Abstract
Background
Polycomb (PcG) and trithorax (trxG) genes encode proteins involved in the maintenance of gene expression patterns, notably Hox genes, throughout development. PcG proteins are required for long-term gene repression whereas TrxG proteins are positive regulators that counteract PcG action. PcG and TrxG proteins form large complexes that bind chromatin at overlapping sites called Polycomb and Trithorax Response Elements (PRE/TRE). A third class of proteins, so-called “Enhancers of Trithorax and Polycomb” (ETP), interacts with either complexes, behaving sometimes as repressors and sometimes as activators. The role of ETP proteins is largely unknown.
Methodology/Principal Findings
In a two-hybrid screen, we identified Cyclin G (CycG) as a partner of the Drosophila ETP Corto. Inactivation of CycG by RNA interference highlights its essential role during development. We show here that Corto and CycG directly interact and bind to each other in embryos and S2 cells. Moreover, CycG is targeted to polytene chromosomes where it co-localizes at multiple sites with Corto and with the PcG factor Polyhomeotic (PH). We observed that corto is involved in maintaining Abd-B repression outside its normal expression domain in embryos. This could be achieved by association between Corto and CycG since both proteins bind the regulatory element iab-7 PRE and the promoter of the Abd-B gene.
Conclusions/Significance
Our results suggest that CycG could regulate the activity of Corto at chromatin and thus be involved in changing Corto from an Enhancer of TrxG into an Enhancer of PcG.
Introduction
In Drosophila, the Bithorax-complex (BX-C) contains the three Hox genes, Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B), that specify the identities of the third thoracic segment (T3) and the eight abdominal segments (A1 to A8) [1]. These genes are expressed in spatially regulated patterns during embryonic development thanks to maternal, gap and pair-rule proteins. Their large cis-regulatory sequences are modular and allow parasegmental regulation. These sequences contain different classes of elements such as initiation elements that respond to early segmentation gene products, insulators and promoter targeting sequences (reviewed in [2]).
Hox expression is maintained in the original pattern during later stages of development by the Polycomb-group (PcG) and trithorax-group (trxG) genes. In mutants of PcG or trxG genes, Hox patterns are established correctly but are not maintained. PcG proteins keep Hox genes silenced whereas TrxG proteins keep Hox genes activated thus counteracting PcG action [3], [4]. PcG and TrxG proteins are required for the maintenance of many gene expression patterns [5]. These maintenance proteins form heteromultimeric complexes that bind to chromatin and alter its structure. Current models propose that PcG complexes lead to compact, transcriptionally inactive chromatin, whereas TrxG complexes maintain chromatin in an open conformation that facilitates transcription. In Drosophila, several PcG and TrxG complexes have been purified so far: the Polycomb Repressive Complex 1 (PRC1), the Polycomb Repressive Complex 2 (PRC2), the PhoRC complex, the Pcl-PRC2 complex, the Trithorax Activating Complex 1 (TAC1) and the Brahma Complex (BRM) also called SWI/SNF complex. They are extremely large complexes that contain several proteins including chromatin modifying enzymes such as histone methyl-transferases, acetyl-transferases or deacetylases [5]–[8].
Although most PcG mutations suppress trxG mutations and vice versa, a large screen to identify modifiers of the trxG gene ash1 allowed isolation of enhancers that were previously identified as PcG [E(z), E(Pc), Asx, Scm, Psc and Su(z)2] [9]. These genes were then called Enhancers of Trithorax and Polycomb (ETPs). Further molecular data showed that some ETPs encode members of PRC complexes, such as E(Z), PSC or SCM, while some do not. Recently, Grimaud et al. proposed to reclassify these maintenance proteins, the label PcG being kept for members of PRC silencing complexes and the label TrxG for members of complexes that counteract PcG-mediated silencing [10]. A third class of proteins would be represented by PcG/TrxG DNA-binding recruiters or specific co-factors. We will keep here the term ETP for those maintenance proteins that play a dual role in PcG and TrxG functions without belonging to any PcG or TrxG complexes identified so far. The GAGA factor, Gaf, encoded by Trithorax-like (Trl), falls into this category. Indeed, it was first described as an activator of Hox genes, and later shown to play a role in the recruitment of PcG complexes without co-purifying with any PRC silencing complexes [11], [12]. The HMG protein DSP1 also meets the criteria to be an ETP: dsp1 mutants exhibit Hox gene loss-of-function phenotypes but DSP1 is also important for PcG recruitment to chromatin [13], [14]. We have previously shown that corto behaves genetically as an ETP. corto mutants present PcG as well as trxG phenotypes and enhance the phenotypes of some PcG, trxG and ETP mutants [15], [16]. Corto directly interacts with Gaf and DSP1 suggesting that ETPs are involved in collaborative processes [16], [17].
