The crystal structure of the SH3 domain of human osteoclast-stimulating factor has been determined and refined to the ultrahigh resolution of 1.07 Å. The structure at atomic resolution provides an accurate framework for structure-based design of its inhibitors.
Keywords: osteoclast-stimulating factor, SH3 domain, atomic resolution, structure-based inhibitor design, osteoporosis
Abstract
Osteoclast-stimulating factor (OSF) is an intracellular signaling protein, produced by osteoclasts themselves, that enhances osteoclast formation and bone resorption. It is thought to act via an Src-related signaling pathway and contains SH3 and ankyrin-repeat domains which are involved in protein–protein interactions. As part of a structure-based anti-bone-loss drug-design program, the atomic resolution X-ray structure of the recombinant human OSF SH3 domain (hOSF-SH3) has been determined. The domain, residues 12–72, yielded crystals that diffracted to the ultrahigh resolution of 1.07 Å. The overall structure shows a characteristic SH3 fold consisting of two perpendicular β-sheets that form a β-barrel. Structure-based sequence alignment reveals that the putative proline-rich peptide-binding site of hOSF-SH3 consists of (i) residues that are highly conserved in the SH3-domain family, including residues Tyr21, Phe23, Trp49, Pro62, Asn64 and Tyr65, and (ii) residues that are less conserved and/or even specific to hOSF, including Thr22, Arg26, Thr27, Glu30, Asp46, Thr47, Asn48 and Leu60, which might be key to designing specific inhibitors for hOSF to fight osteoporosis and related bone-loss diseases. There are a total of 13 well defined water molecules forming hydrogen bonds with the above residues in and around the peptide-binding pocket. Some of those water molecules might be important for drug-design approaches. The hOSF-SH3 structure at atomic resolution provides an accurate framework for structure-based design of its inhibitors.
1. Introduction
The osteoclast (OCL) is the primary bone-resorbing cell responsible for degradation of bone matrix (Roodman, 1996 ▶). Factors produced by OCL play an important role in regulating OCL formation and activity (Reddy & Roodman, 1998 ▶). A novel 28 kDa intracellular protein termed osteoclast-stimulating factor (OSF), highly expressed in OCL, indirectly enhances OCL formation and bone resorption through a cellular signal transduction cascade, possibly through its interaction with c-Src or other c-Src-related proteins (Reddy et al., 1998 ▶). OSF contains a short proline-rich N-terminal region, an SH3 domain, three ankyrin repeats and an aspartate-rich C-terminal region (Reddy et al., 1998 ▶), suggesting that it is potentially involved in protein–protein interactions. Its SH3 domain was found to interact with the 40 kDa spinal muscular atrophy disease-determining gene product, survival motor neuron (SMN), to stimulate OCL formation (Kurihara et al., 2001 ▶), indicating that OSF SH3–SMN interaction may play an important role in congenital bone fractures associated with type I spinal muscular atrophy disease. Inhibition of the interaction of the OSF-SH3 domain with its protein partners might lead to reduced bone resorption by OCL and thus prevent the bone loss that is associated with many bone diseases, such as periodontal disease, osteoporosis, estrogen deficiency, Paget’s disease, inflammatory bone loss, bone malignancy and hyperparathyroidism.
As part of a structure-based anti-bone-loss drug-design program, we have determined a high-resolution X-ray structure of the recombinant human OSF SH3 domain (hOSF-SH3). The structure provides an accurate framework for structure-based studies of SH3-domain inhibitors.
2. Materials and methods
2.1. Cloning and expression
The gene for hOSF (GenBank accession No. NM_012383) was obtained from OpenBiosystem’s IMAGE clone collection (ID BC007459). A truncated fragment of OSF coding for residues 12–72 (SH3 domain) was PCR subcloned using primers 5′-GACAGCTAGCGGGCAAGTTAAAGTCTTCAGAGC-3′ and 5′-CGCGGATCCTTAGGATTCTGCCTGCTCAGCCAC-3′ into the NheI/BamHI restriction sites of pET28b (Novagen). The pET28-OSF-SH3 vector containing a six-His tag was transformed into Escherichia coli BL21(DE3) (Novagen) and the overproduction of the protein was induced at an A 600nm of ∼1.0 with a final concentration of 1 mM IPTG at 291 K. The cells grew for an additional 12 h and were harvested by centrifugation and frozen at 253 K.
