The isolation, purification, crystallization and molecular-replacement solution of mitochondrial type II peroxiredoxin from P. sativum is reported.
Keywords: mitochondrial type II peroxiredoxin, Pisum sativum
Abstract
A cDNA encoding an open reading frame of 199 amino acids corresponding to a type II peroxiredoxin from Pisum sativum with its transit peptide was isolated by RT-PCR. The 171-amino-acid mature protein (estimated molecular weight 18.6 kDa) was cloned into the pET3d vector and overexpressed in Escherichia coli. The recombinant protein was purified and crystallized by the hanging-drop vapour-diffusion technique. A full data set (98.2% completeness) was collected using a rotating-anode generator to a resolution of 2.8 Å from a single crystal flash-cooled at 100 K. X-ray data revealed that the protein crystallizes in space group P1, with unit-cell parameters a = 61.88, b = 66.40, c = 77.23 Å, α = 102.90, β = 104.40, γ = 99.07°, and molecular replacement using a theoretical model predicted from the primary structure as a search model confirmed the presence of six molecules in the unit cell as expected from the Matthews coefficient. Refinement of the structure is in progress.
1. Introduction
Plants perform photosynthesis and assimilatory processes in a continuously changing environment that needs precise regulation. The simultaneous presence of strong oxidants and strong reductants is the basis for such regulation and reactive oxygen species (ROS) play a central role. At low concentrations, ROS behave as secondary messengers in signal transduction, while at high concentration they can trigger apoptosis and damage macromolecules and membranes. Consequently, plants have evolved efficient and specific defence systems that rapidly detoxify dangerous oxidants such as peroxide and superoxide.
Mitochondria are a significant site of ROS production in plant cells. About 2% of the total consumed oxygen is converted into ROS and the main sites of ROS production are complex I and complex II of the mitochondrial electron-transport chain (Moller, 2001 ▶). Therefore, mitochondria have an ensemble of antioxidant enzymes such as superoxide dismutase, ascorbate peroxidase, glutathione peroxidase, glutathione reductase and peroxiredoxins (Prxs) that neutralize these ROS.
Prxs are ubiquitous enzymes involved in peroxide detoxification and signal transduction pathways and may function as redox sensors (Wood, Poole et al., 2003 ▶). They are found in all organisms and exist in multiple isoforms (Dietz, 2003 ▶; Rhee et al., 2005 ▶) that share a conserved cysteine at the N-terminal end which is oxidized by peroxide. In animal cells, Prxs are involved in complex cellular processes including cell proliferation (Prosperi et al., 1993 ▶), differentiation (Rabilloud et al., 1995 ▶), apoptosis (Zhou et al., 2000 ▶) and cancer (Nonn et al., 2003 ▶). In plants, Prxs are grouped into four families on the basis of the number and position of the conserved cysteine residues: 2-Cys Prxs, type II Prxs, Prx Qs and 1-Cys Prxs (Horling et al., 2003 ▶). The type II Prxs are enzymes with varying molecular weights, isoelectric points and subcellular localization and have been proposed as primary sensors for H2O2 (Wood, Poole et al., 2003 ▶; Wood, Schröder et al., 2003 ▶). Plant mitochondrial Prxs (PrxII F) belong to the type II Prxs and have been reported as being essential for the maintainance of redox homeostasis (Finkemeier et al., 2005 ▶).