PcG, TrxG and ETP proteins bind DNA sequences called PRE/TRE that carry the information for the active or silent state of the gene they control (reviewed in [18]). Some PRE/TRE have been shown to maintain this transcriptional state throughout cellular divisions in absence of the initial activator or repressor [19], [20]. Despite massive efforts towards identification of PcG complex targets at genome scale [21]–[23], the mechanism by which the active or inactive state of PRE/TRE is conserved throughout several cell cycles remains still largely unknown. Many PcG and ETP mutants [Asx, corto, E(z), Pc, ph, Psc, Su(z)2, Trl] exhibit proliferation defects as well as chromosome condensation and segregation defects. This suggests that maintenance proteins play a general role in cell cycle control [24]–[28]. An attractive hypothesis is that ETPs are critical to maintain the correct association of PcG or TrxG complexes with chromatin during the cell cycle.
In a two-hybrid screen using Corto as bait, we isolated Cyclin G (CycG), the Drosophila homologue of the mammalian Cyclin G1 and G2 (CycG1, CycG2). Vertebrate CycG1 is a transcriptional target of the tumor suppressor p53 [29], [30]. It is possibly involved in cell proliferation as it is overexpressed in certain cancer cells [31], [32]. However, CycG1 induces G2/M arrest and cell death in response to DNA damage [33]–[35]. Vertebrate CycG2 acts as a negative regulator of cell cycle, as shown by its high level in cells in which G1/S arrest has been induced by growth inhibitory signals [36], [37].
Here, we address the interactions between Corto and CycG both in vitro and in vivo. We show that CycG plays an essential role during development. Moreover, we show that CycG is targeted to many sites on polytene chromosomes where it co-localizes partially with Corto and with the PcG factor PH. As an ETP, corto maintains Abd-B repression in embryos. This could be achieved by association between Corto and CycG since both proteins bind to Abd-B regulatory elements, including the iab-7 PRE and the promoter.
Results
Drosophila Cyclin G interacts with Corto
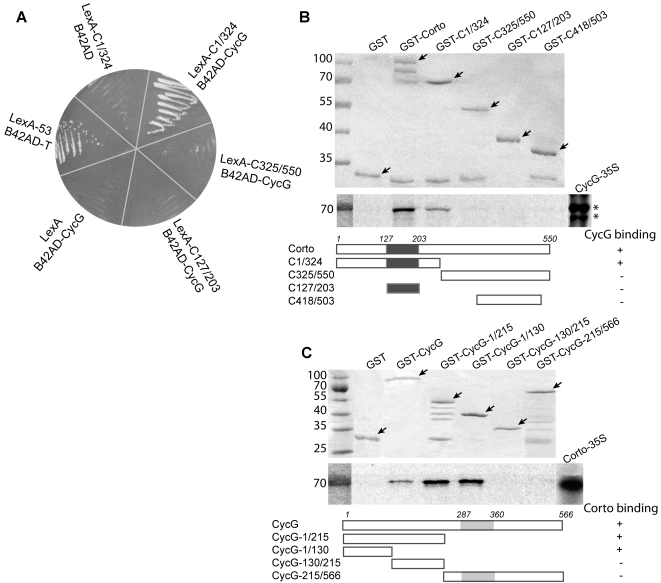
To further investigate Corto function, we performed a two-hybrid screen for potential Corto partners. As bait, we used the amino-terminal half of Corto containing a chromodomain [16]. A positive clone spanning almost the full-length CG11525 cDNA (positions 43 to 2263; Accession number NM 079870) encoding Cyclin G (CycG) was isolated. Subsequent two-hybrid assays showed that the chromodomain was not sufficient for interaction with CycG, and that CycG did not interact with the C-terminal half of Corto (figure 1A).
