Recombinant D. radiodurans TrxR with a His tag at the N-terminus was expressed in Escherichia coli and purified by metal-affinity chromatography. The protein was crystallized using the sitting-drop vapour-diffusion method in the presence of 35% PEG 4000, 0.2 M ammonium acetate and citric acid buffer pH 5.1 at 293 K.
Keywords: thioredoxin reductase, Deinococcus radiodurans
Abstract
Deinococcus radiodurans, a Gram-positive bacterium capable of withstanding extreme ionizing radiation, contains two thioredoxins (Trx and Trx1) and a single thioredoxin reductase (TrxR) as part of its response to oxidative stress. Thioredoxin reductase is a member of the family of pyridine nucleotide-disulfide oxidoreductase flavoenzymes. Recombinant D. radiodurans TrxR with a His tag at the N-terminus was expressed in Escherichia coli and purified by metal-affinity chromatography. The protein was crystallized using the sitting-drop vapour-diffusion method in the presence of 35% PEG 4000, 0.2 M ammonium acetate and citric acid buffer pH 5.1 at 293 K. X-ray diffraction data were collected on a cryocooled crystal to a resolution of 1.9 Å using a synchrotron-radiation source. The space group was determined to be P3221, with unit-cell parameters a = b = 84.33, c = 159.88 Å. The structure of the enzyme has been solved by molecular-replacement methods and structure refinement is in progress.
1. Introduction
Reactive oxygen species (ROS) are generated and degraded by all aerobic organisms as part of normal cell function. ROS include hydroxyl radical (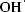 ), hydrogen peroxide (H2O2), superoxide (
), hydrogen peroxide (H2O2), superoxide (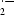 ) and organic hydroperoxides (OHPs). However, excessive production of ROS can lead to oxidative stress and cause damage to cellular macromolecules, including DNA and proteins (Nordberg & Arner, 2001 ▶; Meunier-Jamin et al., 2004 ▶). To maintain their integrity, cells have evolved enzymatic systems to minimize the damage caused by ROS. These enzymatic systems scavenge ROS before they can cause damage or repair macromolecules after ROS damage (Atichartpongkul et al., 2001 ▶; Meunier-Jamin et al., 2004 ▶). One such enzymatic system is the thioredoxin system, which scavenges ROS (Arner & Holmgren, 2000 ▶).
) and organic hydroperoxides (OHPs). However, excessive production of ROS can lead to oxidative stress and cause damage to cellular macromolecules, including DNA and proteins (Nordberg & Arner, 2001 ▶; Meunier-Jamin et al., 2004 ▶). To maintain their integrity, cells have evolved enzymatic systems to minimize the damage caused by ROS. These enzymatic systems scavenge ROS before they can cause damage or repair macromolecules after ROS damage (Atichartpongkul et al., 2001 ▶; Meunier-Jamin et al., 2004 ▶). One such enzymatic system is the thioredoxin system, which scavenges ROS (Arner & Holmgren, 2000 ▶).
Thioredoxin reductases work in conjunction with thioredoxin to form an important cellular disulfide reductase system. All thioredoxin reductases are homodimeric proteins, with each monomer containing a flavin adenine dinucleotide (FAD) binding domain, a nicotinamide adenine dinucleotide phosphate (NADP) binding domain and an active site containing a redox-active disulfide. Electrons are transferred to FAD from NADPH, then from FAD to the active-site disulfide and finally from thioredoxin reductase to thioredoxin. Thioredoxin in turn reduces other proteins such as thioredoxin-dependent thiol peroxidase (TPx) and ribonucleotide reductase. Thioredoxin peroxidase eliminates ROS (H2O2), whereas ribonucleotide reductase is required for DNA synthesis. The thioredoxin system thus plays an important regulatory role in addition to scavenging ROS (Mustacich & Powis, 2000 ▶; Arner & Holmgren, 2000 ▶).