Typically, Prxs are dimers that through further interactions may yield higher oligomerization states which are redox-sensitive and relevant to regulation of the activity (Wood, Schröder et al., 2003 ▶). A toroid decamer (a pentamer of dimers) built up from five dimeric units has been reported for the reduced form of the 2-Cys Prxs from organisms as phylogenetically distant as the bacteria Salmonella typhimurium (Wood et al., 2002 ▶), the trypanosome Crithidia fasciculata (Alphey et al., 2000 ▶), the mammal Bos taurus (Gourlay et al., 2003 ▶) and the plant Hordeum vulgare (König et al., 2003 ▶). However, a recent crystallographic study on the AhpC peroxiredoxin from Mycobacterium tuberculosis revealed the existence of a toroid oligomer consisting of six dimers (Guimaraes et al., 2005 ▶), although its physiological relevance remains unclear. In plants, oligomerized 2-Cys Prxs have been found at tylakoid membranes and a link between oligomerization and tylakoid attachment has been proposed, leading to a hypothesis that 2-Cys Prx may act as a structural redox sensor (König et al., 2003 ▶). The in vitro analysis of conditions of oligomerization of the 2-Cys Prx from Pisum sativum found that the dimer–decamer transition takes place between pH 7.5 and 8.5 and is influenced by dithiothreitol, suggesting that changes in the quaternary structure of the enzyme may be expected during the dark–light transition (Bernier-Villamor et al., 2004 ▶).
Despite its importance, only one structure of plant peroxiredoxin is available in the PDB and it corresponds to a cytosolic form (Echalier et al., 2005 ▶). In this communication, we report the isolation, overexpression, crystallization and molecular replacement of a mature mitochondrial type II peroxiredoxin from P. sativum. The unit cell consists of six molecules and the interactions between them may provide some insight into the oligomerization of Prxs.
2. Materials and methods
2.1. Cloning and expression
The primary structures of PrxII F from Arabidopsis thaliana (accession No. At3g06050) and Oryza sativa (accession No. AU070430) were used to elucidate the sequence of pea PrxII F. A fragment of cDNA enconding the pea mature type II Prx was isolated by RT-PCR with sense 5′-CAC/CATGGCAAAGGTTGCAACTGGAACTG-3′ and antisense 5′-CCG/GATCCTCAAATTTGTCCCAAAATGGT-3′ primers. The PCR-amplified fragment was digested with NcoI and BamHI, cloned into the pET3d vector (Novagen) and overexpressed in Escherichia coli BL21(DE3). Transformed bacteria were incubated at 310 K for 6 h and expression was induced by 0.4 mM IPTG. Cells were harvested by centrifugation at 3000g, resuspended in 20 mM Tris–HCl pH 8.0 and disrupted with a French press.
2.2. Purification
Recombinant PrxII F was purified by ammonium sulfate fractionation (40–95% saturation) in 25 mM Tris–HCl pH 8.0, 150 mM NaCl and centrifugation at 27 000g for 15 min followed by two sequential chromatographic steps on FPLC (Amersham Biosciences, Uppsala, Sweden): (i) Sephacryl S-200 gel filtration in 25 mM Tris–HCl pH 8.0, 150 mM NaCl and (ii) Mono Q HR 5/5 ion exchange in 25 mM Tris–HCl pH 8.0 and an NaCl gradient from 0 to 1000 mM, the protein being eluted at about 200 mM NaCl. The success of each step was monitored by DTT-dependent peroxidase assay (Thurman et al., 1972 ▶) and SDS–PAGE/silver stain. The protein was concentrated to 10 mg ml−1 in 20 mM Tris–HCl pH 7.5, 150 mM NaCl by ultrafiltration using Centricon 10 concentrators (Millipore, Watford, England).
2.3. Crystallization
Crystals were grown by the hanging-drop vapour-diffusion method at 293 K in 24-well VDX plates (Hampton Research). Crystallization conditions were screened using sparse-matrix screens (Jancarik & Kim, 1991 ▶) based on Crystal Screen I (Hampton Research). After optimization, the best crystals were obtained from droplets containing 3 µl of 10 mg ml−1 PrxII F in 20 mM Tris–HCl pH 7.5, 150 mM NaCl and 3 µl reservoir solution [5 mM DTT, 20%(w/v) PEG 2000 and 1%(v/v) 2-propanol in 100 mM citrate buffer pH 5.6] equilibrated against 500 µl reservoir solution.
