The expression, purification and preliminary X-ray diffraction analysis of a catalytic domain of a chitinase from P. furiosus is reported.
Keywords: hyperthermophilic chitinases, catalytic domain, archaea, Pyrococcus furiosus
Abstract
The crystallization and preliminary X-ray diffraction analysis of a catalytic domain of chitinase (PF1233 gene) from the hyperthermophilic archaeon Pyrococcus furiosus is reported. The recombinant protein, prepared using an Escherichia coli expression system, was crystallized by the hanging-drop vapour-diffusion method. An X-ray diffraction data set was collected at the undulator beamline BL44XU at SPring-8 to a resolution of 1.50 Å. The crystals belong to space group P212121, with unit-cell parameters a = 90.0, b = 92.8, c = 107.2 Å.
1. Introduction
Chitin is the second most abundant polysaccharide in the biosphere. N-Acetyl-d-glucosamine (GlcNAc) and its oligosaccharides derived by hydrolysis of chitin are used as food additives and medicines (Dahiya et al., 2006 ▶). Chitinase (EC 3.2.1.14) catalyzes the hydrolysis of the β-1,4-linkages between GlcNAcs in chitin. However, the industrial utilization of this biopolymer on a large scale needs an enzyme that can hydrolyze crystalline chitin without pretreatment with hydrochloric acid. These enzymes may help to reduce and eliminate the large amounts of waste generated by the food industry.
We have found in the genome database of Pyrococcus furiosus (http://gib.genes.nig.ac.jp) that two adjacent open reading frames (PF1234 and PF1233), separated by 37 bp, are homologous to the first and second half of a chitinase from Thermococcus kodakaraensis (TK-ChiA; Oku & Ishikawa, 2006 ▶). We have combined them into one gene by adjusting the frame and the gene product yielded a recombinant chitinase homologous to TK-ChiA (Oku & Ishikawa, 2006 ▶). Surprisingly, this artificial recombinant chitinase (referred to as PF-ChiA) exhibited hydrolytic activity not only towards colloidal chitin but also degraded crystalline chitins at high temperature. Sequence comparisons with TK-ChiA revealed that PF-ChiA contains two chitin-binding domains (ChBD1PF-ChiA and ChBD2PF-ChiA) and two active (catalytic) domains (AD1PF-ChiA and AD2PF-ChiA). AD1PF-ChiA and AD2PF-ChiA are classified into glycoside hydrolase family 18 (CAZy database; http://afmb.cnrs-mrs.fr/CAZY/; Coutinho & Henrissat, 1999 ▶). PF-ChiA is the only known member of the family to contain two catalytic domains. As there is no structural information concerning archaeal chitinases, we are also attempting to elucidate the tertiary structure and structure–function relationship of these enzymes. Therefore, we focused on structural studies of AD2PF-ChiA (residues 409–717, the residue numbering being that for the PF1233 amino-acid sequence), which exhibits homology to the residues 898–1215 of the TK-ChiA amino-acid sequence (http://gib.genes.nig.ac.jp; TK1765). Here, we describe the expression, purification and preliminary X-ray diffraction studies of the active domain AD2PF-ChiA of PF-ChiA.
2. Materials and methods
2.1. Construction of an AD2PF-ChiA expression vector
The AD2PF-ChiA gene was cloned into the pET32 expression vector (pET32_AD2PF-ChiA) by the LIC method (Novagen) using the following primers: primer 1, 5′-GACGACGACAAGATCCTGGA AGTTCTGTTCCAGGGGCCCAATGCAAATCCAATACCAG-3′, and primer 2, 5′-GAGGAGAAGCCCGGTTTATGTTGGAACACTAGCTTCGCG-3′. A plasmid containing the PF1233 gene (Oku & Ishikawa, 2006 ▶) was used as a template. The region of primer 1 in bold corresponds to the PreScission protease (Amersham Biosciences) recognition sequence (PSsequence). Therefore, AD2PF-ChiA has two additional residues (H2N-Gly-Pro) derived from the PSsequence at its N-terminus. As a result, the expression plasmid encoded the fusion protein thioredoxin-His6tag-PSsequence-AD2PF-ChiA. Fig. 1 ▶ shows the amino-acid sequence of AD2PF-ChiA which was crystallized in this study. Escherichia coli Rosetta (DE3) (Novagen) cells harbouring the pET32_AD2PF-ChiA plasmid were cultivated in LB medium containing 50 µg ml−1 ampicillin at 310 K. When the OD600 reached 0.6, expression was induced with 1 mM isopropyl 1-thio-β-d-galactopyranoside for 4 h at 310 K. The cells were harvested by centrifugation (6000g for 20 min) and then stored at 193 K.