Figure 1. Corto and CycG interact in vitro.
A. Two-hybrid tests. Three Corto sub-fragments comprising amino-acids 1 to 324 (C1/324), 325 to 550 (C325/550), and 127 to 203 (C127/203) (chromodomain) were fused with LexA DNA binding domain. B42AD-CycG was isolated from an embryonic library using LexA-C1/324 as bait. Interaction was observed only between LexA-C1/324 and B42-CycG. Negative controls: LexA versus B42AD-CycG and LexA-C1/324 versus B42AD. Positive control: LexA53 (LexA/p53 fusion) versus B42AD-T (B42AD/large T-antigen fusion). B. GST pull-down assays. Top: Coomassie staining of fusion proteins (shown by arrows). Middle: Autoradiography. Input (CycG-35S) shows two isoforms (asterisks). Bottom: schematic representation of Corto full-length and truncated forms (black box: chromodomain). C. GST pull-down assays. Top: Coomassie staining of the fusion proteins (shown by arrows). Middle: Autoradiography. Input (Corto 35S). Bottom: schematic representation of CycG full-length and truncated forms (grey box: cyclin domain).
Then, we performed GST pull-down assays. In vitro translated CycG protein was retained on GST-Corto beads containing the full-length protein and on GST-C1/324 beads containing the amino-terminal half of Corto, but not on the other GST-Corto fusion proteins tested (figure 1B). Reciprocally, in vitro translated Corto protein was retained on GST-CycG beads which contained the full-length CycG protein. Corto was not retained on GST-CycG-215/566 which contains the cyclin domain but was bound by the N-terminal CycG-region (figure 1C). These results corroborate the two-hybrid results and indicate that the amino-terminal half of Corto interacts with the amino-terminal end of CycG.
CycG is an essential gene in flies
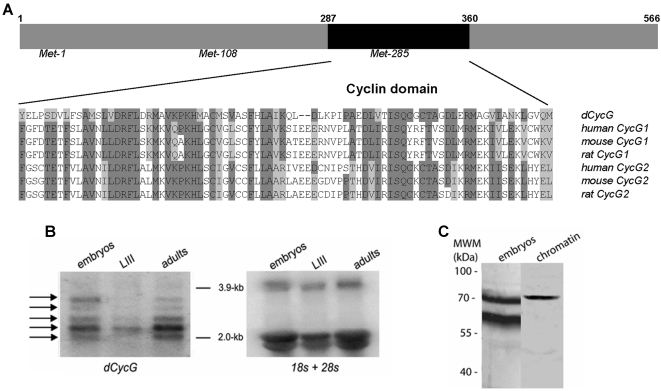
The cyclin domain of Drosophila CycG is highly similar to the cyclin domains of vertebrate CycG1 and CycG2 (42% and 46% identity, respectively; figure 2A). In agreement with genome annotations, Northern blot analysis revealed 5 different mRNAs ranging between 2.0 and 3.5 kb. All transcripts were found throughout development although notably less abundant in third instar larvae (figure 2B). Antibodies were raised against the N-terminal part of CycG. Two isoforms of 68 kDa and 60 kDa were revealed in total embryonic extracts whereas only the 68 kDa species was found in chromatin extracts (figure 2C). Three translation start sites are predicted in CycG using ATGpr software [38], resulting in putative proteins with molecular weights of 63, 50 and 30 kDa, respectively (figure 2A). Our antisera do not allow testing for the presumptive 30 kDa isoform. However, the two isoforms we detect probably correspond to the two larger predicted proteins.
Figure 2. Alignment of cyclin domains of Drosophila and mammalian CycG proteins.
