Crystals adequate for X-ray diffraction analysis have been prepared from L. interrogans ferredoxin-NADP+ reductase.
Keywords: ferredoxin-NADP+ reductase, Leptospira interrogans
Abstract
Ferredoxin-NADP+ reductase (FNR) is an FAD-containing enzyme that catalyzes electron transfer between NADP(H) and ferredoxin. Here, results are reported of the recombinant expression, purification and crystallization of FNR from Leptospira interrogans, a parasitic bacterium of animals and humans. The L. interrogans FNR crystals belong to a primitive monoclinic space group and diffract to 2.4 Å resolution at a synchrotron source.
1. Introduction
Ferredoxin-NADP+ reductase (FNR) is an enzyme present in a wide variety of organisms and tissues that plays a distinct role in maintenance of metabolism (Carrillo & Ceccarelli, 2003 ▶). In chloroplasts, the enzyme is responsible for the NADP+ reduction involved in photosynthetic electron transport. In mitochondria, FNR participates in flavodoxin reduction and is also important in steroid hydroxylation and fatty-acid desaturation (Shin & Arnon, 1965 ▶; Ziegler & Schulz, 2000 ▶).
The first crystal structure obtained of an FNR-family member was that of spinach FNR (Karplus et al., 1991 ▶). It revealed a structural architecture composed of two domains of equal size, each containing approximately 150 residues. The C-terminal domain accommodates NADP(H) and the cleft between the domains, surrounded by two tyrosine residues, serves as the FAD ligand-binding pocket. One of the tyrosines and the isoalloxazine portion of the FAD are coplanar and maximize π-orbital overlap. Crystal structures of FNRs, including those from chloroplasts (Deng et al., 1999 ▶), cyanobacteria (Serre et al., 1996 ▶), Escherichia coli (Ingelman et al., 1997 ▶), Azobacter vinelandii (Prasad et al., 1998 ▶) and most recently Rhodobacter capsulatus (Nogués et al., 2005 ▶), have revealed a conserved tertiary structure, despite the low level of sequence homology between these different forms of the enzyme.
The biological catalytic activity of FNR includes reversible electron transfer between NADP(H) and the iron-containing protein ferredoxin as described in the pathway (Ceccarelli et al., 2004 ▶)
In this pathway, the electron transfer from reduced Fd to NADP+ follows an ordered reaction involving the formation of a ternary complex with the pyridine nucleotide acting as the leading substrate. On a molecular level, it is known that NADP+ reduction proceeds through hydride transfer from the N5 atom of the flavin prosthetic group (Deng et al., 1999 ▶).
The crystal structure of maize Fd isoform I (FdI) in complex with FNR revealed for the first time that binding of Fd induces conformational changes in the active site of FNR which may be involved in modulation of its enzymatic activity (Kurisu et al., 2001 ▶). Furthermore, the X-ray structure of Equisetum arvense ferredoxin isoform II (FdII) provided evidence for putative differences in the binding modes of FdI and FdII to FNR (Kurisu et al., 2005 ▶).
Recently, phylogenetic analysis has assigned the FNR from Leptospira interrogans (LepFNR) to the plastidic FNR class (Ceccarelli et al., 2004 ▶). L. interrogans is a parasitic bacterium that infects humans and mammals, including cattle, dogs, pigs, horses and some wild animals such as rodents, which are the normal carrier hosts. LepFNR belongs to a monophyletic group composed entirely of obligate parasites. Hence, the inclusion of LepFNR among the plastidic forms of the enzyme is surprising and suggests that it may have arisen as the result of lateral gene transfer. Incorporation of a high-efficiency FNR could have provided adaptive advantages to the bacterium, becoming an important element of its life and thus representing a potential pharmaceutical target to interfere with the infection of animals and humans. LepFNR has neither been cloned nor purified to date and its functional and structural properties are still waiting to be uncovered. Although LepFNR belongs to the plastidic FNR class, it shares low primary structure homology with the members of this family, therefore turning this protein into a particularly interesting structure to analyze. Moreover, structural analysis of a greater number of members of the ferredoxin-NADP+ reductase family could shed more light on the precise molecular mechanism of their action and on the structural implications of the evolutionary changes in this enzyme.