2.4. Data collection and molecular replacement
Data collection was carried out at 100 K from a single crystal cryoprotected by soaking for a couple of minutes in a solution containing 20%(v/v) glycerol, 100 mM citrate pH 5.6, 5 mM DTT, 20%(w/v) PEG 2000, 1%(v/v) 2-propanol. X-ray diffraction data were recorded on a Platinum 200 CCD detector mounted on a four-circle Kappa goniometer with Cu Kα radiation from a Microstar micro-focus rotating-anode generator (Bruker AXS) located at Laboratorio de Estudios Cristalográficos, CSIC–Universidad de Granada. Data were processed with SAINT and scaled with XPREP (Bruker, Madison, WI, USA). Molecular replacement was carried out with AMoRe (Navaza, 1994 ▶) using the coordinates generated by EasyPred3D Web Server 1.0 (Lambert et al., 2002 ▶) as a search model.
3. Results and discussion
A variable sequence homology has been found between the six known isoforms of peroxiredoxins. Each one is unique in terms of subcellular localization, expression pattern, catalytic mechanism (dependence on thioredoxin and/or glutathione) and oligomerization state. However, despite their physiological significance, only one structure from a plant is available (PDB code 1tp9). The facts that it corresponds to the cytosolic peroxiredoxin from poplar (Populus trichocarpa; Echalier et al., 2005 ▶) and that mitochondrial type II peroxiredoxin F is essential for redox homeostasis (Finkemeier et al., 2005 ▶) have stimulated us to address the isolation and crystallization of the mitochondrial type II peroxiredoxin from pea.
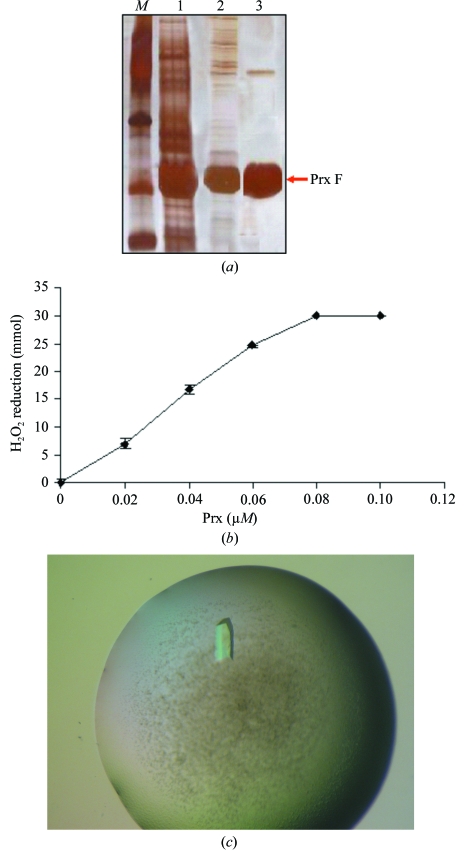
The isolation of this enzyme was approached by RT-PCR and heterologous primers based on the sequences from Arabidopsis and rice. It yielded a 600 bp cDNA fragment encoding an open reading frame (ORF) of 199 amino acids that was deposited in the EMBL/GeneBank/DDBJ databases with accession No. AJ717306. The analysis of the ORF reveals that it comprises a 28-amino-acid N-terminal signal peptide that targets the 171-amino-acid mature protein to the mitochondria (Fig. 1 ▶). Since the structure is the purpose of this work and the unique oligomerization state of the enzyme has a physiological relevance, the mature protein was cloned into a conventional pET3d vector and purified by standard chromatographic techniques instead of using a His-tag approach. The recombinant protein showed peroxidase activity and its analysis by SDS–PAGE and silver staining confirmed the purity of the sample despite the presence of a faint band corresponding to a dimer of peroxiredoxin (Fig. 2 ▶). The formation of dimers in SDS–PAGE has been reported for several hydrophobic domains (Lemmon et al., 1992 ▶) and this is likely to be the case in our protein as indicated by the requirement for 6 M urea to elute it from a phenyl Superose column.
Figure 1.
Primary structure of the pea mitochondrial type II peroxiredoxin and alignment with its cytosolic counterpart from poplar. The signal peptide is coloured violet and the common residues are in yellow.
