystals at 1.10 Å resolution, the crystal belongs to space group P21, with unit-cell parameters a = 50.79, b = 48.79, c = 66.87 Å, β = 99.31°. Preliminary analysis indicates there is one chitinase molecule in the asymmetric unit, with a solvent content of 43.4%.
1. Introduction
Chitin, derived primarily from the exoskeletons of insects and crustaceans, is a β-1,4-linked insoluble linear polymer of N-acetylglucosamine (GlcNAc). It is not only the second most abundant organic compound on our planet after cellulose (Cohen-Kupiec & Chet, 1998 ▶), but is also the major source of amino sugars in nature. Chitin can be hydrolyzed by the chitinases produced by diverse organisms (Flach et al., 1992 ▶) and the hydrolyzed product is used as a nutrient source. Chitinases (EC 3.2.1.14) are found in a broad range of organisms, including bacteria, fungi and higher plants, and play different roles depending on their origin (Flach et al., 1992 ▶; Graham & Sticklen, 1994 ▶; Felse & Panda, 1999 ▶). Plant chitinases are involved in a mechanism to defend against infection by phytopathogenic fungi (Felse & Panda, 1999 ▶), while fungal chitinases are required for hyphal growth (Takaya et al., 1998 ▶) and bacterial chitinases are considered primarily to digest and utilize chitin as a carbon and nitrogen nutrient (Cohen-Kupiec & Chet, 1998 ▶).
Bacillus cereus is a large Gram-positive endospore-forming bacterium that is common in soil and plants (Brunel et al., 1994 ▶). In the control of plant diseases, B. cereus UW85, which is capable of producing two antibiotics responsible for suppression of disease (Silo-Suh et al., 1994 ▶), has been proven to be a reliable agent for biocontrol of Phytophthora damping off and root rot of soybean (Emmert & Handelsman, 1999 ▶). An endophytic B. cereus strain 65 producing a chitobiosidase has also been found to be effective against Rhizoctonia solani in cotton (Pleban et al., 1997 ▶). However, the role of chitobiosidase in the antagonism of B. cereus strain 65 toward fungal pathogens is not clearly understood. A chitin-degrading Bacillus strain, designated NCTU2, was screened from soil and identified. An extracellular chitinase was purified from the culture filtrate. The sequence of the first 15 N-terminal amino acids of the enzyme was determined to be ANNLGSKLLVGYWHN, which is highly homologous to that of ChiA from B. cereus. The corresponding full gene from the genomic DNA of Bacillus NCTU2 encodes a signal peptide (27 amino acids) and a mature protein (333 amino acids) (Wen & Li, 2002 ▶), which is consistent with the calculated value for the mature protein, indicating that the signal peptide can be recognized in Escherichia coli.
The chitinase, designated as ChiNCTU2, from B. cereus NCTU2 belongs to glycosyl hydrolase family 18 (Henrissat & Bairoch, 1996 ▶) and is composed of an N-terminal signal peptide and a C-terminal catalytic domain (CaD). A multiple alignment of family 18 glycosyl hydrolase sequences (including those of B. cereus ChiNCTU2, B. cereus ChiA, Serratia marcescens ChiA and six types of bacterial chitinase) shows the conservation of a (DN)-G-(LIVMF)-(DN)-(LIVMF)-(DN)-X-E motif (residues 308–315 in S. marcescens ChiA). The sequence alignment also showed that a tyrosine residue (Tyr390 in S. marcescens ChiA) is highly conserved and that Glu145 and Tyr227 are potential residues that mediate the catalytic function of ChiNCTU2. To elucidate the catalytic mechanism of ChiNCTU2, we have cloned the cDNA encoding chitinase and established successful protein expression and purification in large quantities for crystallization and the study of its structure and function. Here, we report the overexpression, purification, crystallization and preliminary high-resolution X-ray diffraction characterization of chitinase from B. cereus ChiNCTU2.
