Rice BGlu1 β-glucosidase was purified from recombinant E. coli and crystallized with and without the inhibitor 2-deoxy-2-fluoro-β-d-glucose. The crystals diffracted to 2.15 and 2.75 Å, respectively.
Keywords: BGlu1 β-glucosidase; 2,4-dinitrophenyl-2-deoxy-2-fluoro-β-d-glucopyranoside; rice
Abstract
Rice (Oryza sativa) BGlu1 β-glucosidase was expressed in Escherichia coli with N-terminal thioredoxin and hexahistidine tags and purified by immobilized metal-affinity chromatography (IMAC). After removal of the N-terminal tags, cation-exchange and S-200 gel-filtration chromatography yielded a 50 kDa BGlu1 with >95% purity. The free enzyme and a complex with 2,4-dinitrophenyl-2-deoxy-2-fluoro-β-d-glucopyranoside inhibitor were crystallized by microbatch and hanging-drop vapour diffusion. Small tetragonal crystals of BGlu1 with and without inhibitor grew in 18%(w/v) PEG 8000 with 0.1 M sodium cacodylate pH 6.5 and 0.2 M zinc acetate. Crystals of BGlu1 with inhibitor were streak-seeded into 23%(w/v) PEG MME 5000, 0.2 M ammonium sulfate, 0.1 M MES pH 6.7 to yield larger crystals. Crystals with and without inhibitor diffracted to 2.15 and 2.75 Å resolution, respectively, and had isomorphous orthorhombic unit cells belonging to space group P212121.
1. Introduction
In plants, β-glucosidases (EC 3.2.1.21) and related glycosidases play roles in many biological processes, including defence, lignification, phytohormone activation and cell-wall modification (Esen, 1993 ▶). Their physiological functions depend upon their location and substrate-specificity.
Most plant β-glucosidases belong to glycosyl hydrolase family 1 (GH1), which also includes myrosinases (thio-β-glucosidases), β-mannosidases, β-galactosidases, phospho-β-glucosidases and phospho-β-galactosidases (Henrissat & Bairoch, 1996 ▶). Three-dimensional structures of several GH1 enzymes have been solved, including β-glucosidases, myrosinases and 6-phospho-β-galactosidase (Barrett et al., 1995 ▶; Sanz-Aparicio et al., 1998 ▶; Burmeister et al., 1997 ▶; Wiesmann et al., 1997 ▶). All have a common (β/α)8- or TIM-barrel structure and two catalytic carboxylate groups located at the C-terminal ends of β-strands 4 and 7. GH1 hydrolases use a double-displacement mechanism in which two carboxylic acids act as a general acid/base and nucleophile (Sinnott, 1990 ▶; McCarter & Withers, 1994 ▶). During glycosylation, the glycoside is protonated by the first carboxylic acid, while the second carboxylate makes a nucleophilic attack at the anomeric carbon to form a covalent intermediate. This intermediate is cleaved during deglycosylation, in which the first carboxylate, as a general base, activates the incoming nucleophile (i.e. water). The covalent intermediate can be trapped by 2-deoxy-2-fluoro-β-d-glycosides with good leaving groups, since the electronegative fluorine destabilizes the carbocation-like transition state of deglycosylation (Withers & Street, 1988 ▶; He & Withers, 1997 ▶).
Although GH1 hydrolases are similar, the amino acids near the active site which determine the substrate specificity vary tremendously. Thus, maize (Zea mays) β-glucosidase 1 and sorghum (Sorghum bicolor) dhurrinase share 72% amino-acid sequence identity, but cannot hydrolyze each other’s substrates and have substrate-binding residues located at different sequence positions (Czjzek et al., 2000 ▶; Verdoucq et al., 2004 ▶).