Figure 1.
The amino-acid sequence of AD2PF-ChiA. The additional GP residues derived from the PSsequence are shown in bold. The amino-acid numbers are shown as subscripts. The residue numbering is that for the PF1233 amino-acid sequence.
2.2. Purification
All of the following procedures were carried out at room temperature unless otherwise noted. The cell pellet (approximately 20 g) was suspended in buffer A (20 mM Tris–HCl pH 8.5, 0.5 M NaCl; 4 ml buffer per gram of cell pellet) and then sonicated. After removal of the cell debris by centrifugation (30 000g for 40 min), the supernatant was loaded onto a HiTrap chelating column (5 ml bed volume; Amersham Biosciences) preloaded with Ni2+ and equilibrated with buffer A. The column was washed with five column volumes of buffer A and the bound protein was then eluted with a 100 ml linear gradient of imidazole (0–0.5 M) in buffer A at a flow rate of 3 ml min−1. The objective protein was eluted at around 200–250 mM imidazole. The eluted peak containing the target protein was dialyzed against buffer B (20 mM Tris–HCl pH 8.5, 25 mM NaCl). To remove the thioredoxin-His6tag portion, PreScission protease (100 units) was added to the dialysate and the resultant solution was incubated at 277 K for 12 h. The solution was again applied onto the HiTrap-chelating column (5 ml bed volume) preloaded with Ni2+ and equilibrated with buffer B; the flowthrough fractions were collected. In this step, the thioredoxin-His6tag portion was effectively bound to the column and removed from the protease-digested solution. The flowthrough fraction was applied onto a HiTrap-Q column (5 ml bed volume; Amersham Biosciences), washed with five column volumes of buffer B and eluted with a 100 ml linear gradient of NaCl in buffer A at a flow rate of 3 ml min−1. AD2PF-ChiA eluted at around 150–200 mM NaCl. The fraction containing AD2PF-ChiA was collected and concentrated using a 3.5 kDa cutoff filter (Millipore). The protein was dialyzed against 20 mM Tris–HCl pH 8.0. A 20 mg ml−1 sample in this buffer was used for crystallization trials. The purity of the protein was confirmed by SDS–PAGE. Protein concentrations were estimated using the calculated molar absorption coefficient at 280 nm (∊280 = 26 600; Edelhoch, 1967 ▶).
2.3. Crystallization
Crystal Screen (Hampton Research) and tissue-culture plates (MP Biomedicals) were used for crystallization trials. Crystals were grown by the hanging-drop vapour-diffusion method. A 1.5 µl protein sample (20 mg ml−1 in 20 mM Tris–HCl pH 8.0) was mixed with an aliquot of reservoir solution in a 1:1 ratio and then left to equilibrate against 300 µl reservoir solution at 298 K for two weeks. A reservoir solution from Crystal Screen comprising 0.1 M MES pH 6.5 and 1.6 M magnesium sulfate produced crystals that were good enough for the following X-ray diffraction experiment.
2.4. Diffraction data collection
The optimum cryoconditions were found by testing the crystal using an R-AXIS VII image-plate detector and Cu Kα radiation from an FR-E rotating-anode generator (Rigaku). 18%(v/v) glycerol in 0.1 M MES pH 6.5, 1.6 M magnesium sulfate was chosen as the cryo-buffer. A single crystal (0.1 × 0.1 × 0.5 mm) was isolated from a drop and then scooped up in a Cryo-Loop (Hampton Research). After dipping the crystal in the cryo-buffer for a few seconds, the crystal was immediately cooled to 100 K in a stream of nitrogen gas. Diffraction data were collected at the undulator beamline BL44XU at SPring-8 (Harima, Japan) equipped with a DIP-6040 image plate (Bruker). The crystal-to-detector distance was 250 mm. The oscillation range was 0.5° per frame. The diffraction data from 482 frames were integrated and scaled with DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶). The detailed experimental conditions are summarized in Table 1 ▶.
Table 1. Experimental conditions and data-collection statistics.