2. Materials and methods
2.1. Protein expression and purification
The ChiNCTU2 gene was PCR-amplified from the genomic DNA of B. cereus NCTU2 (Wen & Li, 2002 ▶). The PCR fragment, including the signal peptide, was cloned into a pET-22b(+) vector and expressed in E. coli BL21 (DE3). A single colony was inoculated into 5 ml LB medium containing 0.1 mg ml−1 ampicillin and cultured at 310 K on a rotary shaker for 12 h. The overnight culture was then transferred into a 2 l Erlenmeyer flask containing 1 l LB medium with 0.1 mg ml−1 ampicillin and 100 mg l−1 IPTG and incubated at 310 K for 15 h. The culture broth was centrifuged at 277 K for 10 min at 7000g. The cell pellet was resupended in 10 ml 20 mM sodium phosphate buffer pH 7.0 and then subjected to cell disruption by ultrasonication. After removal of the cell debris on centrifugation, the supernatant (∼10 ml) was loaded onto a hydrophobic interaction column (HIC; 2.4 × 20 cm, high-performance Phenyl Sepharose; Amersham Bioscience, Uppsala, Sweden) which was pre-equilibrated with 20 mM sodium phosphate buffer pH 7.0 containing 1 M (NH4)2SO4. Elution was maintained at a flow rate of 2 ml min−1 with a linear gradient of (NH4)2SO4 from 1000 to 0 mM (at a decreasing rate of 16.67 mM min−1). Fractions with chitinase activity that eluted at 0 mM (NH4)2SO4 (Wen & Li, 2002 ▶) were collected and loaded onto an anion-exchange (5 ml HiTrap Q) column (Amersham Bioscience, Uppsala, Sweden) pre-equilibrated with 20 mM sodium phosphate buffer pH 7.0. Chromatography was performed using isocratic elution with 20 mM sodium phosphate buffer pH 7.0 at a flow rate of 2.0 ml min−1. The active fractions, eluted at 20–40 min, were pooled for further study. All purification steps were performed at ambient temperature (298 K). The protein sample was concentrated on Amicon (10 000 MWCO; Millipore) and the buffer was changed to 20 mM Tris buffer pH 7.0. The yield of protein was approximately 10 mg; the purity was greater than 95% according to analysis with 15% SDS–PAGE and UV–Vis spectra.
2.2. Crystallization
Prior to crystallization trials, the protein sample was ultrafiltrated to a concentration of 10 mg ml−1 in 100 mM sodium cacodylate buffer pH 6.0. Crystallization was performed by the hanging-drop vapour-diffusion method at 291 K using 24-well VDX plates. Small crystals were observed from a condition using 18%(w/v) PEG 8000, 200 mM zinc acetate dihydrate and 100 mM sodium cacodylate buffer pH 6.5 within 2 d of initial screening using the Crystal Sceeen kit (Hampton Research). This condition was refined to produce larger chitinase crystals using 2 µl hanging drops containing equal volumes of protein solution and reservoir solution equilibrated against 700 µl reservoir solution consisting of 22%(w/v) PEG 8000, 50 mM zinc acetate dihydrate and 100 mM sodium cacodylate buffer pH 6.0. Crystals of diffraction quality were used for collection of X-ray data.
2.3. X-ray data collection and processing
The protein crystals were initially screened and characterized using synchrotron radiation as an X-ray source at the SPXF beamline BL13B1 equipped with a CCD detector (Q315, ADSC) at National Synchrotron Radiation Research Center (NSRRC), Taiwan. Data collection was completed at the Taiwan-contracted protein crystallographic beamline BL12B2 equipped with a CCD detector (Quantum-4R, ADSC) at SPring-8 in Japan. The crystal was transferred from a crystallization drop into 10 µl cryoprotectant solution containing 18%(w/v) PEG 8000, 200 mM zinc acetate dihydrate, 20%(v/v) glycerol and 0.1 M sodium cacodylate buffer pH 6.0 for a few seconds, mounted on a nylon loop (0.1–0.2 mm, Hampton Research) and then flash-cooled in liquid nitrogen at 100 K. For complete data collection, a 360° rotation with 1.0° oscillations was measured using an X-ray wavelength of 1.00 Å with 15 s exposure duration and a crystal-to-detector distance of 180 mm at 110 K in a nitrogen stream using an X-Stream cryosystem (Rigaku/MSC Inc.). All data were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
The gene corresponding to mature ChiNCTU2 was initially constructed in the pRSET vector and overexpressed in E. coli strain BL21 (DE3). The recombinant enzyme was exhibited as an inclusion body. Unfortunately, efforts to refold the inactive protein were unsuccessful. The full gene (containing the signal peptide) was thus reconstructed into the pET-22b(+) vector. Two oligonucleotides were designed and used as primers for PCR cloning. The primers had sites for NdeI and SacI in sense and XhoI in antisense. The PCR fragment was subsequently cloned into the pET-22b(+) vector using NdeI and XhoI cutting sites. The chiNCTU2/pET-22b(+) was further transformed into E. coli BL21 (DE3) strain for protein expression.