The ability of Deinococcus radiodurans to withstand extreme ionizing radiation has made the bacterium a subject of intense study, with potential applications in remediation of radioactive mixed waste sites. D. radiodurans sustains the same amount of damage at high irradiation doses compared with other bacteria. It is therefore hypothesized that to survive ionizing radiation D. radiodurans must have an exceptional ability to repair double-stranded DNA breaks in its genome. This is probably achieved by homologous recombination after the occurrence of double-stranded DNA breaks (Battista, 2000 ▶). In addition to DNA-repair genes, D. radiodurans contains several genes that code for proteins that scavenge ROS and thus prevent DNA damage (White et al., 1999 ▶). It is therefore possible that prevention of DNA damage supplements DNA repair to make D. radiodurans tolerant of ionizing radiation (Meunier-Jamin et al., 2004 ▶).
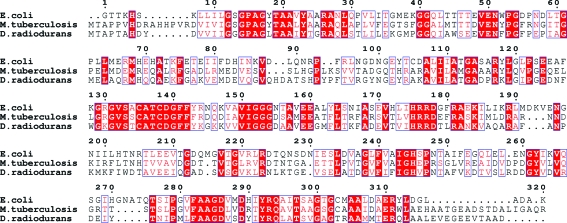
D. radiodurans has two thioredoxin genes and a single thioredoxin reductase gene which form part of its defence against oxidative damage. The TrxR homologue from D. radiodurans shows 44% identity to Escherichia coli TrxR and high identity to many other TrxRs (Fig. 1 ▶). The complete D. radiodurans genome of strain R1 is available and a putative peroxidase (TPx) has been identified (White et al., 1999 ▶); thus, the following electron-transfer path is plausible: NADPH→TrxR→Trx→TPx-H2O2. Thioredoxin reductase is a part of the enzymatic systems that protect D. radiodurans against ROS damage and may be important in understanding the extremophilic nature of D. radiodurans. We have overexpressed, purified and crystallized thioredoxin reductase from D. radiodurans in order to determine its three-dimensional structure. This paper describes the preliminary X-ray crystallographic characterization of TrxR from D. radiodurans.
Figure 1.
Sequence alignment of thioreodixn reductase from E. coli, M. tuberculosis and D. radiodurans. Identical residues are highlighted in red, while similar residues are boxed in blue. Figure created using ESPript (Gouet et al., 1999 ▶).
2. Materials and methods
2.1. Cloning and overexpression
The TrxR gene (DR1982) was obtained by the polymerase chain reaction (PCR) using D. radiodurans strain R1 genomic DNA as a template (ATCC). The following were used as forward and reverse oligonucleotide primers for PCR, respectively: 5′-GCG CCA TGG GTA TGA CGG CAC CTA CTG-3′ and 5′-GCG GGA TCC TCA GTC GGC AGCC-3′.
The resulting amplified 1 kbp fragment was purified by gel extraction, digested with NcoI and BamHI and cloned into a modified pET-30b vector (thrombin cleavage site replaced with a TEV protease site) kindly provided by Dr H. Liu (St Andrews University, Scotland). For cloning, Escherichia coli bacterial strain NovaBlue (Novagen, USA) was used. The cloned gene was verified by DNA sequencing (Plant Biotechnology Institute, Saskatoon). The resulting construct was transformed into E. coli expression strain Rosetta (Novagen, USA). Growth took place on LB agar plates supplemented with 50 µg ml−1 kanamycin. Single colonies were selected and grown overnight in LB media with 50 µg ml−1 kanamycin. The primary culture was used to inoculate 4 l of LB medium and the cells were grown at 310 K to an OD600 of 0.6. Protein overexpression was then induced by 0.4 mM IPTG. The cells were grown for 4 h after induction and harvested by centrifugation (20 min, 8000g, 277 K) and the pellets stored at 193 K.