Opassiri et al. (2003 ▶) cloned the cDNA for a GH1 β-glucosidase (BGlu1) highly expressed in rice (Oryza sativa) flowers and germinating shoots. Rice BGlu1 and barley (Hordeum vulgare) BGQ60/ β-II β-glucosidase have ∼66% amino-acid sequence identity, hydrolyze glucooligosaccharides with a preference for 1,3-linked dimers and longer 1,4-linked oligosaccharides and have binding subsites for at least six β-1,4-linked glucose residues (Hrmova et al., 1998 ▶; Opassiri et al., 2004 ▶). However, rice BGlu1 hydrolyzes β-glucoside ten times faster than β-mannoside and cellotriose faster than cellobiose, while barley shows the opposite preferences. To determine the subsites for oligosaccharide binding and the structural basis for the differences in substrate binding, BGlu1 produced in Escherichia coli was purified with removal of its fusion tag and protein crystallization and diffraction studies were initiated.
2. Materials and methods
2.1. Recombinant production and purification of BGlu1 protein
BGlu1 was produced as an N-terminal thioredoxin/His-tag fusion protein from the bglu1 cDNA (accession No. U28047) cloned in pET32a(+), and expressed in E. coli Origami (DE3) (Opassiri et al., 2003 ▶). In this construct, five amino acids, AMADV, are found between the enterokinase site, for removal of the N-terminal thioredoxin, His6 and S tags, and Val29 of BGlu1 (Genbank Accession AAA84906), which is the predicted N-terminus of the mature protein. The fusion protein was purified by immobilized metal-affinity chromatography (IMAC) at 277 K; fractions were assayed for β-glucosidase activity with p-nitrophenol β-d-glucopyranoside and those containing activity were pooled as described by Opassiri et al. (2003 ▶). Imidazole was removed and the buffer changed to 20 mM Tris–HCl pH 8.0 in a 30 kDa molecular-weight cutoff (MWCO) Centricon centrifugal filter (Millipore). To remove the N-terminal fusion tag, BGlu1 fusion protein was cleaved with 0.006 µg enterokinase (New England Biolabs) per milligram of protein in 20 mM Tris–HCl pH 7.2, 200 mM calcium chloride at 296 K for 16 h. To remove the cleaved fusion tag and uncleaved fusion protein from the tag-free BGlu1, the digest was incubated with 1 ml Ni–NTA Superflow resin per milligram of protein for 1 h at 277 K with gentle shaking. The solution of unbound protein was collected and the resin was washed with 5 and 10 mM imidazole in equilibration buffer (150 mM NaCl, 20 mM Tris–HCl pH 8.0). The unbound and wash fractions containing BGlu1 lacking fusion tag (tag-free BGlu1) were combined, the buffer was changed to 50 mM phosphate pH 7.0 and the protein was concentrated using a 10 kDa MWCO Centricon centrifugal filter. The tag-free protein was loaded onto a 5 ml HiTrap SP Sepharose Fast Flow cation-exchange column (GE Healthcare) equilibrated with five volumes of 50 mM phosphate buffer pH 7.0. Tag-free BGlu1 was eluted with a 0–150 mM NaCl gradient in the same buffer at a flow rate of 1.5 ml min−1. Fractions containing β-glucosidase activity were pooled and concentrated to 10 mg ml−1 in a 10 kDa MWCO Centricon centrifugal filter. The protein was then passed through a Superdex 200 gel-filtration column (HR10/30, GE Healthcare) equilibrated and eluted with 150 mM NaCl, 20 mM Tris–HCl pH 8.0 at a flow rate of 0.25 ml min−1.
2.2. Protein crystallization of BGlu1 without inhibitor
The purified protein was concentrated using a 10 kDa MWCO Centricon centrifugal filter at 4000g and was adjusted to 10–20 mg ml−1 with 150 mM NaCl, 20 mM Tris–HCl pH 8.0. Before crystallization, the protein solution was filtered though a Ultrafree-MC 0.22 µm filter (Millipore) at 4000g for 5 min to eliminate dust, microcrystals and precipitated protein. Screening for crystallization conditions was performed with JBScreen HTS I and II (Jena Bioscience) and Crystal Screen High Throughput HR2-130 (Hampton Research) kits, as well as a systematic screen of monovalent and divalent salts (0.2 M) and pH (in the range 4.5–8.5 at 0.1 M concentration) in the presence of 25%(w/v) PEG 4000. Initially, microbatch screening was performed with 96-well plastic plates (Hampton Research) and all crystallization trials were performed at 288 K. Each drop contained 1.0 µl of 10 mg ml−1 protein and 1.0 µl precipitant solution under 10 µl oil (paraffin, highly liquid; Merck).