2.2. Protein purification
The cell pellet was resuspended in Ni buffer A (20 mM Tris pH 8.0, 500 mM NaCl, 5 mM imidazole), lysed by sonication and centrifuged at 20 000g for 20 min at 277 K. The soluble crude lysate was filtered through a 0.45 µm filter and applied onto a Chelating Sepharose column (Pharmacia) which had been previously charged with 50 mM NiSO4 and equilibrated with Ni buffer A. The column was then washed with Ni buffer A to remove unbound proteins. The His6-SH3 fusion protein was eluted with a 5–500 mM imidazole linear gradient in 20 mM Tris–HCl pH 8.0, 500 mM NaCl and then dialyzed against high-salt thrombin cleavage buffer (20 mM Tris–HCl pH 8.4, 500 mM NaCl, 2.5 mM CaCl2) overnight at 277 K. One unit of thrombin protease (Novagen) was added per milligram of fusion protein and the His tag was cleaved for 4 h at room temperature. The thrombin cleavage reaction was terminated by a 1:10 dilution with 20 mM Tris–HCl pH 8.0 and the mixture was applied onto a Q Sepharose column (Pharmacia) that had been previously equilibrated with Q buffer A (20 mM Tris–HCl pH 8.0, 50 mM NaCl). SH3 was eluted from Q Sephrose with a 50–1000 mM NaCl linear gradient in 20 mM Tris–HCl pH 8.0. Fractions containing SH3 were pooled and concentrated by ultrafiltration.
2.3. Crystallization and data collection
The stock protein solution used for crystallization contained 20 mM Tris–HCl buffer pH 8.0 and 200 mM sodium chloride with a protein concentration of 10 mg ml−1. Crystals were grown at 277 K by the sitting-drop vapor-diffusion method with 100 mM sodium HEPES buffer pH 7.5, 2%(v/v) polyethylene glycol (PEG) 400, 2 M ammonium sulfate as the reservoir solution. The crystallization drops were prepared by mixing 5 µl protein solution with 4 µl reservoir solution and were equilibrated against 500 µl reservoir solution. Useful crystals appeared within a week and grew to a typical size of about 0.15 × 0.07 × 0.03 mm. Crystals formed in space group C2221, with unit-cell parameters a = 36.81, b = 80.48, c = 49.16 Å, and contained one monomer in the asymmetric cell. X-ray diffraction data to 1.07 Å resolution were collected at beamline 22-ID in the facilities of the South East Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory, USA. The program package HKL-2000 ▶ was used to process the diffraction data (Otwinowski & Minor, 1997 ▶). The statistics for data collection and processing are summarized in Table 1 ▶.
Table 1. Summary of data collection, phasing and refinement.
Values in parentheses are for the highest resolution shell.