Values in parentheses are for the highest resolution shell.
| X-ray source | BL44XU, SPring-8 |
| Wavelength (Å) | 0.7 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 90.0, b = 92.8, c = 107.2 |
| Resolution range (Å) | 50–1.5 (1.55–1.50) |
| Rsym† (%) | 10.7 (26.6) |
| 〈I/σ(I)〉 | 18.0 (6.8) |
| Total reflections | 1109463 |
| Unique reflections | 142710 (14039) |
| Redundancy | 7.8 (5.9) |
| Completeness (%) | 99.5 (98.8) |
R
sym = 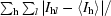
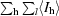 , where I
l is the lth observation of reflection h and 〈I
h〉 is the weighted average intensity for all observations l of reflection h.
, where I
l is the lth observation of reflection h and 〈I
h〉 is the weighted average intensity for all observations l of reflection h.
3. Results and discussion
As the level of expression of AD2PF-ChiA alone was very low (less than 0.1 mg from 1 l culture), we expressed AD2PF-ChiA as a thioredoxin-fused form. Using this system, a sufficient amount and purity of AD2PF-ChiA were obtained, with a yield of 10 mg from 1 l culture.
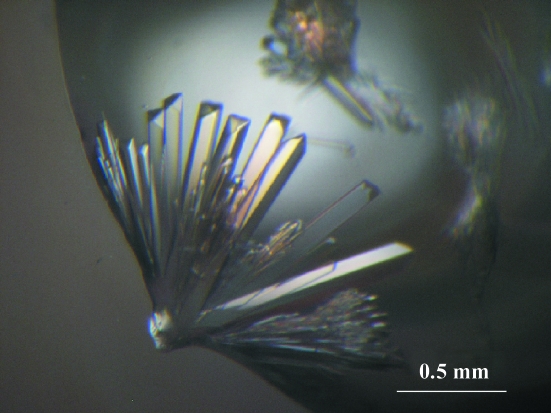
Crystals of AD2PF-ChiA appeared after a week of vapour diffusion and stopped growing within two weeks (Fig. 2 ▶). Diffraction data were collected at 100 K to a resolution of 1.50 Å. The data-collection statistics are summarized in Table 1 ▶. Assumption of the presence of two molecules per asymmetric unit gives a crystal volume per protein weight (V M; Matthews, 1968 ▶) of 3.20 Å3 Da−1 and a solvent content of 61%.
Figure 2.
Photograph of AD2PF-ChiA crystals grown by the hanging-drop vapour-diffusion method. A crystal of 0.1 × 0.1 × 0.5 mm in size was used for X-ray analyses.
Crystallographic studies of AD2PF-ChiA, together with those of ChBD2PF-ChiA (Nakamura et al., 2005 ▶), should provide insight into the enzymatic utilization of the abundant crystalline chitin. We have recently succeeded in obtaining a selenomethionyl derivative of AD2PF-ChiA in order to solve its crystal structure using the MAD method.
Acknowledgments
We wish to thank Emeritus Professor N. Yasuoka, Himeji Institute of Technology for helpful discussions on crystallization and X-ray diffraction analysis. We also thank Ms C. Yoshikawa-Kageyama for performing part of the molecular-biological work. We gratefully acknowledge the assistance of Dr M. Yoshimura during the collection of X-ray data on beamline BL44XU at SPring-8. We thank Dr A. Kobayashi, National Institute of Advanced Industrial Science and Technology, for being so kind and helpful in revising all important scientific aspects of this project. This work was supported by the National Project on Protein Structural and Functional Analyses. The X-ray diffraction experiments were carried out with the approval of the Japan Synchrotron Radiation Research Institute (proposal No. 2005AC05A44XU-7201-N).
References
- Coutinho, P. M. & Henrissat, B. (1999). Recent Advances in Carbohydrate Bioengineering, pp. 3–12. Cambridge: The Royal Society of Chemistry.
- Dahiya, N., Tewari, R. & Hoondal, G. S. (2006). In the press.
- Edelhoch, H. (1967). Biochemistry, 6, 1948–1954. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Ishikawa, K., Hagihara, Y., Oku, T., Nakagawa, A., Inoue, T., Ataka, M. & Uegaki, K. (2005). Acta Cryst. F61, 476–478. [DOI] [PMC free article] [PubMed]
- Oku, T. & Ishikawa, K. (2006). In the press.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]