2. Materials and methods
2.1. Protein expression and purification
The FNR from L. interrogans was overexpressed in E. coli BL21(DE3)pLys cells transformed with the pET32JO-LepFNR vector as a fusion with Trx-His6 protein containing a thrombin-recognition site between the Trx-His6 and the LepFNR. The pET32JO-LepFNR expression vector was constructed by inserting the coding sequence for L. interrogans FNR into pET32JO, a modified pET32b vector (Novagen; Rial et al., 2000 ▶). The coding sequence for LepFNR was amplified by PCR using as primers the oligonucleotides LepFNRup, 5′-AACTGCAGGTATGCATTCGCTCATGAAACCGACTAGA-3′, and LepFNRlw, 5′-CGGAATTCTCAATATGTTTCCACAAATAATTGATGGGCTC-3′, and the genomic DNA from L. interrogans serovar Lai 56601, kindly provided by Dr Xiao-Kui Guo of the Department of Microbiology, Shanghai Second Medical University, Shanghai, People’s Republic of China, as a template. The amplification product was introduced into plasmid pTOPO (Invitrogen) and was then incorporated into pET32JO as a PstI/SacI fragment (1010 bp), finally leading to pET32JO-LepFNR. For functional expression, bacteria were grown at 310 K in 1 l LB medium supplemented with ampicillin (100 µg ml−1) and chloramphenicol (40 µg ml−1). Expression was induced at an OD600 of 0.8 by the addition of IPTG to a final concentration of 1 mM for 3 h at 303 K. Subsequent Ni–NTA chromatography of the His-tagged protein was performed according to the instructions of the manufacturer (Qiagen). After dialysis against a solution containing 50 mM Tris–HCl pH 8, 150 mM NaCl, the fusion protein was digested with thrombin using a 1:80 protease:protein ratio for 4 h at 297 K. The Trx-His6 was separated from LepFNR with Ni–NTA resin. Finally, the reductase was concentrated using Centriprep-10 (Amicon) to a final concentration of 29 mg ml−1.
2.2. Crystallization
Initial attempts to crystallize LepFNR were performed using the sparse-matrix screening method by hanging-drop vapour diffusion using Crystal Screens 1 and 2 (Hampton Research) and PEG 3350 over a wide pH range. Hanging drops containing 1 µl protein at 10 mg ml−1 in 50 mM Tris buffer pH 8.0 were mixed with equal amounts of reservoir solution and equilibrated against 500 µl reservoir solution. Clusters of crystals grew in about 10 d in a condition containing 30% PEG 3350, 0.3 M ammonium fluoride pH 6.5. This condition was then refined and LepFNR crystals suitable for X-ray diffraction were finally grown in 0.25 M ammonium fluoride and 27% PEG 3350 in 50 mM Tris buffer pH 7.0. Hanging drops contained 1 µl protein solution mixed with an equal amount of reservoir solution; the volume of the reservoir was 500 µl.
2.3. Data collection and processing
A single crystal was harvested using a nylon loop (Hampton Research) and transferred from the crystallization drop to 5 µl of a cryogenic solution containing 1 µl ethylene glycol mixed with 4 µl reservoir buffer for a few seconds. The crystal was then flash-cooled to 100 K in a nitrogen stream and used for data collection. 138 images were collected using the oscillation method with a range of 1° per image on a MAR CCD detector using synchrotron radiation at the MX-1 beamline (Polikarpov, Oliva et al., 1997 ▶; Polikarpov et al., 1998 ▶) of the National Synchrotron Light Laboratory (LNLS, Campinas, Brazil). The energy of electrons in the LNLS ring is 1.37 GeV and the critical synchrotron-radiation wavelength is equal to 6.0 Å. Therefore, the LepFNR diffraction data set was collected at 1.42 Å wavelength in order to optimize both the diffraction efficiency of the protein crystal and the synchrotron-radiation flux at this medium-energy synchrotron (Polikarpov, Teplyakov et al., 1997 ▶; Teplyakov et al., 1998 ▶). The data set was reduced and merged using MOSFLM (Leslie, 1992 ▶) and SCALA (Collaborative Computational Project, Number 4, 1994 ▶) programs.
3. Results and discussion
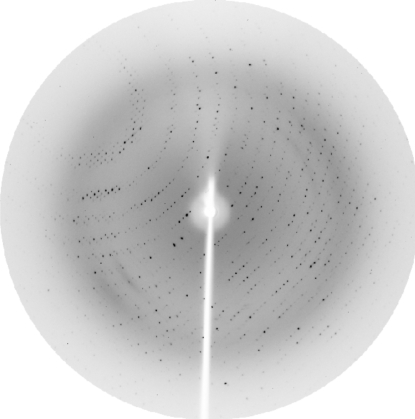
Initial attempts to crystallize FNR lead to highly mosaic crystals that were unsuitable for data collection. Reduction of the crystallization temperature did not improve the quality of the crystals. X-ray quality crystals were grown by reducing the precipitating agent concentration during the PEG concentration optimization grid (Fig. 1 ▶). Diffraction data collected at the synchrotron source extended to 2.4 Å resolution (Fig. 2 ▶ and Table 1 ▶). Initial analysis of the crystal solvent content using the Matthews coefficient (Matthews, 1968 ▶) suggested that the asymmetric unit contains four molecules with 53.5% solvent content or three molecules with 65.1% solvent content. Molecular replacement using the X-ray structure of maize-root FNR (PDB code 1jb9; 35% amino-acid sequence identity), improved by CHAINSAW (Collaborative Computational Project, Number 4, 1994 ▶), as a search model was carried out with Phaser (McCoy et al., 2005 ▶). Phaser simulations converged to a clear solution with four molecules in the asymmetric unit with a Z score of 18.7 in the translation function after placing the last molecule. Structural refinement is in progress.