2.2. Purification
The frozen cell pellets were thawed and resuspended in sonication buffer (50 mM Tris–HCl pH 8.0, 1 mM AEBSF, 20 µg ml−1 DNase and 20 µg ml−1 lysozyme). The thawed cells were mechanically disrupted by sonication and cell debris was removed by centrifugation. The resulting supernatant was loaded onto a POROS MC50 metal-chelation column (Applied Biosystems,USA) pre-equilibrated with a buffer containing 5 mM imidazole, 0.5 M NaCl, 50 mM Tris–HCl pH 8.0. The column was washed with a 60 mM imidazole, 0.5 M NaCl, 50 mM Tris–HCl pH 8.0 buffer to remove non-specific binding and the protein eluted with a 0–1 M imidazole gradient. Fractions (10 ml) were collected and the purity of the protein was checked on Coomassie-stained SDS–PAGE. Those fractions showing high level of purity were pooled and dialyzed against crystallization buffer (50 mM Tris–HCl pH 8). Since the protein was considered to be greater than 98% pure after the metal-chelation purification step, the His tag was not cleaved from the protein.
3. Crystallization, data collection and processing
3.1. Crystallization
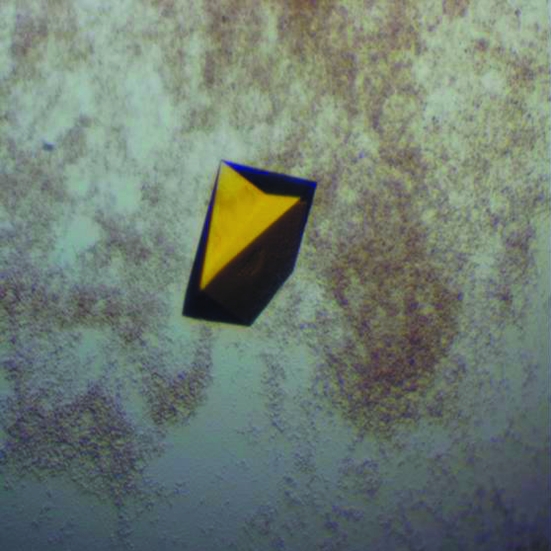
The protein was concentrated to 10 mg ml−1 and used for crystallization trials at 293 K. Initial screening was performed by the sitting-drop vapour-diffusion method using broad screens from Qiagen. A 4 µl drop consisting of 2 µl well buffer and 2 µl protein solution was equilibrated against 100 µl reservoir buffer. Crystals appeared in several conditions after 3 d. After optimization, the best crystals were obtained in 35%(w/v) PEG 4000, 0.2 M ammonium acetate, 0.1 M citric acid buffer pH 5.1, with dimensions of ∼0.3 × 0.3 × 0.5 mm (Fig. 2 ▶).
Figure 2.
D. radiodurans TrxR crystal grown using 35% PEG 4000 and citric acid buffer pH 5.1.
3.2. Data collection and processing
The crystals were frozen in a nitrogen cold steam (100 K) without supplementing the crystallization solution with additional cryoprotectants. Diffraction data were collected using an ADSC Q315 detector on beamline 14-BM-C at BioCARS, Advanced Photon Source (APS), Chicago, IL, USA. The crystal-to-detector distance was maintained at 275 mm with an oscillation range per image of 0.5°, covering a total oscillation range of 90°. The resulting intensity data were indexed and integrated using MOSFLM (Leslie, 1992 ▶), scaled and merged using SCALA (Collaborative Computational Project, Number 4, 1994 ▶) and converted to structure factors using TRUNCATE (Collaborative Computational Project, Number 4, 1994 ▶). The space group was initially determined to be P3; however, based on systematic absences the space group was predicted to be one of P3x, P3x1x or P3x21. Using R merge values at 1.9 Å resolution and the self-rotation function from MOLREP (Vagin & Teplyakov, 1997 ▶), the space group was determined to be P3221. Matthews coefficient calculations (Matthews, 1968 ▶) suggested the presence of a dimer in the asymmetric unit of the crystal (V M = 2.3 Å3 Da−1, with 47.2% solvent content). A summary of crystallographic data statistics is shown in Table 1 ▶.
Table 1. Data-collection parameters and crystallographic data statistics.