| Data and phasing statistics | |
| Space group | C2221 |
| Unit-cell parameters (Å) | a = 36.81, b = 80.48, c = 49.16 |
| Wavelength (Å) | 1.00 |
| Resolution (Å) | 1.07 (1.11–1.07) |
| Wilson B factor (Å2) | 9.5 |
| Reflections (total/unique) | 142748/28252 |
| Completeness (%) | 86.3 (49.9) |
| I/σ(I) | 17.4 (2.5) |
| Rmerge(I) (%) | 9.8 (24.1) |
| Molecular-replacement model | Grb2 SH3 domain |
| Refinement statistics | |
| Resolution range (Å) | 10–1.07 |
| No. of reflections | 27365 |
| No. of atoms | |
| Protein | 477 |
| Water | 130 |
| R factors | |
| Rwork (%) | 15.8 |
| Rfree (%) | 21.1 |
| R.m.s. deviation of bonds | |
| Lengths (Å) | 0.014 |
| Angle distances (°) | 0.030 |
| Average B factor (Å2) | 20.8 |
2.4. Structure determination and refinement
The orientation and position of hOSF-SH3 in the crystal unit cell were determined using the molecular-replacement protocols in the program CNS (Brünger et al., 1998 ▶) with the structure of Grb2 (PDB code 1gri) as the search model. The composite OMIT map was calculated to guide electron-density fitting of the model. Energy-restrained crystallographic refinement was carried out with the maximum-likelihood algorithms implemented in CNS. Refinement proceeded through several cycles in combination with manual checking with the program O (Jones et al., 1991 ▶). The addition of 100 water molecules and refinement to 1.07 Å resulted in R and R free values of 0.243 and 0.245, respectively. Further refinement was continued with SHELX97 (Sheldrick & Schneider, 1997 ▶), first subjecting the structure to cycles of isotropic conjugate-gradient least-squares refinement; tightly restrained anisotropic displacement parameters were then introduced and refined. No bulk-solvent model (SHELXL SWAT option) was used during the refinement as it did not substantially lower the R factors. The final refinement cycle resulted in R and R free values of 0.158 and 0.211, respectively. The final model contains residues 12–69 and 130 water molecules. The phasing and refinement statistics are summarized in Table 1 ▶.
2.5. Protein-fold analysis
Secondary-structure elements were defined by the hydrogen-bonding patterns in combination with visual inspection. The DALI algorithm of comparing protein domain structures by alignment of distance matrices was used to search for structural homologs of the hOSF-SH3 domain and was also used for structure-based sequence alignment (Holm & Sander, 1993 ▶, 1998 ▶). Ribbon diagrams were prepared using the program MOLSCRIPT (Kraulis, 1991 ▶).
3. Results and discussion
3.1. Structure determination
The hOSF-SH3 domain included residues 12–72 of the hOSF sequence (Reddy et al., 1998 ▶). hOSF-SH3 was crystallized and its structure was determined by the molecular-replacement method (Table 1 ▶). The structure was refined against 1.07 Å resolution data, making it one of the highest resolution SH3-domain structures reported to date (Berisio et al., 2001 ▶; Wisniewska et al., 2005 ▶; Hoelz et al., 2006 ▶). The C-terminal residues 70–72 were disordered and could not be seen in the 2F o − F c electron-density map. The final refined model, which includes residues 12–69 and 130 ordered water molecules, has a working R value of 0.158 and a free R value of 0.211. The stereochemistry is excellent, with r.m.s. deviations for bond lengths and angle distances of 0.014 and 0.030 Å, respectively (Table 1 ▶). The Ramachandran plot statistics showed that 95.9% of the backbone dihedral angles were in most favored regions, 4.1% were in additional allowed regions and none of the non-glycine residues were in disallowed regions.
3.2. Overall structure
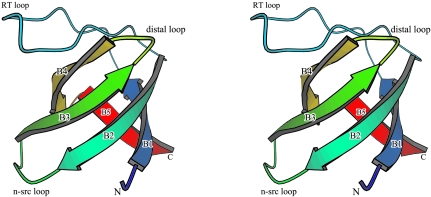
Like the structures of other SH3 domains, hOSF-SH3 adopts a classical SH3 fold consisting of five antiparallel β-strands arranged as two orthogonal β-sheets that form a β-barrel (Fig. 1 ▶). Each β-sheet consists of three β-strands with one long strand (B2; residues 37–44) contributing different segments to each sheet. One of the sheets is formed by β-strands B2, B1 (residues 14–20) and B5 (residues 65–69), while the other is formed by B2, B3 (residues 48–54) and B4 (residues 57–63). The first two β-strands (B1 and B2) are linked by the RT loop (residues 21–36, named for the RT residues found in the c-Src SH3 domain), which forms a hairpin-like structure. The second β-strand B2 is linked to the third β-strand B3 by a short n-Src loop (residues 45–47). The third loop, linking β-strands B3 and B4, is known as the distal loop (residues 55–56). Residue Asn64 links the last two β-strands B4 and B5.
Figure 1.
