Two structures of the enzyme phosphomannomutase/phosphoglucomutase in complex with ribose 1-phosphate and xylose 1-phosphate are described. Despite structural differences from the enzyme’s preferred phosphohexose substrates, both ligands trigger the interdomain rotation required for catalysis.
Keywords: phosphomannomutase/phosphoglucomutase, ribose 1-phosphate, xylose 1-phosphate
Abstract
Two complexes of the enzyme phosphomannomutase/phosphoglucomutase (PMM/PGM) from Pseudomonas aeruginosa with a slow substrate and with an inhibitor have been characterized by X-ray crystallography. Both ligands induce an interdomain rearrangement in the enzyme that creates a highly buried active site. Comparisons with enzyme–substrate complexes show that the inhibitor xylose 1-phosphate utilizes many of the previously observed enzyme–ligand interactions. In contrast, analysis of the ribose 1-phosphate complex reveals a combination of new and conserved enzyme–ligand interactions for binding. The ability of PMM/PGM to accommodate these two pentose phosphosugars in its active site may be relevant for future efforts towards inhibitor design.
1. Introduction
The enzyme PMM/PGM is a member of the α-d-phosphohexomutase enzyme superfamily (Shackelford et al., 2004 ▶). In the bacterium Pseudomonas aeruginosa, this enzyme participates in the biosynthesis of three virulence factors: alginate, lipopolysaccharide and rhamnolipid (Govan & Deretic, 1996 ▶; Olvera et al., 1999 ▶; Rocchetta et al., 1999 ▶). It has dual substrate specificity, catalyzing the reversible conversion of either glucose 6-phosphate to glucose 1-phosphate (G1P) or mannose 6-phosphate to mannose 1-phosphate, depending on the biosynthetic pathway in which it is operating. The reaction requires a phosphorylated serine on the enzyme (residue 108) and proceeds via the formation of a bisphosphorylated sugar intermediate (e.g. glucose 1,6-bisphosphate).
Previous characterization of PMM/PGM has shown that it is a heart-shaped protein with 463 residues comprising four domains (Regni et al., 2002 ▶). Structural studies of PMM/PGM in complex with its preferred substrates and products, as well as the reaction intermediate glucose 1,6-bisphosphate, have revealed key details of its ligand-binding specificity and new insights into the enzyme mechanism (Regni et al., 2004 ▶, 2006 ▶). In particular, these studies have shown how the enzyme accommodates 1-phosphosugars and 6-phosphosugars in two distinct orientations in the same active site.
Here, we describe two structures of P. aeruginosa PMM/PGM in complex with a slow substrate, ribose 1-phosphate (R1P; Ye et al., 1994 ▶), and the inhibitor xylose 1-phosphate (X1P). These two structures show that phosphosugars other than the preferred hexose substrates can still trigger the interdomain rotation required for formation of a high-affinity ligand-binding site. This result emphasizes the importance of the phosphate group of the ligand as a key determinant of binding. In the case of R1P, small rearrangements of residues in the active site allow the enzyme to maintain a number of conserved structural interactions to the ligand, despite the structural differences from its preferred phosphohexose substrates.
2. Materials and methods
2.1. Sample preparation and data collection
R1P and X1P were purchased from Sigma. Purification and crystallization of the PMM/PGM–ligand complexes was carried out as previously described (Naught & Tipton, 2001 ▶; Regni et al., 2000 ▶). Briefly, full-length wild-type PMM/PGM was expressed in Escherichia coli from the pET-3a vector, purified via ion-exchange and hydrophobic interaction chromatography and concentrated to ∼5 mg ml−1 in 10 mM MOPS pH 7.4. Crystals of apo-PMM/PGM were grown by the hanging-drop vapour-diffusion method from well buffer consisting of 50–60% saturated sodium/potassium tartrate and 100 mM Na HEPES pH 7.5; drops contained 2 µl protein solution and 2 µl well buffer. Seeding was generally used to speed crystal growth and improve morphology (Regni et al., 2000 ▶). These crystals contain the phosphorylated form of the enzyme, as purified from E. coli, but are catalytically inactive owing to substitution of Zn2+ in the Mg2+-binding site. For complex formation, crystals were transferred quickly into solutions of 79%(w/w) PEG 4000, 100 mM sodium HEPES pH 7.4 and 25 mM ligand and were allowed to soak for several minutes.