A. Alignment of the cyclin domains of Drosophila CycG (NM 524609), human CycG1 (NP 954854), mouse CycG1 (NP 033961), rat CycG1 (NP 037055), human CycG2 (NM 004345), mouse CycG2 (NP 031661) and rat CycG2 (NP 446451) (dark grey: identical amino-acids, light grey: similar amino-acids). B. Developmental Northern blot. Poly(A+) RNA from w1118 embryos, third instar larvae (LIII) and adult flies were probed with 32P-labelled CycG cDNA (left). Arrows point to five mRNA species ranging from 2.0 to 3.5 kb in size that may correspond to CG11525 RA-RE transcripts predicted in Flybase. The same filter was hybridized with 32P-labelled 18S and 28S probes (right) for loading control. C. Total protein (left) and chromatin (right) extracts were prepared from w1118 0–14 h embryos and analysed using rabbit anti-CycG antibodies.
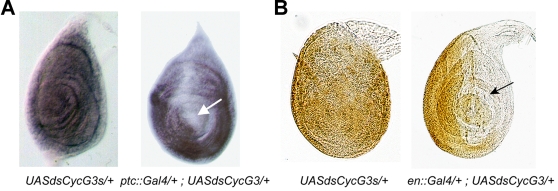
Since no mutant of CG11525/CycG was available, we designed a P{UAS::dsCycG} construct to inactivate the gene by RNA-interference (RNAi) as described previously [39]. Tissue specific RNAi using various Gal4 driver lines resulted in a considerable downregulation of CycG activity as visualized by reduction of CycG mRNA and CycG protein levels (figure 3), respectively. Ubiquitous downregulation of CycG (da::Gal4; UAS::dsCycG or Act::Gal4; UAS::dsCycG) animals led to lethality of late third instar larvae or pharates. However, the percentage of dead animals varied depending on the transgenic line: lethality of da::Gal4>UAS::dsCycG2 was estimated to 31% in females and 56% in males whereas no da::Gal4>UAS::dsCycG3 or Act::Gal4>UAS::dsCycG3 adults were obtained (table 1). Lethality was complete in Act::Gal4>UAS::dsCycG2 males and reached 86% in Act::Gal4>UAS::dsCycG2 females. We observed that males of this genotype never undergo metamorphosis and stop their development as third instar larvae, dying after a few days, whereas most females die as late pharates. Thus, these two lines interfere with CycG activity to different degrees, indicative of partial inactivation. Overexpression of CycG with an ubiquitous driver (da::Gal4>UAS::CycG or Act::Gal4>UAS::CycG) suppressed the lethality induced by UAS::dsCycG3 suggesting that it was linked to specific inactivation of CycG and not to Off-target effects (table 1). Taken together, these results show that CycG plays an essential role during development.
Figure 3. CycG is an essential gene in Drosophila.
A. Validation of RNAi efficiency by in situ hybridization. Leg imaginal discs from ptc::Gal4>UAS::dsCycG3 third instar larvae were hybridized using CycG as a probe. Expression of the CycG mRNA was specifically reduced in the ptc expression domain along the antero-posterior border (arrow). B. Validation of RNAi efficiency by immuno-labelling. Leg imaginal discs from en::Gal4>UAS::dsCycG3 were immuno-labelled using rabbit anti-CycG antibodies. The expression of CycG was specifically reduced in the en expression domain i.e. in the posterior compartment (arrow), whereas it was ubiquitous in the control (UAS::dsCycG3).
Table 1. Lethality of RNAi CycG flies.