2.3. Screening and optimization of BGlu1 crystallization with inhibitor
A 10 mg ml−1 protein solution was mixed with 1.0 mM 2,4-dinitrophenyl 2-deoxy-2-fluoro-β-d-glucopyranoside (DNP2FG, Sigma Chemical Corp.; 1:5 molar ratio) and then filtered and screened as described for BGlu1 without inhibitor. Promising conditions were optimized by hanging-drop vapour diffusion in 24-well plates. Primary variables were precipitant concentration, pH, protein concentration and the ratios of protein:inhibitor and protein:precipitant, while secondary variables included addition of coprecipitant, salt, glycerol and buffer type. Initial drops of 1 µl protein solution and 1 µl reservoir solution were equilibrated against a 500 µl reservoir.
2.4. Microseeding with BGlu1 with inhibitor
Crystal clusters were removed from their initial drop, repeatedly washed with fresh mother liquor and then crushed in a microcentrifuge tube containing 1 ml mother liquor. This microcrystal stock was serially diluted with mother liquor and dilutions were streaked with a cat whisker into pre-equilibrated hanging drops and then incubated at 288 K.
2.5. Data collection and processing
Crystals of BGlu1 with and without inhibitor were mounted in nylon loops (Hampton Research), soaked in cryoprotectant containing 18%(v/v) glycerol in precipitant solution [18%(w/v) PEG 8000 with 0.1 M sodium cacodylate pH 6.5 and 0.2 M zinc acetate for free enzyme or 23%(w/v) PEG MME 5000, 0.2 M ammonium sulfate, 0.1 M MES pH 6.7 for inhibitor-bound enzyme] for 5 s and flash-cooled in a nitrogen stream at 105 K generated by an X-Stream 2000 low-temperature system (Rigaku/MSC). X-ray diffraction data were collected in-house using an RU-H3R rotating-anode X-ray generator (Rigaku/MSC) running at 50 kV and 100 mA. Diffraction data from a single crystal were recorded on an R-AXIS IV++ image-plate system (Rigaku/MSC) and were processed with the CrystalClear/d*TREK program suite (Pflugrath, 1999 ▶).
3. Results and discussion
3.1. Protein purification
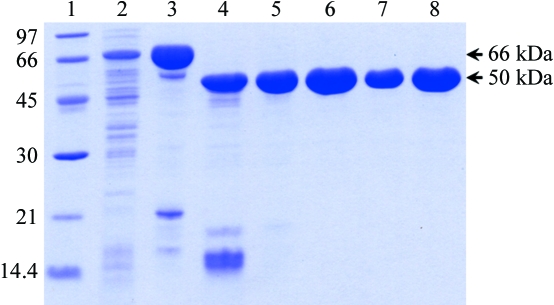
Expression of BGlu1 N-terminally thioredoxin and hexahistidine-tagged fusion protein in E. coli Origami (DE3) and purification by IMAC yielded 8–9 mg protein per litre of E. coli culture. The 66 kDa fusion protein appeared to be approximately 90% pure, with prominent protein bands at 66, ∼50 and ∼20 kDa on SDS–PAGE (Fig. 1 ▶). After removal of the N-terminal tag by cleavage and adsorption of the fusion tag to IMAC resin and purification by cation-exchange and S-200 gel-filtration chromatography, about 2.6 mg of homogeneous 50 kDa protein was obtained per litre of cell culture, with >95% purity (Fig. 1 ▶).
Figure 1.
SDS–PAGE of purified BGlu1 fractions from the purification steps for crystallization. Lane 1, Bio-Rad low molecular-weight markers (kDa); lane 2, soluble extract of E. coli cells; lane 3, fusion protein after initial IMAC; lane 4, enterokinase digest; lane 5, BGlu1 after subtractive IMAC; lane 6, BGlu1 after SP-Sepharose, lanes 7 and 8; BGlu1 after the final S200. 5 µg of protein was loaded in all lanes, except lane 7, in which 2.5 µg was loaded.