Crystals were flash-cooled without further cryoprotection for data collection at 100 K. Data sets from the R1P and X1P complexes were collected at beamlines 14-BMC and SBC 19-ID at the Advanced Photon Source, Argonne National Laboratory. Data sets of 270 and 447 frames were collected for the X1P and R1P complexes, respectively, using oscillation steps of 0.5°. Diffraction data were processed with DENZO and merged with SCALEPACK (Otwinowski & Minor, 1997 ▶) (Table 1 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell: 1.73–1.79 Å for the X1P complex and 2.40–2.49 Å for the R1P complex.
| X1P | R1P | |
|---|---|---|
| Data collection | ||
| X-ray source | SBC 19-ID | 14-BMC |
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å) | a = 71.00, b = 74.55, c = 86.09 | a = 70.71, b = 74.10, c = 84.34 |
| Wavelength (Å) | 0.979 | 1.0398 |
| Resolution (Å) | 50.0–1.73 | 50.0–2.40 |
| Total no. of reflections | 202946 | 70265 |
| Unique reflections | 46057 | 16595 |
| Redundancy | 4.4 | 4.2 |
| Rmerge (%) | 6.3 (38.5) | 10.0 (49.2) |
| I/σ(I) | 18.4 (2.5) | 14.3 (2.1) |
| Reflections with I > 3σ(I) (%) | 75.0 (37.7) | 69.3 (38.3) |
| Completeness (%) | 96.1 (90.6) | 91.7 (83.9) |
| Mosaicity (°) | 0.97 | 1.89 |
| Refinement statistics | ||
| Resolution (Å) | 50.0–1.7 | 43.8–2.4 |
| Rwork/Rfree | 0.18/0.21 | 0.19/0.26 |
| Non-H atoms | 4034 | 3599 |
| Waters | 530 | 144 |
| 〈B〉 (Å2) | ||
| Protein atoms | 23.7 | 22.2 |
| Waters | 37.8 | 32.7 |
| Ligand | 19.4 | 38.0 |
| R.m.s.d. bonds (Å) | 0.010 | 0.009 |
| R.m.s.d. angles (°) | 1.23 | 1.19 |
| Ramachandran statistics (%) | ||
| Most favored | 91.9 | 90.9 |
| Additionally allowed | 7.5 | 8.6 |
| Luzzati coordinate error (Å) | 0.194 | 0.300 |
2.2. Structure solution, refinement and analysis
The complex of PMM/PGM with G1P (without ligand, waters or multiple conformations) was used as a starting point for rigid-body refinement of the X1P and R1P complexes. Electron density for the ligands in each complex appeared as nearly continuous positive peaks in F o − F c maps contoured at 3.0σ, allowing unambiguous placement of the ligands. The structures were refined to convergence through iterative cycles of positional refinement using REFMAC v.5.0 (Murshudov et al., 1999 ▶) and manual rebuilding with O (Jones et al., 1991 ▶) or COOT (Emsley & Cowtan, 2004 ▶). The progress of the refinement was monitored by following R free. 5% of each data set was set aside for cross-validation prior to refinement; the same reflections in the R1P and X1P data sets were flagged as the test set as in the starting model (PDB code 1p5d). Water molecules were placed automatically by WATPEAK (Collaborative Computational Project, Number 4, 1994 ▶) in peaks >3.0σ in F o − F c maps and within hydrogen-bonding distance of N or O atoms; water molecules without electron density at 1.0σ in 2F o − F c maps and with B factors above 60 Å2 were removed from the model.
The final models (Table 1 ▶) extend from residue 9 to residue 463. Each model contains the following heteroatoms: phosphoserine 108, Zn2+, phosphosugar and solvent. The X1P complex has five residues with two conformations, while the R1P complex has one side chain and one atom (O5) of the ligand modelled in two conformations. The R1P complex has three residues whose side chains were truncated to alanine; the X1P complex has nine. All models were validated with the programs WHATIF (Hooft et al., 1996 ▶), SFCHECK (Vaguine et al., 1999 ▶) and PROCHECK (Laskowski et al., 1993 ▶).
For structural analyses, the pocket surface areas and volumes were calculated using CAST (Liang et al., 1998 ▶), the solvent-accessible surface areas of the ligands were calculated using AREAIMOL (Collaborative Computational Project, Number 4, 1994 ▶), domain rotations were obtained using DYNDOM (Hayward & Berendsen, 1998 ▶) and Cα superpositions were performed using TOP (Lu, 2000 ▶). Potential enzyme–ligand hydrogen bonds with favorable geometry as determined by CONTACT (Collaborative Computational Project, Number 4, 1994 ▶) are compiled in Table 2 ▶. Figures were prepared with PyMOL (DeLano, 2002 ▶).