Values in parentheses are for the highest resolution shell.
| Temperature (K) | 100 |
| Wavelength (Å) | 0.98 |
| Space group | P3221 |
| Unit-cell parameters (Å, °) | a = b = 84.33, c = 159.88, α = β = 90, γ = 120 |
| Resolution limits (Å) | 80.06–1.90 (2.00–1.90) |
| Total No. of reflections | 311938 (43600) |
| No. of unique reflections | 49077 (6865) |
| Completeness (%) | 93.6 (90.6) |
| Rmerge (%) | 6.9 (33.2) |
| Multiplicity | 6.4 (6.4) |
| I/σ(I) | 21.6 (4.5) |
| Solvent content (%) | 47.2 |
| No. of molecules in ASU | 2 |
| Matthews coefficient (Å3 Da−1) | 2.3 |
3.3. Structure solution
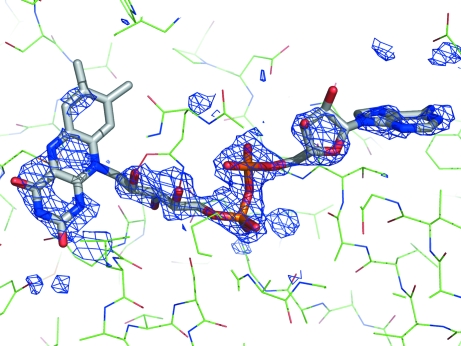
Structure solution was carried out using MrBUMP (Keegan & Winn, unpublished work), an automated scheme for molecular replacement which will be part of the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶). MrBUMP (using MOLREP) obtained a solution using the Mycobacterium tuberculosis TrxR crystal structure as a search model (PDB code 2a87; Akif et al., 2004 ▶). M. tuberculosis TrxR has 45% sequence identity to D. radiodurans TrxR (Fig. 1 ▶). Curiously, only one monomer gave a solution (2a87_B); using the other monomer as a search model gave no clear solution. The dimer was regenerated as part of MrBUMP. Initial restrained refinement carried out using REFMAC5 (Winn et al., 2001 ▶) as part of MrBUMP resulted in an R factor of 0.431 from an initial R factor of 0.547. The resulting model lacked the FAD cofactor, but the initial F o − F c map showed clear density for FAD (Fig. 3 ▶).
Figure 3.
Difference map (F o − F c) generated from the molecular-replacement solution from MrBUMP. The map (coloured blue) is contoured at 2.5σ. The actual model is shown as green lines, while the FAD (not included in initial model, but inserted to show location) is shown as a stick model. The figure was generated using PyMOL (DeLano, 2002 ▶).
Refinement of the structure and further structural and enzymatic studies involving TrxR and the other components of the D. radiodurans thioredoxin system are currently under way.
Acknowledgments
We would like to thank E. A. Sieminska and I. Kourinov for their assistance during data collection and M. Winn for his helpful discussions regarding MrBUMP. This work was funded in part by a Saskatchewan Health Research Grant (SHRF) grant to DARS and an NSERC USRA grant (SAB). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health.
References
- Akif, M., Chauhan, R. & Mande, S. C. (2004). Acta Cryst. D60, 777–779. [DOI] [PubMed] [Google Scholar]
- Arner, E. S. & Holmgren, A. (2000). Eur. J. Biochem.267, 6102–6109. [DOI] [PubMed] [Google Scholar]
- Atichartpongkul, S., Loprasert, S., Vattanaviboon, P., Whansuk, W., Helmann, J. D. & Mongkolsuk, S. (2001). Microbiology, 147, 1775–1782. [DOI] [PubMed] [Google Scholar]
- Battista, J. R. (2000). Curr. Biol.10, R204–R205. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Delano, W. L. (2002). The PyMOL Molecular Visualization System. http://www.pymol.org.
- Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Meunier-Jamin, C., Kapp, U., Leonard, G. & McSweeney, S. (2004). Acta Cryst. D60, 920–922. [DOI] [PubMed] [Google Scholar]
- Mustacich, D. & Powis, G. (2000). Biochem. J.346, 1–8. [PMC free article] [PubMed] [Google Scholar]
- Nordberg, J. & Arner, E. S. J. (2001). Free Radic. Biol. Med.31, 1287–1312. [DOI] [PubMed] [Google Scholar]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]
- White, O. et al. (1999). Science, 286, 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–123. [DOI] [PubMed] [Google Scholar]