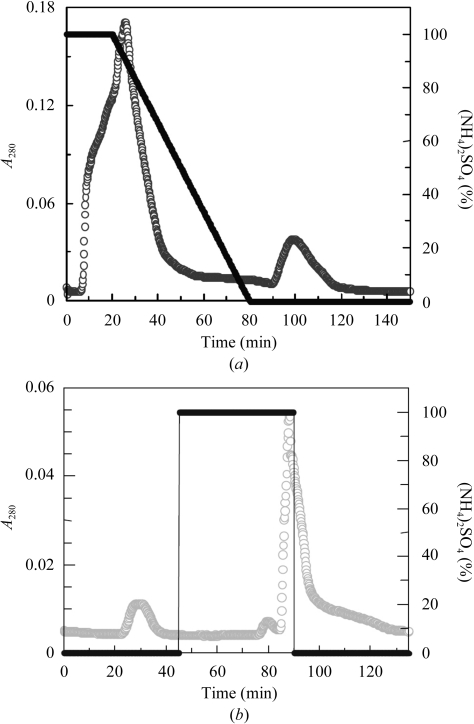
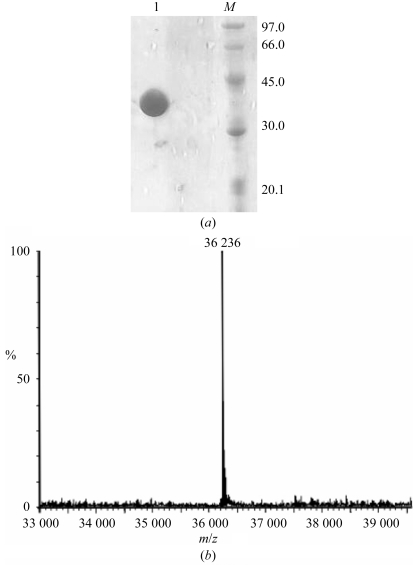
The chitinase was obtained intracellularly from E. coli BL21 (DE3) cells (cultured for 15 h) harbouring ChiNCTU2/pET-22b(+). Using the crude intracellular enzyme as starting material permitted the purification of chitinase by hydrophobic interaction chromatography (Fig. 1 ▶ a). The fractions with chitinase activity (90–120 min) were collected and applied onto a HiTrap Q ion-exchange column for further purification. During ion-exchange column chromatography chitinase did not bind to the Q-column and was eluted directly (20–24 min) without NaCl (Fig. 1 ▶ b). SDS–PAGE showed that the fraction with chitinase activity contained a highly pure protein with a molecular weight about 36 000 Da under SDS-denaturating and reducing conditions (Fig. 2 ▶ a). ESI–MS analysis gave a more precise molecular weight of 36 236 Da (Fig. 2 ▶ b), which is highly consistent with the calculated molecular weight (36 235 Da) of the mature protein, indicating that the signal peptide can be recognized in E. coli.
Figure 1.
(a) HIC column chromatography separation of ChiNCTU2 from crude enzyme [empty circles, A 280; filled circles, % (NH4)2SO4]. The column was pre-equilibrated with 1 M ammonium sulfate buffer pH 7.0. (b) Q-column chromatographic separation of the fraction containing ChiNCTU2 [empty circles, A 280; filled circles, % (NH4)2SO4].
Figure 2.
Molecular-weight analysis of the chitinase. (a) SDS–PAGE analysis. M, protein markers; lane 1, purified enzyme from an ion-exchange column. (b) ESI–MS analysis.
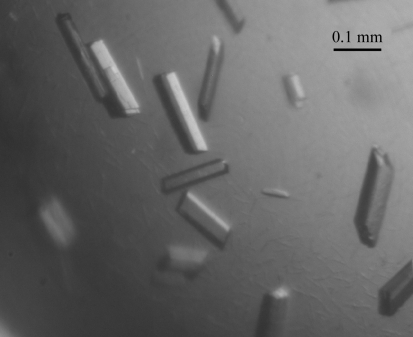
Protein crystals of rectangular shape appeared after 3 d and continued to grow to final dimensions of 0.06 × 0.06 × 0.2 mm within one week in an incubator at 291 K (Fig. 3 ▶). The protein crystals were sentitive to changes in precipitant concentration during transfer to the cryoprotectant solution containing 20% glycerol. Several crystals were tested prior to data collection as many had a mosaicity of >1° that caused overlap of diffraction spots in the high-resolution regions. Radiation damage was observed on protracted exposure during data collection, which caused a decrease in I/σ(I) and an increase in R sym. Analysis of the diffraction pattern indicates that the crystals exhibit monoclinic symmetry and systematic absences indicated the space group to be P21. Assuming the presence of one chitinase molecule per asymmetric unit, the Matthews coefficient is estimated to be 2.26 Å3 Da−1, corresponding to a solvent content of 43.4% (Matthews, 1968 ▶), which is within the normal range for protein crystals. Details of data statistics are summarized in Table 1 ▶.
Figure 3.
Single crystals of the chitinase grown by the hanging-drop method.
Table 1. Diffraction data statistics for ChiNCTU2.