Figure 2.
(a) Purification of mitochondrial type II Prx (lane 1, crude extract; lane 2, after Sephacryl S-200; lane 3, pure protein after MonoQ chromatography). (b) Peroxidase activity of the enzyme. (c) Crystal of recombinant mature pea mitochondrial type II peroxiredoxin.
Screening of crystallization conditions revealed that the protein is prone to precipitate in many combinations of precipitants and buffers. Several rounds of experiments led to conditions that despite promoting some precipitation yield crystals that are suitable for X-ray diffraction (Fig. 2 ▶ c). Typically, crystals grow from a precipitate within 5 d and diffract to beyond 2.4 Å on a rotating-anode generator, although suitable completeness for structural purposes was only achieved at 2.8 Å (Table 1 ▶). The space group was determined as P1, with unit-cell parameters a = 61.88, b = 66.40, c = 77.23 Å, α = 102.90, β = 104.40, γ = 99.07°. The Matthews probability calculator (Kantardjieff & Rupp, 2003 ▶) estimates V M as 2.43 Å3 Da−1 with a probability of 0.87, which corresponds to six monomers per unit cell and 49.44% solvent content. The symmetry and unit-cell content was confirmed by molecular replacement using a model generated by EsyPre3D (Lambert et al., 2002 ▶) as a search model since peroxiredoxin from poplar (PDB code 1tp9) shares 36% identity with pea mitochondrial type II peroxiredoxin.
Table 1. Summary of the main features of the data set.
Values in parentheses correspond to the highest resolution shell.
| Data collection | |
| X-ray source | Microstar micro-focus rotating-anode generator |
| Wavelength | 1.5418 (Cu Kα) |
| No. of crystals | 1 |
| Temperature of data collection (K) | 100 |
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 61.88, b = 66.40, c = 77.23, α = 102.90, β = 104.40, γ = 99.07 |
| Data set | |
| Resolution range (Å) | 15–2.8 (2.9–2.8) |
| Measured reflections | 92649 |
| Unique reflections | 25983 |
| Rsym† (%) | 6.6 (9.92) |
| Completeness (%) | 92.8 (66) |
| Completeness [I/σ(I) > 2] (%) | 79.0 (60) |
| Mean I/σ(I) | 15.71 (19.16) |
R
sym = 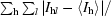
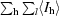 , where I
hl is the lth observation of reflection h and 〈I
h〉 is the weighted average intensity for all observations l of reflection h.
, where I
hl is the lth observation of reflection h and 〈I
h〉 is the weighted average intensity for all observations l of reflection h.
Refinement is currently in progress and the structure and interactions between the molecules present in the unit cell will provide an insight into the nature of oligomerization of mitochondrial Prxs. The biochemical characterization results show that unlike 2-Cys Prxs, pea mitochondrial type II peroxiredoxin is prone to oligomerization in the absence of reductants (manuscript in preparation). The presence of three dimers in the the P1 unit cell suggests an arrangement that might imply a new quaternary structure or an intermediate state such as those observed for AhpC Prxs from M. tuberculosis (Guimaraes et al., 2005 ▶ and S. typhimurium (Wood et al., 2002 ▶). The oligomerization plays an important role in the attachment of plant 2-Cys Prxs to the membrane (König et al., 2003 ▶) that may be shared by mitochondrial Prxs although under different redox conditions and probably with different quaternary structures.
Acknowledgments
We thank Laboratorio de Estudios Cristalográficos, CSIC–Universidad de Granada for access to the X-ray facility and Francisca Castro for her excellent technical assistance. FJL-J is indebted to Professor P. Aranda, A. Osuna and F. Santoyo for their support. This work was supported by Dirección General de Enseñanza Superior e Investigación Científica (Ministerio de Educación y Cultura; Project BFI 2002-03207).
