The PIN domain of human EST1A was expressed, purified and crystallized by the sitting-drop vapour-diffusion method.
Keywords: EST1A, PIN domains
Abstract
Human EST1A (ever shorter telomeres 1A) is associated with most or all active telomerase in cell extracts and is involved either directly or indirectly in telomere elongation and telomere capping. The C-terminal region of EST1A contains the PIN (PilT N-terminus) domain, a putative nuclease domain. The PIN domain of human EST1A was expressed, purified and crystallized by the sitting-drop vapour-diffusion method. The crystals belonged to space group C2, with unit-cell parameters a = 107.3, b = 51.6, c = 100.5 Å, β = 119.3°, and diffracted X-rays to 1.8 Å resolution. The asymmetric unit contained two molecules of the PIN domain and the solvent content was 57%.
1. Introduction
In many eukaryotes, telomere lengths are properly maintained through telomerase-dependent reverse transcription (reviewed in Vega et al., 2003 ▶; Cech, 2004 ▶; Smogorzewska & de Lange, 2004 ▶). The telomerase core generally consists of two components: a reverse transcriptase (TERT) and an RNA serving as a template (TR). In Saccharomyces cerevisiae, Est1p, a third telomerase subunit, recruits the telomerase to the telomere ends and/or activates the telomerase at the telomere ends (Evans & Lundblad, 1999 ▶; Pennock et al., 2001 ▶; Taggart et al., 2002 ▶). In humans, three Est1p homologues, EST1A, EST1B and EST1C, have been identified (Reichenbach et al., 2003 ▶; Snow et al., 2003 ▶). Like yeast Est1p, EST1A and EST1B are associated with telomerase activity in human cell extracts (Reichenbach et al., 2003 ▶; Snow et al., 2003 ▶). Overexpression of EST1A alone results in telomere shortening, whereas co-overexpression of EST1A and TERT results in telomere lengthening (Snow et al., 2003 ▶). These results suggest that the telomere shortening observed with overexpression of EST1A alone is attributed to titration of TERT. In addition, overexpression of EST1A alone induces anaphase bridges owing to chromosome-end fusions, suggesting that EST1A is involved in telomere capping (Reichenbach et al., 2003 ▶). Both EST1A and EST1B contain C-terminal PIN (PilT N-terminus) domains, which are approximately 180 amino acids in length.
To date, four crystal structures of PIN-domain proteins from Pyrobaculum aerophilum (PAE2754; Arcus et al., 2004 ▶), Archaeoglobus fulgidus (AF0591; Levin et al., 2004 ▶), Pyrococcus horikoshii (PH0500; Jeyakanthan et al., 2005 ▶) and Pyrococcus furiosus (Pfu-367848-001; PDB code 1y82) have been determined. These archaeal PIN-domain proteins are approximately 140 amino acids in length. The protein PAE2754 possesses exonuclease activity, indicating a DNA- or RNA-editing role (Arcus et al., 2004 ▶).
The PIN domain of human EST1A contains additional inserts of about 40 amino acids compared with archaeal PIN-domain proteins. Here, we describe the expression, purification, crystallization and preliminary X-ray diffraction analysis of the PIN domain of human EST1A.
2. Protein expression and purification
A cDNA clone (KIAA0732; NCBI accession No. AB018275) coding for human EST1A was obtained from the Kazusa DNA Research Institute (Chiba, Japan). The region encoding the PIN domain (residues 1239–1419) was inserted into the pGEX-4T-1 vector (Amersham Biosciences) to express a GST-PIN fusion protein. Escherichia coli Rosetta (DE3) was transformed with the plasmid. The E. coli cells were grown in Luria–Bertani broth containing ampicillin (50 µg ml−1) until mid-log phase and protein expression was induced by the addition of 1.0 mM isopropyl β-d-thiogalactopyranoside (IPTG) at 293 K. The cells were harvested by centrifugation at 5000g for 15 min and resuspended in lysis buffer containing 200 mM NaCl, 20 mM Tris–HCl pH 8.0, 1.0 mM DTT (dithiothreitol) and 0.5 mM EDTA.