Stereo ribbon representation of the hOSF-SH3 structure. A rainbow ramp color coding of blue to red is used to mimic the chain trace from the N-terminus to the C-terminus. Both termini and the secondary-structure elements are labeled. Positions of the loops are also indicated.
3.3. Structure comparison and the putative peptide-binding site
The overall structure of hOSF-SH3 is very similar to several other SH3 domains despite having a relatively low sequence homology (Table 2 ▶). Among these, hOSF-SH3 has the highest sequence identity with the SH3 domains of signal transduction adaptor Grb2 (44% over 54 equivalent positions) and tyrosine kinase c-Src (44% over 55 equivalent positions). The structural similarity Z scores (Holm & Sander, 1993 ▶, 1998 ▶) range from 11.4 to 9.6, with r.m.s. deviations of equivalent positions in the range 0.7–1.4 Å. Structure-based sequence alignment of these SH3 domains reveals that the putative proline-rich peptide-binding site of hOSF-SH3 consists of residues that are highly conserved in the SH3-domain family (Figs. 2 ▶ and 3 ▶), including residues Tyr21, Phe23, Trp49, Pro62, Asn64 and Tyr65. Tyr21 and Phe23 come from the RT loop, Trp49 from the β-strand B3, Pro62 from the β-strand B4 and Tyr65 from the β-strand B5, while Asn64 links the β-strands B4 and B5. These residues form the hydrophobic binding pocket. In addition, residues that are less conserved and/or even specific to hOSF are also part of the peptide-binding site (Fig. 4 ▶), including residues Thr22, Arg26, Thr27, Glu30, Asp46, Thr47, Asn48 and Leu60, which might be key to designing specific inhibitors for hOSF SH3 domain to fight osteoporosis and related bone-loss diseases.
Table 2. Alignment statistics of hOSF-SH3 with structurally similar SH3 domains.
| Protein | PDB code | Z† | R.m.s.d.‡ (Å) | LALI§ | LSEQ2¶ | IDE†† (%) | Reference |
|---|---|---|---|---|---|---|---|
| hOSF-SH3 domain | 1zlm | 15.3 | 0.0 | 58 | 58 | 100 | This paper |
| Oncogene c-Crk | 1cka | 11.4 | 1.0 | 55 | 56 | 33 | Wu et al. (1995 ▶) |
| Neutrophil cytocol factor 4 | 1w6x | 11.1 | 1.0 | 54 | 56 | 31 | Massenet et al. (2005 ▶) |
| Tyrosine kinase Lck | 1lck | 10.8 | 1.0 | 54 | 164 | 37 | Eck et al. (1994 ▶) |
| Actin-binding protein Abp1 | 1jo8 | 10.5 | 1.0 | 54 | 58 | 37 | Fazi et al. (2002 ▶) |
| Signal transduction adaptor Grb2 | 1gri | 10.0 | 0.7 | 54 | 211 | 44 | Maignan et al. (1995 ▶) |
| Tyrosine kinase Abl | 1bbz | 9.7 | 1.4 | 54 | 58 | 26 | Pisabarro et al. (1998 ▶) |
| Anti-oncogene 53BP2 | 1ycs | 9.6 | 1.2 | 55 | 193 | 31 | Gorina & Pavletich (1996 ▶) |
| Tyrosine kinase c-Src | 1fmk | 9.6 | 1.4 | 55 | 438 | 44 | Xu et al. (1997 ▶) |
Z score, strength of structural similarity in standard deviations above expected.
Positional root-mean-square deviation of superimposed Cα atoms.
Total number of equivalent residues.
Length of the entire chain of the equivalent structure.
Percentage sequence identity over equivalent positions.
Figure 2.
Structure-based sequence alignment of hOSF-SH3 with the most similar SH3 domains. The sequence numbers for the first and the last residues are indicated. The positions of the β-strands in hOSF-SH3 are indicated by underlining the sequence and are labeled. Residues that are highly conserved in the SH3-domain family are indicated in bold type. Residues that are known to form the proline-rich ligand-binding pocket are marked with an asterisk. See Table 2 ▶ for protein PDB codes and references.