Table 2. Potential enzyme–ligand hydrogen-bond contacts in the R1P and X1P complexes with PMM/PGM.
All hydrogen bonds are <3.3 Å with favorable geometry. New contacts relative to previously characterized complexes are shown in bold.
| Ligand atom | Protein atom | X1P (Å) | R1P (Å) |
|---|---|---|---|
| Phosphate contacts | |||
| O1P | Arg421 NH1 | 2.79 | 2.73 |
| Water | 2.65† | ||
| O2P | Ser423 OG | 2.67 | 2.54 |
| Thr425 OG1 | 2.66 | 2.58 | |
| Water | 2.75 | ||
| O3P | Tyr17 OH | 2.64 | 2.57 |
| Arg421 NH2 | 2.93 | 2.90 | |
| Asn424 N | 2.84 | 2.72 | |
| Sugar contacts | |||
| O2 | Lys285 NZ | 3.27 | 3.03 |
| Water | 3.20† | 3.20 | |
| Water | 2.98 | ||
| O3 | Glu325 OE1 | 2.56 | 2.66 |
| Ser327 OG | 2.69 | ||
| Water | 2.83 | 2.91† | |
| O4 | His308 N | 2.94 | |
| Glu325 OE2 | 2.75 | ||
| O5 | Arg20 NH2 | 3.19 | |
| Ser108 O1P | 2.87 | ||
| Ser108 O2P | 2.86 | ||
| Water | 2.69 | 2.54† |
The same water molecule contacts both residues.
3. Results and discussion
3.1. Overall structures
The structures of apo-PMM/PGM, of four complexes with its preferred substrates and of two with intermediates have previously been reported (Regni et al., 2002 ▶, 2004 ▶, 2006 ▶). In the enzyme–substrate complexes, the protein was shown to adopt a ‘closed’ conformation, burying the ligands in the central active-site cleft. This cleft contains key residues for catalysis, including the catalytic phosphoserine 108, which is the site of phosphoryl transfer, and also a metal-binding loop which coordinates the Mg2+ ion necessary for activity. The active site also encompasses the conserved phosphate-binding site formed by residues from domains 1 and 4 of the enzyme, which interacts with the phosphate group of the substrates, regardless of whether they are 1- or 6-phosphosugars.
The X1P and R1P complexes with PMM/PGM are quite similar in overall structure to the enzyme–substrate complexes. For the purposes of discussion, we cite comparisons with the G1P complex (PDB code 1p5d), since this ligand is structurally the most similar to X1P (Fig. 1 ▶). The X1P and R1P complexes have Cα root-mean-square deviations (r.m.s.d.) of 0.17 and 0.26 Å, respectively, with the G1P complex. As seen in the enzyme–substrate complexes, a rotation of domain 4 of the enzyme by ∼9° occurs relative to the apoprotein structure, enclosing the bound ligands in the active site (Figs. 2a and 2b). Both X1P and R1P are highly buried in this pocket, with less than 10% of their total surface area exposed to solvent.
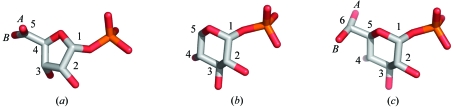
Figure 1.
Structures of (a) ribose 1-phosphate (R1P), (b) xylose 1-phosphate (X1P) and (c) glucose 1-phosphate (G1P). Ligands are shown as bound in the active site of PMM/PGM, using superpositions of the enzyme Cα atoms. Numbers refer to the sugar C atoms. In the case of R1P and G1P, the O5 and O6 hydroxyls, respectively, were modeled in two conformations at 50% occupancy (labeled as A and B).
3.2. Enzyme–ligand interactions
The preferred substrates of PMM/PGM are the six-carbon phosphosugars glucose and mannose. The two ligands used in this study, xylose and ribose, are both pentose phosphosugars. However, xylose forms a six-membered pyranose ring, while ribose forms a five-membered furanose ring (Fig. 1 ▶). X1P is structurally identical to the substrate G1P except that it lacks C6 and the O6 hydroxyl and therefore cannot participate in phosphoryl transfer. In contrast, since R1P has a furanose ring, this ligand shows significant structural differences (e.g. overall dimensions, ring size and number of sugar hydroxyls) compared with the phosphohexose substrates of PMM/PGM. However, as shown in Fig. 1 ▶, both X1P and R1P adopt an overall binding orientation in the active site similar to that of G1P, with their phosphate groups occupying the conserved phosphate-binding site.