The cells were disrupted by sonication and the insoluble materials were removed by centrifugation at 40 000g for 60 min. The supernatant containing the GST-PIN fusion protein was applied onto a glutathione-Sepharose 4 Fast Flow column (Amersham Biosciences). The column was washed with 200 mM NaCl, 20 mM Tris–HCl pH 8.0, 1.0 mM DTT and 0.5 mM EDTA and the fusion protein was then cleaved on the column by incubating with thrombin (Amersham Biosciences) for 15 h in buffer containing 200 mM NaCl, 20 mM Tris–HCl pH 8.0, 1.0 mM DTT and 0.5 mM EDTA. The PIN domain was eluted with 200 mM NaCl, 20 mM Tris–HCl pH 8.0, 1.0 mM DTT and 0.5 mM EDTA and was further purified using a HiLoad 26/60 Superdex 75 gel-filtration column (Amersham Biosciences). The thrombin was removed from the PIN-domain sample when the PIN domain was eluted from the gel-filtration column. The fraction containing the PIN domain was dialyzed against 20 mM Tris–HCl pH 8.0 and 1.0 mM DTT and concentrated to 15 mg ml−1 using an Apollo ultrafiltration concentrator (Orbital Biosciences). The protein concentration was determined using the Bradford Protein Assay (Bio-Rad).
3. Crystallization
The sparse-matrix screening kits Crystal Screens I and II (Hampton Research) and Wizard I and II (Emerald Biostructures) were used for initial crystallization trials using the sitting-drop vapour-diffusion method. Drops consisting of 2 µl protein solution and 2 µl reservoir solution were equilibrated against 500 µl reservoir solution in Cryschem plates (Hampton Research) and incubated at 278 K.
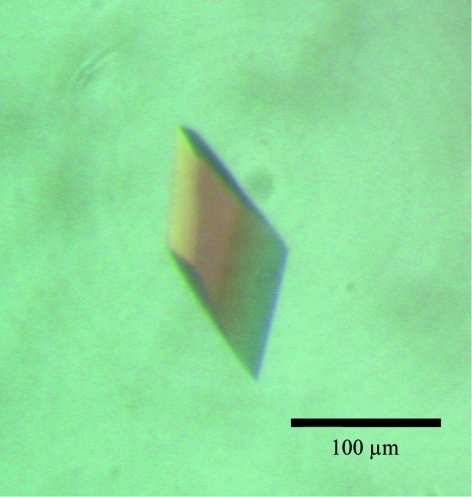
Single crystals were obtained with reservoir solution containing 1.26 M (NH4)2SO4, 0.1 M CHES–NaOH pH 9.5 and 0.2 M NaCl. Subsequently, reservoir solutions of varying pH and precipitant concentration were tested, but no additional improvement in crystal quality for X-ray diffraction was observed. The crystals used for data collection had typical dimensions of 80 × 100 × 120 µm (Fig. 1 ▶).
Figure 1.
A crystal of the PIN domain with approximate dimensions of 80 × 100 × 120 µm.