Figure 3.
Structure comparison and the peptide-binding model. Stereoview of structural superposition of hOSF-SH3 (black) with c-Crk-SH3 (red; PDB code 1cka) in complex with the proline-rich peptide PPPALPPKK (green). Residues involved in peptide binding are shown as stick models and the rest as Cα traces. The peptide is also shown with stick models. For clarity, only the termini and the key residues of hOSF-SH3 are labeled.
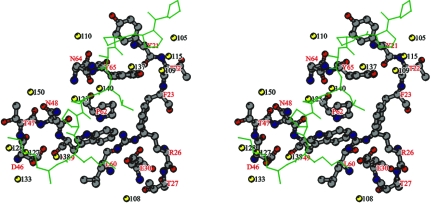
Figure 4.
Stereoview of the peptide-binding site showing the water network in and around the binding pocket. To help define the binding pocket, a proline-rich peptide PPPALPPKK (green) was modeled into the proposed peptide-binding site. Residues of hOSF-SH3 forming the peptide-binding pocket are shown as ball-and-stick models and their atoms are colored according to atom types: C atoms, grey; N atoms, blue; O atoms, red. The peptide is shown as a thin-stick model. Water molecules are shown as yellow balls and labeled with black numbers.
There are a total of 13 well defined water molecules forming hydrogen bonds with the hOSF SH3 residues in and around the peptide-binding pocket (Table 3 ▶; Fig. 4 ▶). Six of those water molecules, 105, 115, 109, 137, 140 and 123, form a long water channel along one end of the binding pocket consisting of hOSF residues Tyr21, Thr22, Phe23, Arg26, Asn48, Trp49, Pro62, Asn64 and Tyr65. Four other water molecules, 127, 128, 133 and 138, form another water network at the other end of the binding pocket consisting of residues Asp46, Thr47, Asn48 and Trp49. Some of those water molecules might be important for drug-design approaches.
Table 3. Hydrogen bonds between OSF and water molecules in the peptide-binding site.
| OSF atom | Water atom | Hydrogen-bond distance (Å) |
|---|---|---|
| Thr22 N | 105 O | 2.83 |
| Thr22 O | 115 O | 2.69 |
| Thr27 OG1 | 108 O | 2.71 |
| Asp46 OD1 | 138 O | 2.40 |
| Asp46 OD2 | 127 O | 2.77 |
| Asp46 OD2 | 133 O | 2.71 |
| Asp46 OD2 | 138 O | 2.98 |
| Thr47 N | 128 O | 2.91 |
| Asn48 O | 123 O | 2.87 |
| Asn48 OD1 | 150 O | 2.69 |
| Asn64 O | 110 O | 2.67 |
| Asn64 OD1 | 123 O | 2.92 |
| Asn64 OD1 | 140 O | 2.51 |
| Asn64 ND2 | 140 O | 2.93 |
| Tyr65 OH | 109 O | 2.60 |
| Tyr65 OH | 137 O | 2.79 |
Recently, an ensemble of 20 NMR structures of hOSF-SH3 was deposited in the Protein Data Bank (PDB code 1x2k). The solution structures have essentially the same arrangement of secondary structure as the X-ray-determined structure reported here, with an overall r.m.s. deviation of around 0.73 Å over 55 equivalent Cα positions (residues 15–69).
3.4. Implications for inhibitor design against osteoclast-mediated bone resorption
OSF induces OCL formation and bone resorption through a cellular signal transduction cascade, possibly through its interactions with c-Src or other Src-related proteins (Reddy et al., 1998 ▶). Like the SH3 domains of other proteins, the SH3 domain of hOSF contains a peptide-binding pocket that is also likely to favor the binding of short proline-rich regions in the ligand proteins (Figs. 3 ▶ and 4 ▶). We are currently working on cocrystallizing the hOSF SH3 domain with proline-rich peptides. Blocking the binding of proline-rich motifs to the pocket of the SH3 domain might disrupt the interactions of hOSF with its signal transduction partners, leading to the inhibition of osteoclast-mediated bone resorption. The atomic resolution crystal structure of hOSF-SH3 as reported here provides an accurate framework for structure-based design of effective SH3-domain inhibitors that might become potential anti-bone-loss drugs.