Residues from multiple domains of PMM/PGM participate in direct interactions with X1P (Fig. 2 ▶ c and Table 2 ▶). They include extensive interactions between the phosphate group of the ligand and residues in domains 1 and 4 of the protein. Also, contacts to three sugar hydroxyls (O2, O3 and O4) are made by residues in domain 3 (Lys285, His308, Glu325 and Ser327). The PMM/PGM complex with X1P is very similar to the previously characterized complex with G1P. This is also consistent with the K d for X1P, which is approximately ∼8 µM (Regni & Henzl, unpublished data). Except for a missing contact between O5 and Arg247, all the direct contacts between the enzyme and X1P are the same as those observed in the G1P complex, with only small changes in bond distances and geometry (Table 2 ▶). As indicated by the low r.m.s.d., most residues in the active site of the X1P and G1P complexes are essentially superimposable: a few exceptions to this are the side chains of several residues that contact O6 in the G1P complex but change conformation in the X1P complex, presumably because the ligand lacks this functional group. Five well ordered water molecules form apparent hydrogen bonds to X1P (Fig. 2 ▶ c); two of these are new relative to the G1P complex. Four of these five waters make bridging interactions between the inhibitor and side chains of the enzyme.
Figure 2.
PMM/PGM in complex with X1P and R1P. The protein is colored by domain, with blue, light blue, pink and magenta for domains 1, 2, 3 and 4, respectively. (a) Ribbon diagram of PMM/PGM with X1P (yellow) and R1P (green) bound in the active site. (b) Superposition of the Cα atoms of the R1P complex with those of apo-PMM/PGM (PDB code 1k2y) shown in gray, demonstrating the rotation of domain 4. Close-up view of the active site of PMM/PGM with bound (c) X1P and (d) R1P. For clarity, only interactions between side chains of the enzyme and ligands are highlighted; water molecules are shown as spheres. (e) For comparison, a superposition of the slow substrate R1P and a preferred substrate G1P (cyan) in the active site of PMM/PGM is shown.
Given the similarities of the protein conformation and enzyme–ligand interactions between the X1P and G1P complexes, it is apparent that contacts made to the O6 hydroxyl are not a critical determinant of ligand binding, even though O6 is one of the sites of phosphoryl transfer during catalysis. Based on the four enzyme–substrate complexes, it was proposed that the extensive contacts observed between the enzyme and phosphate group of the ligands are a primary determinant of substrate recognition (Regni et al., 2004 ▶). The PMM/PGM complex with X1P supports this proposal; binding of this ligand produces the same structural changes in the enzyme, including the creation of a domain 1–4 interface critical for sequestering ligands in the active site. Although O6 is apparently not essential for binding or conformational change of the protein, our structural studies do not address the potential energetic importance of contacts to this hydroxyl, which remain to be determined.
In contrast to X1P, which is incapable of undergoing phosphoryl transfer, R1P is a slow substrate for PMM/PGM, although the specific activity of the enzyme with R1P is only about 1% of that with G1P (Ye et al., 1994 ▶). As seen in other complexes, multiple contacts are made between PMM/PGM and the phosphate group of the ligand (Fig. 2 ▶ d and Table 2 ▶), primarily by residues in domain 4 and also by Tyr17 from domain 1. Two sugar hydroxyls of R1P (O2 and O3) also participate in direct hydrogen bonds with two residues from domain 3 (Lys285 and Glu325, respectively). Several novel interactions, including contacts to O5 from residues in domain 1 (Arg20 and phosphoserine 108), are also observed in the complex with R1P.
Many of the observed enzyme–R1P interactions are similar to those seen in previous complexes, despite the distinct structural differences between R1P and the preferred substrates of PMM/PGM (Fig. 2 ▶ e). In particular, all of the conserved enzyme contacts to the phosphate group of the ligand are retained. However, the bond distances and angles of these phosphate contacts vary more than those seen in other complexes, where only tiny differences were observed regardless of ligand identity. In addition, some key contacts to the sugar hydroxyls are maintained, including those from Lys285 and Glu325 (Fig. 2 ▶ d). Unlike the contacts made to its phosphohexose substrates, however, Glu325 does not make a bidentate interaction with the ligand, but rather makes a second hydrogen bond to a water molecule that occupies a position similar to that of the O4 hydroxyl to glucose. Some other interactions conserved in the substrate complexes are missing, including those from Ser327 and His308. Generally, small differences in the conformations of several side chains, including Glu325 and Lys285, are seen relative to the G1P complex. In combination, however, these changes are sufficient to position the ligand appropriately for phosphoryl transfer, at least at low efficiency. Structural superpositions show that O5 of R1P comes within 1.0 Å of the O6 of G1P, depending on which rotamer of these hydroxyls is compared (Fig. 2 ▶ e).