Values in parentheses are for the highest resolution shell (1.16–1.10 Å).
| Wavelength (Å) | 1.00 |
| Temperature (K) | 100 |
| Resolution range (Å) | 30.0–1.10 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 50.79, b = 48.79, c = 66.87, β = 99.31 |
| Unique reflections | 123057 |
| Completeness (%) | 95.6 (91.4) |
| 〈I/σ(I)〉 | 20.9 (2.18) |
| Average redundancy | 5.5 (3.8) |
| Rsym† (%) | 7.5 (26.5) |
| Mosaicity | 0.41 |
| No. of molecules per ASU | 1 |
| Matthews coefficient (Å3 Da−1) | 2.26 |
| Solvent content (%) | 43.4 |
R
sym = 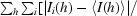
 , where I
i is the ith measurement and 〈I(h)〉 is the weighted mean of all measurements of I(h).
, where I
i is the ith measurement and 〈I(h)〉 is the weighted mean of all measurements of I(h).
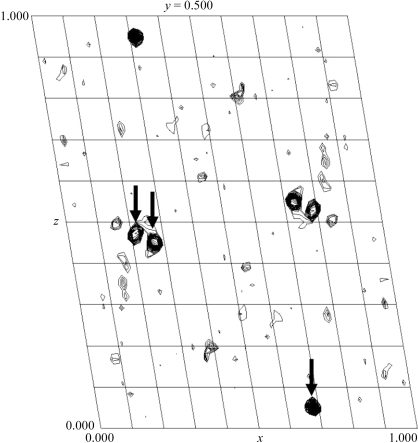
Initial attempts to solve the crystal structure of chiNCTU2 by the molecular-replacement method provided no clear solution. Because of the presence of zinc ions in the crystallization solution, we then attempted to use a zinc multiple-wavelength anomalous diffraction (MAD) method to solve the crystal structure. The X-ray energy scan and fluorescence measurements on the crystal showed a typical zinc anomalous scattering pattern near the edge. The Harker sections of an anomalous difference Patterson map generated by the program PHASES-95 (Furey & Swaminathan, 1997 ▶) using a peak wavelength of 1.28228 Å show three clear major sites for Zn atoms for phase calculation using the zinc-MAD method (Fig. 4 ▶). Completion of the structure determination will be described in a separate paper.
Figure 4.
Zn anomalous difference Patterson map (20–2.5 Å) showing a clear solution for zinc sites indicated by arrows at the Harker section of space group P21.
Acknowledgments
We are grateful to our colleagues Drs Yuch-Cheng Jean, Chun-Shiun Chao and supporting staff for technical assistance and discussion of the synchrotron-radiation X-ray facility during data collection at BL13B1 of NSRRC in Taiwan and Drs Yu-San Huang and Kuan-Li Yu at BL12B2 of SPring-8 in Japan. We also thank Professor Wen-guey Wu for helpful discussions. This study was supported in part by the National Synchrotron Radiation Research Center 944RSB02 and National Science Council NSC 94-2321-B-213-001 in Taiwan to C-JC and NSC 93-2113-M-009-001 and MOE-ATU program to Y-KL.
References
- Brunel, B., Périssol, C., Fernandez, M., Boeufgras, J. M. & Le Petit, J. (1994). FEMS Microbiol. Ecol.14, 331–342.
- Cohen-Kupiec, R. & Chet, I. (1998). Curr. Opin. Biotechnol.9, 270–277. [DOI] [PubMed] [Google Scholar]
- Emmert, E. A. B. & Handelsman, J. (1999). FEMS Microbiol. Lett.171, 1–9. [DOI] [PubMed] [Google Scholar]
- Flach, J., Pilet, P.-E. & Jollès, P. (1992). Experientia, 48, 701–716. [DOI] [PubMed] [Google Scholar]
- Felse, P. A. & Panda, T. (1999). Appl. Microbiol. Biotechnol.51, 141–151. [DOI] [PubMed] [Google Scholar]
- Furey, W. & Swaminathan, S. (1997). Methods Enzymol.277, 590–620. [DOI] [PubMed]
- Graham, L. S. & Sticklen, M. B. (1994). Can. J. Bot.72, 1057–1083.
- Henrissat, B. & Bairoch, A. (1996). Biochem. J.316, 695–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pleban, S., Chernin, L. & Chet, I. (1997). Lett. Appl. Microbiol.25, 284–288. [DOI] [PubMed] [Google Scholar]
- Silo-Suh, L. A., Lethbridge, B. J., Raffel, S. J., He, H., Clardy, J. & Handelsman, J. (1994). Appl. Environ. Microbiol.60, 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya, N., Yamazaki, D., Horiuchi, H., Ohta, A. & Takagi, M. (1998). Microbiology, 144, 2647–2654. [DOI] [PubMed] [Google Scholar]
- Wen, C.-M. & Li, Y.-K. (2002). Biotechnol. Appl. Biochem.35, 213–219. [DOI] [PubMed] [Google Scholar]