4. Preliminary X-ray analysis
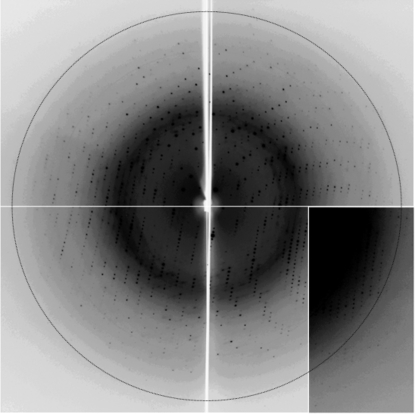
X-ray diffraction experiments were performed at beamline NW12 of the Photon Factory-Advanced Ring (PF-AR), Tsukuba, Japan. The crystals were transferred into a cryoprotectant solution consisting of 0.9 M (NH4)2SO4, 0.07 M CHES–NaOH pH 9.5 and 0.14 M NaCl and 25% glycerol for approximately 30 s. The crystals were then mounted in nylon loops (Hampton Research) and flash-cooled in a nitrogen stream at 95 K. Diffraction data were collected using a Quantum 210 CCD X-ray detector (Area Detector Systems Corporation) in 1.0° oscillation steps and the crystal diffracted X-rays to 1.8 Å resolution (Fig. 2 ▶). The data set was processed and scaled using the program HKL2000 (Otwinowski & Minor, 1997 ▶). The crystals belonged to space group C2, with unit-cell parameters a = 107.3, b = 51.6, c = 100.5 Å, β = 119.3°. The data-collection statistics are shown in Table 1 ▶. When the asymmetric unit contains two molecules of the PIN domain, the calculated solvent content is 57% and the Matthews coefficient is 2.9 Å3 Da−1 (Matthews, 1968 ▶). Crystals of the SeMet derivative were obtained and experimental phasing using the crystals is currently under way.
Figure 2.
A diffraction image (1.0° oscillation) of the PIN-domain crystal. The diffraction extends to 1.8 Å resolution (indicated by the circle).
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| X-ray source | PF-AR NW12 |
| Wavelength (Å) | 1.000 |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 107.3, b = 51.6, c = 100.5, β = 119.3 |
| Resolution range (Å) | 50.0–1.80 (1.86–1.80) |
| Observed reflections | 139704 |
| Unique reflections | 45150 |
| Completeness (%) | 96.0 (88.0) |
| Rmerge† (%) | 5.6 (19.6) |
| 〈I〉/〈σ(I)〉 | 22.8 (4.6) |
R
merge = 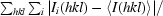
 , where I
i(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl) is its average.
, where I
i(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl) is its average.
Acknowledgments
We thank Dr Takahiro Nagase (Kazusa DNA Research Institute) for giving us the human KIAA0732 cDNA clone. We also thank the beamline staff at the Photon Factory for their kind help during data collection. The synchrotron-radiation experiments at beamline NW12 at PF-AR were performed with the approval of the Photon Factory, KEK (Proposal No. 05G079). This work was supported in part by the National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology of Japan and by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Arcus, V. L., Bäckbro, K., Roos, A., Daniel, E. L. & Baker, E. N. (2004). J. Biol. Chem.279, 16471–16478. [DOI] [PubMed] [Google Scholar]
- Cech, T. R. (2004). Cell, 116, 273–279. [DOI] [PubMed] [Google Scholar]
- Evans, S. K. & Lundblad, V. (1999). Science, 286, 117–120. [DOI] [PubMed] [Google Scholar]
- Jeyakanthan, J., Inagaki, E., Kuroishi, C. & Tahirov, T. H. (2005). Acta Cryst. F61, 463–468. [DOI] [PMC free article] [PubMed]
- Levin, I. et al. (2004). Proteins, 56, 404−408. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pennock, E., Buckley, K. & Lundblad, V. (2001). Cell, 104, 387–396. [DOI] [PubMed] [Google Scholar]
- Reichenbach, P., Höss, M., Azzalin, C. M., Nabholz, M., Bucher, P. & Lingner, J. (2003). Curr. Biol.13, 568–574. [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A. & de Lange, T. (2004). Annu. Rev. Biochem.73, 177–208. [DOI] [PubMed] [Google Scholar]
- Snow, B. E., Erdmann, N., Cruickshank, J., Goldman, H., Gill, R. M., Robinson, M. O. & Harrington, L. (2003). Curr. Biol.13, 698–704. [DOI] [PubMed] [Google Scholar]
- Taggart, A. K., Teng, S. C. & Zakian, V. A. (2002). Science, 297, 1023–1026. [DOI] [PubMed] [Google Scholar]
- Vega, L. R., Mateyak, M. K. & Zakian, V. A. (2003). Nature Rev. Mol. Cell. Biol.4, 948–959. [DOI] [PubMed]