3.2. Crystallization and X-ray diffraction
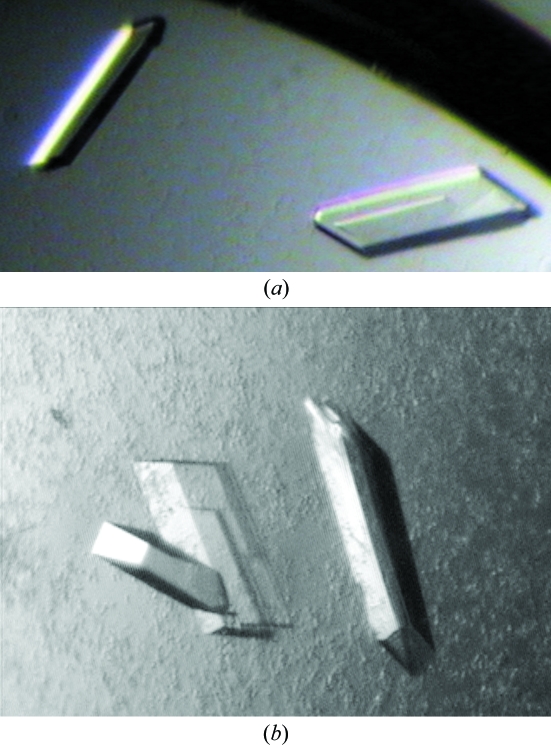
For BGlu1 without inhibitor, small tetragonal crystals appeared after 3–5 d under several conditions, with PEGs ranging in molecular weight from 3350 to 8000 as the most successful precipitants in microbatch screening. After 1–2 months in 18%(w/v) PEG 8000 with 0.1 M sodium cacodylate pH 6.5 and 0.2 M zinc acetate, larger crystals developed. A crystal with dimensions of 120 × 60 × 16 µm (Fig. 2 ▶ a) was used to collect a preliminary data set to at least 2.75 Å resolution. The crystal of BGlu1 with DNP2FG inhibitor crystallized within one week as clusters of thin plates in microbatch screening under the same conditions. Optimization produced crystal clusters and slow-growing crystals, so streak-seeding with a 100-fold dilution of crushed crystal clusters from 20%(w/v) PEG 8000, 0.1 M sodium cacodylate pH 6.5 and 0.2 M zinc acetate was performed in hanging drops. An orthorhombic single crystal formed within 5 d in a drop containing 2 µl 5 mg ml−1 protein and 1 µl 23%(w/v) PEG MME 5000, 0.2 M ammonium sulfate, 0.1 M MES pH 6.7 precipitant, which was equilibrated with the reservoir for 2 h before seeding. After 26 d, the crystal dimensions were 320 × 140 × 20 µm (Fig. 2 ▶ b). A 140 × 140 × 20 µm fragment of this crystal diffracted X-rays to 2.15 Å resolution. BGlu1 crystals with and without inhibitor both belong to the P212121 space group with almost the same unit-cell parameters, as summarized in Table 1 ▶ together with other data-collection statistics.
Figure 2.
Crystals of BGlu1 with and without inhibitor crystallized in an orthorhombic crystal form. (a) Plate-shaped tetragonal crystal of BGlu1 with dimensions 120 × 60 × 16 µm grown in 18%(w/v) PEG 8000, 0.2 M zinc acetate dehydrate and 0.1 M sodium cacodylate pH 6.5 by microbatch screening. (b) BGlu1 protein incubated with DNP2FG crystallized as tetragonal crystals, one with dimensions of 320 × 140 × 20 µm, in a hanging-drop vapour-diffusion plate. The precipitant was 23%(w/v) PEG MME 5000, 0.2 M ammonium sulfate, 0.1 M MES pH 6.7.
Table 1. Data-collection statistics for BGlu1 and BGlu1 with inhibitor.