| Genotype | Lethality observed (%) | Emerged flies |
| X/X ; UAS::dsCycG2/+ ; da::Gal4/+ | 31 | 230 |
| X/Y ; UAS::dsCycG2/+ ; da::Gal4/+ | 56 | 185 |
| X/X ; UAS::dsCycG3/da::Gal4 | 100 | - |
| X/Y ; UAS::dsCycG3/da::Gal4 | 100 | - |
| Act::Gal4/X ; UAS::dsCycG2/+ | 86 | 18 |
| Act::Gal4/Y ; UAS::dsCycG2/+ | 100 | - |
| Act::Gal4/X ; UAS::dsCycG3/+ | 100 | - |
| Act::Gal4/Y ; UAS::dsCycG3/+ | 100 | - |
| X/X ; UAS::CycG/+ ; da::Gal4/UAS::dsCycG3 | 0 | 145 |
| X/Y ; UAS::CycG/+ ; da::Gal4/UAS::dsCycG3 | 0 | 153 |
| Act::Gal4/X ; UAS::CycG/+ ; UAS::dsCycG3/+ | 0 | 168 |
| Act::Gal4/Y ; UAS::CycG/+ ; UAS::dsCycG3/+ | 0 | 112 |
Corto and CycG interact in vivo and co-localize at multiple sites on polytene chromosomes
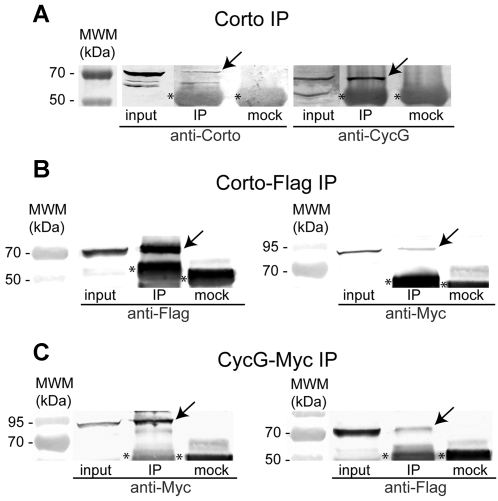
In vivo physical interactions between Corto and CycG were first analysed by co-immunoprecipitation of total embryonic protein extracts (figure 4A). Only the 68 kDa and not the 60 kDa CycG isoform co-immunoprecipitated with Corto. We cannot exclude that the latter may be hidden by co-migrating IgG. To confirm this result, we co-transfected Schneider S2 cells with pAct::Corto-Flag and pAct::Myc-CycG and probed for interaction using anti-Flag and anti-Myc antibodies. Myc-CycG co-immunoprecipitated with Corto-Flag and conversely (figure 4B–C).
Figure 4. Corto and CycG interact in vivo.
A. CycG co-immunoprecipitates with Corto in embryonic extracts. Protein extracts from 0–14 h embryos were incubated with rabbit anti-Corto antibodies (IP) or rabbit preimmune serum (mock). Western blot analysis was performed using rat anti-Corto antibodies (left) or rat anti-CycG antibodies (right). Note specific precipitation of Corto (70 kDa) and the 68 kDa CycG species (arrows). The asterisks label unspecific IgG signals in all panels. B. Myc-CycG co-immunoprecipitates with Corto-Flag in S2 cell extracts. S2 cells were co-transfected with pAct::Corto-Flag and pAct::Myc-CycG. Proteins extracts were incubated with either mouse anti-Flag antibodies (IP) or mouse anti-HA antibodies (mock). Western blot analysis was performed using mouse anti-Flag antibodies (left) or mouse anti-Myc antibodies (right). Specific precipitation of Corto-Flag (left) or Myc-CycG (right) is indicated by arrows. C. Corto-Flag co-immunoprecipitates with Myc-CycG in S2 cell extracts. S2 cells were co-transfected with pAct::Corto-Flag and pAct::Myc-CycG. For immunoprecipitation we used either mouse anti-Myc antibodies (IP) or mouse anti-HA antibodies (mock), and for detection, mouse anti-Myc antibodies (left) or mouse anti-Flag antibodies (right). Specific precipitation of Myc-CycG (left) or Corto-Flag (right) is indicated by arrows. 4% of the starting material used in each IP (input) and 50% of the immunoprecipitated material were loaded onto the gel in all our assays.
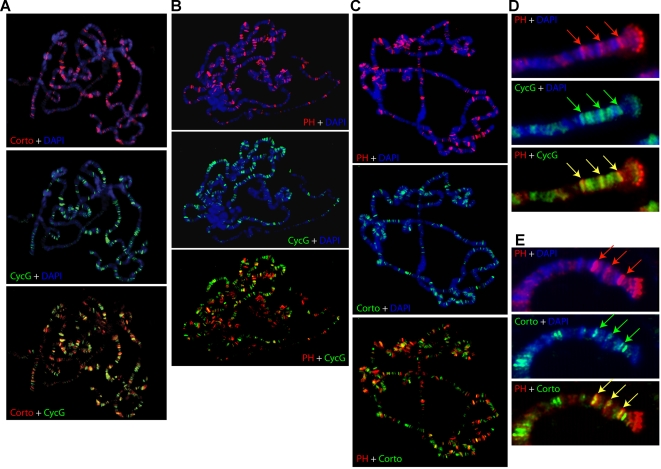
Corto was previously shown to bind polytene chromosomes at multiple discrete loci suggesting that it might participate in the regulation of many genes [17]. Using CycG antisera, we first showed that CycG is ubiquitously expressed in embryos and larvae (not shown). To test whether interactions between Corto and CycG could take place on chromatin, we explored the binding of CycG to polytene chromosomes (figure 5A). We detected CycG at multiple discrete sites. 30 to 40% of these sites overlapped with Corto binding sites suggesting that Corto and CycG could indeed interact on chromatin.
