The structure of porcine pancreatic elastase at 1.5 Å resolution has a unique conformation induced by tris(hydroxymethyl)aminomethane as a buffer component.
Keywords: porcine pancreatic elastase, unit-cell contraction, crystal packing, native structure
Abstract
Porcine pancreatic elastase (PPE) was crystallized under new sulfate-free conditions containing 0.3 M NaCl and 50 mM tris(hydroxymethyl)aminomethane–HCl at pH 7.0. The crystal structure determined at 1.5 Å resolution had a unique conformation in four regions which contained loop portions. A chloride ion was bound near the catalytic triad instead of the sulfate ion in PDB entry 1qnj, a typical PPE crystal structure. However, the chloride ion did not affect the configuration of the catalytic triad. A tris(hydroxymethyl)aminomethane (Tris) molecule was bound to the S4 and S5 subsites in place of the adjacent molecule in the 1qnj crystal and played a significant role in the structural change of the region. The distortion in this region may subsequently have induced conformational changes in the other three regions. The fact that Tris and these four regions make a diagonal line in the ac plane may have affected the crystal-packing contraction along the a and c axes in the crystal compared with the typical crystal.
1. Introduction
Porcine pancreatic elastase (EC 3.4.21.36; PPE) is a serine protease classified into the chymotrypsin family and consists of 240 amino acids. Because of the significance of the serine proteases in studies on enzyme catalysis and the medicinal importance of their inhibition, the details of their catalytic mechanisms remain of interest.
PPE can be crystallized easily and is therefore used for general structural studies. There were 62 entries and 22 entries waiting for release for PPE in its canonical space group P212121 in the PDB on 6 February 2006. The backbones of these PPE structures are rigid, although many of the inhibitors in the respective complexes induce movements in the side chains (Kinoshita et al., 2005 ▶). In addition to inhibitor-binding complexes, the native state is important for structural studies. The atomic structure of native-state PPE at 1.1 Å resolution has been reported (Würtele et al., 2000 ▶). However, the structure may not reflect the inherent configuration of the catalytic triad consisting of Ser195, His57 and Asp102, since a large ion such as sulfate binds in the vicinity. Recently, the sulfate-free native form has been crystallized using sodium acetate buffer, sodium citrate and calcium chloride, but acetate occupies the sulfate-binding region in the crystals (Weiss et al., 2002 ▶).
As a result of attempts to obtain PPE crystals from sulfate- and acetate-free solutions, we found new crystallization conditions containing sodium chloride and tris(hydroxymethyl)aminomethane (Tris). The resulting PPE structure reveals some insights into the catalytic triad in the native state and the structural flexibility in its backbone. Furthermore, we found evidence of contraction of the crystal lattice.
2. Materials and methods
PPE was purchased from Elastin Product Co. Inc. and concentrated to 30 mg ml−1 in 50 mM Tris–HCl buffer pH 7.0. Initial crystals were obtained by the sitting-drop vapour-diffusion method from condition No. E2 in a Crystal Screen HT kit (Hampton Research) consisting of 0.5 M sodium chloride, 10 mM magnesium chloride. The crystallization conditions were refined by the addition of Tris–HCl buffer. The best crystals were obtained using 4 µl protein solution and 4 µl reservoir containing 0.3 M sodium chloride and 50 mM Tris–HCl pH 7.0.
Crystals for PPE were dipped into cryosolutions including 20% glycerol, 0.5 M sodium chloride and 50 mM Tris–HCl pH 7.0 and mounted on a nylon loop (Hampton Research). The crystals were then flash-cooled to 100 K in a nitrogen-gas stream. Diffraction data sets were collected at beamline BL41XU at the SPring-8 synchrotron-radiation facility using a Quantum 315 CCD detector (ADSC). Data integration, scaling and merging were performed using the program HKL-2000 (Otwinowski & Minor, 1997 ▶).