Values in parentheses are for the outer shell.
| BGlu1 | BGlu1 with DNP2FG | |
|---|---|---|
| Wavelength (Å) | 1.5418 | 1.5418 |
| Resolution (Å) | 55.72–2.75 (2.85–2.75) | 53.82–2.15(2.23–2.15) |
| Completeness (%) | 99.5 (99.9) | 96.5 (83.1) |
| Rmerge (%) | 14.6 (35.4) | 11.0 (30.7) |
| 〈I/σ(I)〉 | 3.6 (1.6) | 4.3 (1.8) |
| Mosaicity (°) | 0.660 | 0.670 |
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å) | a = 78.854, b = 100.337, c = 127.097 | a = 79.369, b = 100.922, c = 127.229 |
| Unit-cell volume (Å3) | 1005589.7 | 1019114.5 |
| No. of unique reflections | 50449 (5078) | 103691 (8925) |
| No. of observed reflections | 97594 (9650) | 213255 (13611) |
| VM (Å3 Da−1) | 2.5 | 2.5 |
| Solvent content (%) | 50.7 | 51.3 |
| No. of molecules per ASU | 2 | 2 |
3.3. Molecular replacement
BGlu1 structures with and without inhibitor were solved by molecular replacement using the AMoRe program (Navaza, 1994 ▶). The cyanogenic β-glucosidase structure from white clover (Trifolium repens; PDB code 1cbg; Barrett et al., 1995 ▶) was used as a search model, since it had the highest sequence similarity (46% identity). Two molecules per asymmetric unit were found in both cases. Model building and refinement of these two structures are in progress. Structural models of rice BGlu1 in the free form and the intermediate generated with DNP2FG should yield a better understanding of how substrates bind to the active site.
Acknowledgments
We thank Robert C. Robinson and Terese Bergfors for advice on crystallization experiments and useful discussions. This work was supported by grants from the Thailand Research Fund (TRF, BRG4780020 and RTA4780006) and Suranaree University of Technology. WC is a Royal Golden Jubilee PhD fellow of the TRF.
References
- Barrett, T., Suresh, C. G., Tolley, S. P., Dodson, E. J. & Hughes, M. A. (1995). Structure, 3, 951–960. [DOI] [PubMed] [Google Scholar]
- Burmeister, W. P., Cottaz, S., Driguez, H., Iori, R., Palmieri, S. & Henrissat, B. (1997). Structure, 5, 663–675. [DOI] [PubMed] [Google Scholar]
- Czjzek, M., Cicek, M., Zamboni, V., Burmeister, W. P., Bevan, D. R., Henrissat, B. & Esen, A. (2000). Proc. Natl Acad. Sci. USA, 97, 13555–13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen, A. (1993). β-Glucosidases: Biochemistry and Molecular Biology, edited by A. Esen, pp. 1–14. Washington, DC: American Chemical Society.
- He, S. & Withers, S. G. (1997). J. Biol. Chem.272, 24864- 24867. [DOI] [PubMed]
- Henrissat, B. & Bairoch, A. (1996). Biochem. J.316, 695–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova, M., MacGregor, E. A., Biely, P., Stewart, R. J. & Fincher, G. B. (1998). J. Biol. Chem.273, 11134–11143. [DOI] [PubMed] [Google Scholar]
- McCarter, J. & Withers, S. G. (1994). Curr. Opin. Struct. Biol.4, 885–892. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Opassiri, R., Hua, Y., Wara-Aswapati, O., Akiyama, T., Svasti, J., Esen, A. & Ketudat Cairns, J. R. (2004). Biochem. J.379, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opassiri, R., Ketudat Cairns, J. R., Akiyama, T., Wara-Aswapati, O., Savasti, J. & Esen, A. (2003). Plant Sci.165, 627–638.
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- Sanz-Aparicio, J., Hermoso, J. A., Martinez-Ripoll, M., Lequerica, J. L. & Polaina, J. (1998). J. Mol. Biol.275, 491–502. [DOI] [PubMed] [Google Scholar]
- Sinnott, M. L. (1990). Chem. Rev.90, 1171–1202.
- Verdoucq, L., Morinière, J., Bevan, D. R., Esen, A., Vasella, A., Henrissat, B. & Czjzek, M. (2004). J. Biol. Chem.279, 31796–31803. [DOI] [PubMed] [Google Scholar]
- Wiesmann, C., Hengstenberg, W. & Schulz, G. E. (1997). J. Mol. Biol.269, 851–860. [DOI] [PubMed] [Google Scholar]
- Withers, S. G. & Street, I. P. (1988). J. Am. Chem. Soc.110, 8551–8553.