Only three well ordered water molecules participate in contacts with R1P bound in the active site (Fig. 2 ▶ d). Thus, a number of otherwise conserved water-mediated contacts are missing, including several to the phosphate O atoms of the ligand. It is tempting to speculate on the functional relevance of this observation; however, it seems likely that the relatively small number of observed waters is primarily a consequence of the moderate resolution of the structure (2.4 Å), which is lower than that of the other PMM/PGM ligand complexes. Curiously, the limited resolution of this complex appears to be ligand-dependent: despite repeated attempts at soaks with R1P, we were unable to produce crystals that diffracted to the typical limit of 2.0 Å or better. Nevertheless, contacts between the enzyme and R1P are still extensive, utilizing nearly all of the functional groups of the ligand except for O4. This result is consistent with earlier observations noting the plasticity of the active site of PMM/PGM, which can accommodate its glucose- and mannose-based substrates in two distinct high-affinity binding orientations (Regni et al., 2004 ▶).
In summary, the two PMM/PGM complexes with the phosphopentoses X1P and R1P demonstrate that considerable structural variation in the sugar portion of the ligand can be tolerated in the active site of the enzyme. Contacts to the phosphate group, on the other hand, are similar in both complexes and thus appear likely to be necessary and perhaps sufficient to trigger the interdomain rearrangement of the enzyme. These structures provide additional information on the plasticity of the active site of PMM/PGM and suggest that a variety of phosphosugar templates may be useful as starting points for efforts at inhibitor design.
Supplementary Material
PDB reference: PMM/PGM–R1P complex, 2h4l, r2h5lsf
PDB reference: PMM/PGM–X1P complex, 2h5a, r2h5asf
Acknowledgments
We thank the staff of beamlines SBC 19-ID and 14-BMC of the Advanced Photon Source of Argonne National Laboratory for synchrotron time and technical expertise. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science under Contract No. W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources under grant No. RR07707.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- DeLano, W. L. (2002). The PyMOL Molecular Visualization System. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Govan, J. R. W. & Deretic, V. (1996). Microbiol. Rev.60, 539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward, S. & Berendsen, H. J. (1998). Proteins, 30, 144–154. [PubMed] [Google Scholar]
- Hooft, R. W., Vriend, G., Sander, C. & Abola, E. E. (1996). Nature (London), 381, 272. [DOI] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291. [Google Scholar]
- Liang, J., Edelsbrunner, H. & Woodward, C. (1998). Protein Sci.7, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, G. (2000). J. Appl. Cryst.33, 176–183. [Google Scholar]
- Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. (1999). Acta Cryst. D55, 247–255. [DOI] [PubMed] [Google Scholar]
- Naught, L. E. & Tipton, P. A. (2001). Arch. Biochem. Biophys.396, 111–118. [DOI] [PubMed] [Google Scholar]
- Olvera, C., Goldberg, J. B., Sanchez, R. & Soberon-Chavez, G. (1999). FEMS Microbiol. Lett.179, 85–90. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Regni, C., Naught, L. E., Tipton, P. A. & Beamer, L. J. (2004). Structure, 12, 55–63. [DOI] [PubMed] [Google Scholar]
- Regni, C., Schramm, A. M. & Beamer, L. J. (2006). J. Biol. Chem.281, 15564–15571. [DOI] [PubMed] [Google Scholar]
- Regni, C., Tipton, P. A. & Beamer, L. J. (2000). Acta Cryst. D56, 761–762. [DOI] [PubMed] [Google Scholar]
- Regni, C., Tipton, P. A. & Beamer, L. J. (2002). Structure, 10, 269–279. [DOI] [PubMed] [Google Scholar]
- Rocchetta, H. L., Burrows, L. L. & Lam, J. S. (1999). Microbiol. Mol. Biol. Rev.63, 523–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford, G. S., Regni, C. A. & Beamer, L. J. (2004). Protein Sci.13, 2130–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed] [Google Scholar]
- Ye, R. W., Zielinski, N. A. & Chakrabarty, A. M. (1994). J. Bacteriol.176, 4851–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: PMM/PGM–R1P complex, 2h4l, r2h5lsf
PDB reference: PMM/PGM–X1P complex, 2h5a, r2h5asf