The initial structures were resolved using AMoRe (Navaza, 1994 ▶). As a starting model for both analyses, PDB entry 1qnj (Würtele et al., 2000 ▶) was used. All ions and water molecules were deleted from the model. After rigid-body refinement and restrained refinements of the protein atoms, difference electron-density maps (2F obs − F calc) were calculated. Model modification and the addition of ions and water molecules were performed by manual inspection of the model using computer graphics. All refinements and electron-density map calculations were performed using the programs DS Modelling and CNX (Accelrys). The coordinates have been deposited with the PDB. Details of the data collections and refinements are summarized in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Na2SO4-soaking form | NaCl/Tris form | |
|---|---|---|
| Data collection | ||
| Crystal-to-detector distance (mm) | 155 | 155 |
| Wavelength (Å) | 0.8 | 1.0 |
| Exposure time (s) | 1 | 3 |
| Total rotation range (°) | 180 | 180 |
| Space group | P212121 | P212121 |
| Unit-cell parameters | ||
| a (Å) | 49.88 | 46.13 |
| b (Å) | 57.85 | 57.29 |
| c (Å) | 74.33 | 72.89 |
| Resolution limits (Å) | 50–1.3 (1.40–1.30) | 30–1.5 (1.55–1.50) |
| Mosaicity | 0.45 | 1.13 |
| Total No. of reflections | 331403 | 195477 |
| Unique reflections | 52552 | 31471 |
| Redundancy | 6.3 (3.4) | 6.2 (2.6) |
| Completeness (%) | 96.1 (87.5) | 98.8 (89.8) |
| I/σ(I) | 14.1 (1.6) | 23.6 (1.5) |
| Rmerge† (%) | 9.0 (33.4) | 5.4 (40.4) |
| Refinement | ||
| Resolution limits (Å) | 50–1.3 (1.31–1.30) | 30–1.5 (1.52–1.50) |
| Reflections in test set (%) | 5 | 5 |
| R‡ (%) | 21.1 (27.4) | 22.3 (30.0) |
| Rfree§ (%) | 22.6 (29.4) | 24.6 (30.8) |
| No. of protein atoms | 1845 | 1845 |
| No. of ion atoms | 10¶ | 9†† |
| No. of water molecules | 292 | 236 |
| R.m.s.d. bond lengths (Å) | 0.013 | 0.007 |
| R.m.s.d. bond angles (°) | 1.6 | 1.8 |
| PDB code | 2de9 | 2de8 |
R
merge = 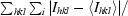
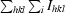 .
.
R = 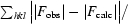
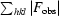 .
.
R free is equivalent to R, but is calculated using a test set of reflections excluded from the final refinement stages.
Two sulfates.
One Cl− and one tris(hydroxymethyl)aminomethane molecule.
3. Results and discussion
3.1. Crystal structure of the NaCl/Tris form
Crystals obtained from the new crystallization conditions (NaCl/Tris form) showed a similar prismatic shape to those from solutions containing sulfate (the typical form). The space group was also conserved between the NaCl/Tris and typical forms. However, the crystal lattice of the NaCl/Tris form is contracted along the a axis and slightly contracted along the c axis compared with that of the starting model 1qnj (Würtele et al., 2000 ▶), which is representative of the typical form and has unit-cell parameters a = 49.91, b = 57.82, c = 74.27 Å.
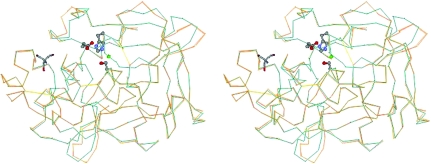
The backbone structure arising from the NaCl/Tris form is conserved compare with that of 1qnj, with an r.m.s. deviation of 0.66 Å for Cα atoms, except for the regions A, B, C and D which include loop portions (Fig. 1 ▶). This conformational change of the backbone is rare for PPE structures. Even large peptide inhibitors do not induce backbone-level movements of PPE (Nakanishi et al., 2000 ▶; Aÿ et al., 2003 ▶; Matern et al., 2003 ▶; Kinoshita et al., 2005 ▶). Regions A (Arg36–Trp38) and D (Leu202–Ala208) protrude into the solvent region and therefore may be flexible. These regions were far from the active site and the recognition site to which substrates and inhibitors bind. On the other hand, regions B (Asp60–Thr64) and C (Asn95–Tyr101) consisted of parts of the recognition site.