Figure 1.
Crystals of LepFNR. Typical dimensions are approximately 0.2 × 0.3 × 0.2 mm.
Figure 2.
Diffraction pattern of a LepFNR crystal collected at the LNLS MX-1 beamline (Polikarpov, Oliva et al., 1997 ▶; Polikarpov et al., 1998 ▶). The maximum resolution at the edge of the image is 2.4 Å.
Table 1. X-ray data-collection statistics.
Values in parentheses are for the highest resolution shell (2.54–2.40 Å).
| Wavelength (Å) | 1.42 |
| Resolution range (Å) | 25.0–2.40 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 65.07, b = 111.87, c = 89.84, β = 92.77 |
| Completeness (%) | 95.0 (73.7) |
| Redundancy | 2.8 (2.5) |
| Rmerge† (%) | 5.1 (12.6) |
| Average I/σ(I) | 9.7 (4.2) |
| Total reflections | 251430 |
| Unique reflections | 48944 |
R
merge = 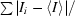
 , where I
i is the intensity of the ith observation and 〈I〉 is the mean intensity of the reflections.
, where I
i is the intensity of the ith observation and 〈I〉 is the mean intensity of the reflections.
Acknowledgments
This work was supported in part by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, Brazil) via grants 04/08070-9 and 06/00182-8, by Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq, Brazil) and by grants from CONICET and ANPCyT, Argentina. The authors also thank the Laboratório Nacional de Luz Sincrotron (LNLS) for technical support during data collection.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carrillo, N. & Ceccarelli, E. A. (2003). Eur. J. Biochem.270, 1900–1915. [DOI] [PubMed] [Google Scholar]
- Ceccarelli, E. A., Arakaki, A. K., Cortez, N. & Carrillo, N. (2004). Biochim. Biophys. Acta, 168, 155–165. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Deng, Z., Aliverti, A., Zanetti, G., Arakaki, A. K., Ottado, J., Orellano, E. G., Calcaterra, N. B., Ceccarelli, E. A., Carrillo, N. & Karplus, P. A. (1999). Nature Struct. Biol.6, 847–853. [DOI] [PubMed] [Google Scholar]
- Ingelman, M., Bianchi, V. & Eklund, H. (1997). J. Mol. Biol.268, 147–157. [DOI] [PubMed] [Google Scholar]
- Karplus, P. A., Daniels, M. J. & Herriot, J. R. (1991). Science, 251, 60–66. [PubMed] [Google Scholar]
- Kurisu, G., Kusunoki, M., Katoh, E., Yamazaki, T., Teshima, K., Onda, Y., Kimata-Ariga, Y. & Hase, T. (2001). Nature Struct. Biol.8, 117–121. [DOI] [PubMed] [Google Scholar]
- Kurisu, G., Nishiyama, D., Kusunoki, M., Fujikawa, S., Katoh, M., Hanke, G. T., Hase, T. & Teshima, K. (2005). J. Biol. Chem.280, 2275–2281. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Nogués, I., Perez-Dorado, I., Frago, S., Bittel, C., Mayhew, S. G., Gómez-Moreno, C., Hermoso, J. A., Medina, M., Cortez, N. & Carrillo, N. (2005). Biochemistry, 44, 11730–11740. [DOI] [PubMed] [Google Scholar]
- Polikarpov, I., Oliva, G., Castellano, E. E., Garratt, R. C., Arruda, P., Leite, A. & Craievich, A. (1997). Nucl. Instrum. Methods A, 405, 159–164.
- Polikarpov, I., Perles, L. A., de Oliveira, R. T., Oliva, G., Castellano, E. E., Garratt, R. & Craievich, A. (1998). J. Synchrotron Rad.5, 72–76. [DOI] [PubMed] [Google Scholar]
- Polikarpov, I., Teplyakov, A. & Oliva, G. (1997). Acta Cryst. D53, 734–737. [DOI] [PubMed] [Google Scholar]
- Prasad, G. S., Kresge, N., Muhlberg, A. B., Shaw, A., Jung, Y. S., Burgess, B. K. & Stout, C. D. (1998). Protein Sci.7, 2541–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial, D. V., Arakaki, A. K., & Ceccarelli, E. A. (2000). Eur. J. Biochem.267, 6239–6248. [DOI] [PubMed] [Google Scholar]
- Serre, L., Vellieux, F. M. D., Medina, M., Gómez-Moreno, C., Fontecilla-Camps, J. C. & Frey, M. (1996). J. Mol. Biol.263, 20–39. [DOI] [PubMed] [Google Scholar]
- Shin, M. & Arnon, D. I. (1965). J. Biol. Chem.240, 1405–1411. [PubMed] [Google Scholar]
- Teplyakov, A., Oliva, G. & Polikarpov, I. (1998). Acta Cryst. D54, 610–614. [DOI] [PubMed] [Google Scholar]
- Ziegler, G. A. & Schulz, G. E. (2000). Biochemistry, 39, 10986–10995. [DOI] [PubMed] [Google Scholar]