Supplementary Material
PDB reference: osteoclast-stimulating factor, 1zlm, r1zlmsf
Acknowledgments
This work was supported in part by NSF-EPSCoR, NASA and a generous gift from an anonymous donor to the Laboratory for Structural Biology, University of Alabama in Huntsville. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract No. W-31–109-Eng-38. We thank J. Looger of UAH for technical assistance in computer programs and staff of the SER-CAT beamline 22-ID at the Advanced Photon Source for help with data collection. We are also grateful to OpenBiosystems Inc. for the free gift of hOSF cDNA.
References
- Berisio, R., Viguera, A., Serrano, L. & Wilmanns, M. (2001). Acta Cryst. D57, 337–340. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Eck, M. J., Atwell, S. K., Shoelson, S. E. & Harrison, S. C. (1994). Nature (London), 368, 764–769. [DOI] [PubMed]
- Fazi, B., Cope, M. J., Douangamath, A., Ferracuti, S., Schirwitz, K., Zucconi, A., Drubin, D. G., Wilmanns, M., Cesareni, G. & Castagnoli, L. (2002). J. Biol. Chem.277, 5290–5298. [DOI] [PubMed]
- Gorina, S. & Pavletich, N. P. (1996). Science, 274, 1001–1005. [DOI] [PubMed]
- Hoelz, A., Janz, J. M., Lawrie, S. D., Corwin, B., Lee, A. & Sakmar, T. P. (2006). J. Mol. Biol.358, 509–522. [DOI] [PubMed]
- Holm, L. & Sander, C. (1993). J. Mol. Biol.233, 123–138. [DOI] [PubMed]
- Holm, L. & Sander, C. (1998). Proteins, 33, 88–96. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kraulis, P. J. (1991). J. Appl. Cryst.24, 946–950. [Google Scholar]
- Kurihara, N., Menaa, C., Maeda, H., Haile, D. J. & Reddy, S. V. (2001). J. Biol. Chem.276, 41035–41039. [DOI] [PubMed]
- Maignan, S., Guilloteau, J. P., Fromage, N., Arnoux, B., Becquart, J. & Ducruix, A. (1995). Science, 268, 291–293. [DOI] [PubMed]
- Massenet, C., Chenavas, S., Cohen-Addad, C., Dagher, M. C., Brandolin, G., Pebay-Peyroula, E. & Fieschi, F. (2005). J. Biol. Chem.280, 13752–13761. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pisabarro, M. T., Serrano, L. & Wilmanns, M. (1998). J. Mol. Biol.281, 513–521. [DOI] [PubMed]
- Reddy, S., Devlin, R., Menaa, C., Nishimura, R., Choi, S. J., Dallas, M., Yoneda, T. & Roodman, G. D. (1998). J. Cell. Physiol.177, 636–645. [DOI] [PubMed]
- Reddy, S. V. & Roodman, G. D. (1998). Crit. Rev. Eukaryot. Gene Expr.8, 1–17. [DOI] [PubMed]
- Roodman, G. D. (1996). Endocr. Rev.17, 308–332. [DOI] [PubMed]
- Sheldrick, G. M. & Schneider, T. R. (1997). Methods Enzymol.227, 319–343. [PubMed]
- Wisniewska, M., Bossenmaier, B., Georges, G., Hesse, F., Dangl, M., Kunkele, K. P., Ioannidis, I., Huber, R. & Engh, R. A. (2005). J. Mol. Biol.347, 1005–1014. [DOI] [PubMed]
- Wu, X., Knudsen, B., Feller, S. M., Zheng, J., Sali, A., Cowburn, D., Hanafusa, H. & Kuriyan, J. (1995). Structure, 3, 215–226. [DOI] [PubMed]
- Xu, W., Harrison, S. C. & Eck, M. J. (1997). Nature (London), 385, 595–602. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: osteoclast-stimulating factor, 1zlm, r1zlmsf