Figure 1.
Cα traces of the NaCl/Tris form (blue) and the 1qnj structure (orange). The catalytic triad, Ser195, His57 and Asp102, is shown in a ball-and-stick representation. A chloride ion (green ball) binds near the catalytic triad and Tris (displayed in stick representation) binds to the S4 and S5 subsites.
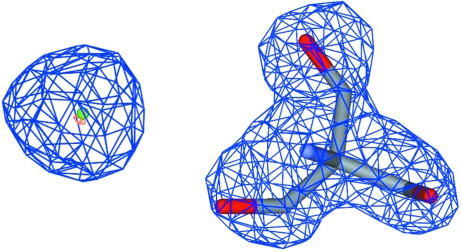
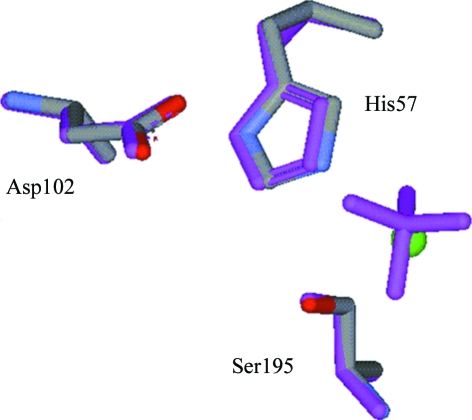
A chloride ion was assigned at high electron density with a well balanced spherical shape (Fig. 2 ▶). The chloride ion occupied the active site corresponding to the sulfate-binding site in the 1qnj structure and made hydrogen bonds with residues responsible for enzyme reaction (Table 2 ▶). Although the chloride ion, which is small in size compared with the sulfate or acetate in the typical form, bound to the active site in the NaCl/Tris form, the relative configuration of the catalytic triad and the individual conformations of the three amino acids were conserved between the two forms (Fig. 3 ▶).
Figure 2.
Chloride (left) and Tris (right) molecules with corresponding sites in the 2F obs − F calc OMIT map. The blue mesh indicates a 1σ height cutoff and the orange mesh a 4.8σ height cutoff.
Table 2. Distances between atoms of the catalytic region of PPE.
| Distance (Å) | |
|---|---|
| Ser195 Oγ⋯His57 N∊2 | 3.3 |
| Ser195 Oγ⋯Cl− | 3.1 |
| His57 Nδ1⋯Asp102 Oδ2 | 2.7 |
| His57 N∊2⋯Cl− | 3.6 |
| Gly193 NH⋯Cl− | 3.4 |
Figure 3.
Configurations of the catalytic triad in the NaCl/Tris form and the 1qnj form (magenta).
Tris could be fitted into a large electron-density envelope near the S4 and S5 subsites (Fig. 2 ▶), the nomenclature of which was defined in detail by Bode et al. (1989 ▶). A Tris molecule tightly fused two PPE molecules via three hydroxyl groups in the NaCl/Tris crystals. Two of the hydroxyl groups formed hydrogen bonds with Ser217 Oγ and Arg217 NH of the first molecule. The other hydroxyl group made a hydrogen bond with Arg61 C=O of the second molecule. In the case of the 1qnj crystal structure, Glu62 O∊1 of the second molecule made a direct hydrogen bond with Arg217 NH of the first molecule. Therefore, Tris plays an important role in inducing conformational change in region B of the NaCl/Tris form.