References
- Alphey, M. S., Bond, C. S., Tetaud, E., Fairlamb, A. H. & Hunter, W. N. (2000). J. Mol. Biol.300, 903–916. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor, L., Navarro, E., Sevilla, F. & Lázaro, J. J. (2004). J. Exp. Bot.55, 2191–2199. [DOI] [PubMed] [Google Scholar]
- Dietz, K. J. (2003). Annu. Rev. Plant Physiol. Plant Mol. Biol.54, 93–107. [DOI] [PubMed]
- Echalier, A., Trivelli, X., Corbier, C., Rouhier, N., Walker, O., Tsan, P., Jacquot, J.-P., Aubry, A., Krimm, I. & Lancelin, J.-M. (2005). Biochemistry, 44, 1755–1767. [DOI] [PubMed] [Google Scholar]
- Finkemeier, I., Goodman, M., Lankemeyer, P., Kandlbinder, A., Sweetlove, L. J. & Dietz, K. J. (2005). J. Biol. Chem.280, 12168–12180. [DOI] [PubMed] [Google Scholar]
- Gourlay, L. J., Bhella, D., Kelly, S. M., Price, N. C. & Lindsay, J. G. (2003). J. Biol. Chem.278, 32631–32637. [DOI] [PubMed] [Google Scholar]
- Guimaraes, B. G., Souchon, H., Honore, N., Saint-Joanis, B., Brosch, R., Shepard, W., Cole, S. T. & Alzari, P. M. (2005). J. Biol. Chem.280, 25735–25742. [DOI] [PubMed] [Google Scholar]
- Horling, F., Lamkemeyer, P., König, J., Finkemeier, I., Kandlbinder, A., Baier, M. & Dietz, K.-J. (2003). Plant Physiol.131, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Kantardjieff, K. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König, J., Lotte, K., Plessow, R., Brockninke, A., Baier, M. & Dietz, K.-J. (2003). J. Biol. Chem.278, 24409–24420. [DOI] [PubMed] [Google Scholar]
- Lambert, C., Leonard, N., De Bolle, X. & Depiereux, E. (2002). Bioinformatics, 18, 1250–1256. [DOI] [PubMed] [Google Scholar]
- Lemmon, M. A., Flanagan, J. M., Treutlein, H. R., Zhang, J. & Engelman, D. M. (1992). Biochemistry, 31, 12719–12725. [DOI] [PubMed] [Google Scholar]
- Moller, I. M. (2001). Annu. Rev. Plant Physiol. Plant Mol. Biol.52, 561–591. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994) Acta Cryst. A50, 157–163.
- Nonn, L., Berggren, M. & Powis, G. (2003). Mol. Cancer Res.1, 682–689. [PubMed] [Google Scholar]
- Prosperi, M. T., Ferbus, D., Karczinski, I. & Goubin, G. (1993). J. Biol. Chem.268, 11050–11056. [PubMed] [Google Scholar]
- Rabilloud, T., Berthier, R., Vinçon, M., Ferbus, D., Goubin, G. & Lawrence, J. J. (1995). Biochem. J.312, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S. G., Chae, H. Z. & Kim, K. (2005). Free Radic. Biol. Med.31, 292–303. [DOI] [PubMed]
- Thurman, R. G., Ley, H. G. & Scholz, R. (1972). Eur. J. Biochem.25, 420–430. [DOI] [PubMed] [Google Scholar]
- Wood, Z. A., Poole, L. B., Hantgan, R. R. & Karplus, P. A. (2002). Biochemistry, 41, 5493–5504. [DOI] [PubMed] [Google Scholar]
- Wood, Z. A., Poole, L. B. & Karplus, P. A. (2003). Science, 300, 650–653. [DOI] [PubMed] [Google Scholar]
- Wood, Z. A., Schröder, E., Harris, J. R. & Poole, L. B. (2003). Trends Biochem. Sci.28, 32–39. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Kok, K. H., Chun, A. C., Wong, C. M., Wu, H. W., Lin, M. C., Fung, P. C., Kung, H. & Jin, D. Y. (2000). Biochem. Biophys. Res. Commun.268, 921–927. [DOI] [PubMed] [Google Scholar]