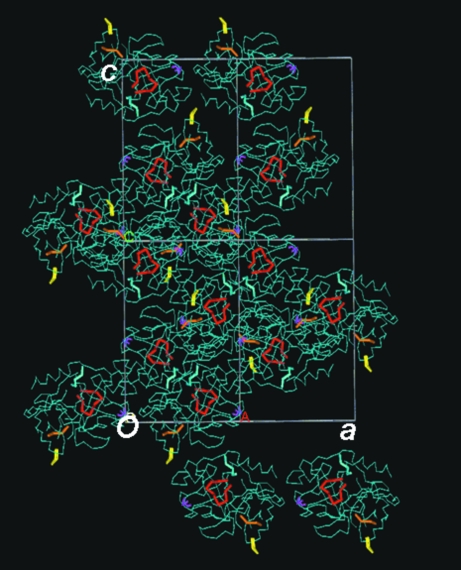
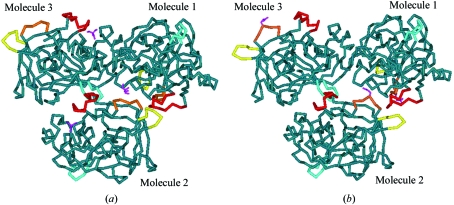
Crystal-packing diagrams of the NaCl/Tris form revealed that the conformational change of region B may have induced that of the other regions A, C and D. Tris and regions A, B, C and D form a diagonal line in the ac plane (Fig. 4 ▶). Region B of the second molecule is in the vicinity of the region A of the second molecule and region C of the first molecule, while region C of the second molecule is close to region D of the third molecule (Fig. 5 ▶). This structural repetition in the NaCl/Tris crystal may have affected the crystal-lattice contraction along the two axes.
Figure 4.
Crystal-packing diagram of the ac plane in the NaCl/Tris crystal. Tris (magenta) and the structurally unique regions A (yellow), B (orange), C (red) and D (blue) form a diagonal line in the ac plane.
Figure 5.
Molecular–molecular interaction diagrams in the NaCl/Tris form (a) and in the typical form (b). Tris (magenta) or Glu50 (magenta) and the structurally unique regions A (yellow), B (orange), C (red) and D (blue) assemble in the centre of the three PPE molecules.
In order to clarify whether Tris affects the contraction in the unit-cell parameters, Tris was removed by a soaking procedure. An NaCl/Tris crystal was dipped into a Tris-free solution containing 0.5 M Na2SO4, 50 mM sodium acetate buffer pH 5.0 and 20% glycerol prior to data collection. The unit-cell parameters of the soaked crystal (the Na2SO4-soaking form) returned to those of 1qnj. The Na2SO4-soaking form did not contain Tris in the S4 and S5 subsites and Glu62 O∊1 of the second molecule made a hydrogen bond directly to Arg217 NH of the first molecule. Additionally, two sulfates occupied the sulfate-binding sites in the 1qnj structure. The backbone structure from the Na2SO4-soaking form had high similarity to that of the 1qnj structure, with an r.m.s. deviation of 0.13 Å for Cα atoms. This observation shows that Tris is essential for structural change in the NaCl/Tris form.
Here, we report the crystal structure of the native PPE obtained under the new crystallization conditions containing NaCl/Tris. The structure reveals novel insights involving the structural rigidity of the catalytic triad, the flexibility of four regions containing loop portions and the binding mode of Tris in the S4 and S5 subsites.
Supplementary Material
PDB reference: porcine pancreatic elastase, 2de9, r2de9sf
PDB reference: 2de8, r2de8sf
References
- Aÿ, J., Hilpert, K., Krauss, N., Schnerider-Mergener, J. & Hohme, W. (2003). Acta Cryst. D59, 247–254. [DOI] [PubMed] [Google Scholar]
- Bode, W., Meyer, E. Jr & Powers, J. C. (1989). Biochemistry, 28, 1951–1963. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Kitatani, T., Warizaya, M. & Tada, T. (2005). Acta Cryst. F61, 808–811. [DOI] [PMC free article] [PubMed]
- Matern, U., Schleberger, C., Jelakovic, S., Weckesser, J. & Schulz, G. E. (2003). Chem. Biol.10, 997–1001. [DOI] [PubMed] [Google Scholar]
- Nakanishi, I., Kinoshita, T., Sato, A. & Tada, T. (2000). Biopolymers, 53, 434–445. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Weiss, M. S., Panjikar, S., Nowak, E. & Tucker, P. A. (2002). Acta Cryst. D58, 1407–1412. [DOI] [PubMed] [Google Scholar]
- Würtele, M., Hahn, M., Hilpert, K. & Hohne, W. (2000). Acta Cryst. D56, 520–523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: porcine pancreatic elastase, 2de9, r2de9sf
PDB reference: 2de8, r2de8sf