Figure 5. Corto, CycG and PH partially co-localize in vivo.
Simultaneous detection of Corto (red) and CycG (green) (A), PH (red) and CycG (green) (B), PH (red) and Corto (green) (C) on polytene chromosomes stained with DAPI (blue). Close-ups of chromosome 3 L ends showing 3 loci simultaneously bound by PH (Red) and CycG (green) (D), and the same 3 loci simultaneously bound by PH (red) and Corto (green) (E). Overlaps appear yellow.
We next asked whether Corto was essential for CycG recruitment to chromatin, and analysed CycG fixation on polytene chromosomes derived from corto07128/Df(3R)6–7 larvae. On the whole, no modification of the CycG binding pattern was observed (data not shown) suggesting that CycG recruitment does not depend on Corto. We have previously shown that Corto shares many sites on polytene chromosomes with PcG proteins ([16], [17] and figure 5C). We observed that CycG also shares many sites with PH (figure 5B). Moreover, some sites were simultaneously occupied by Corto, CycG and PH, suggesting that the interaction between Corto and CycG could be related to PcG function (figure 5D, E).
corto participates in the regulation of Abd-B expression in embryos
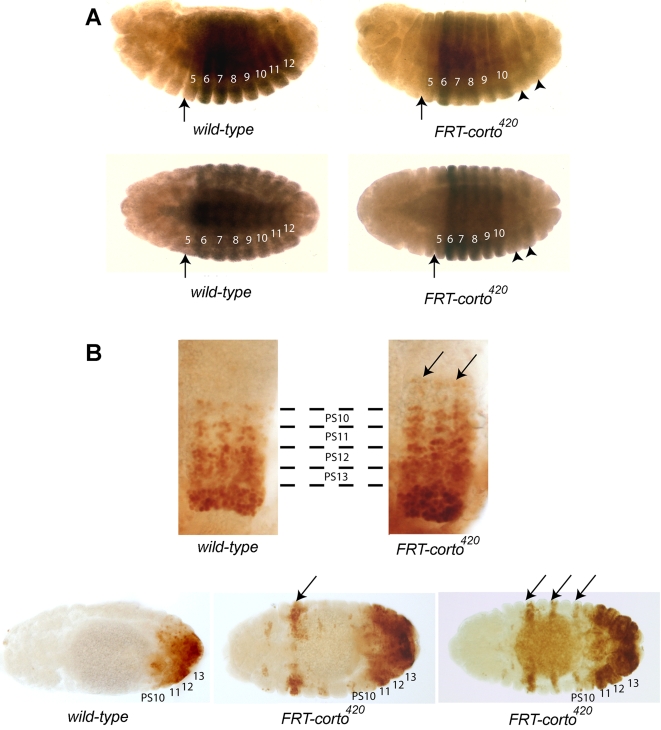
Previous findings indicate that corto is involved in the regulation of Hox genes such as Scr or Ubx in larvae [15], [17]. Interestingly, in corto germinal clone embryos, Ubx was strongly down-regulated in parasegments (PS) 11–12 whereas normally expressed in more anterior segments ([15] and figure 6A). In wild-type embryos, Abd-B expression domain extends from PS 10 to 13 [40], [41]. In light of the posterior prevalence phenomenon, Ubx down-regulation could be due to up-regulation of Abd-B in the same segments. Indeed, in corto germinal clone embryos, the expression of Abd-B not only increased in the normal Abd-B expression domain but also extended more anteriorly than PS10 (figure 6B). Moreover, discontinuous ectopic expression of Abd-B occurred in some anterior parasegments indicative of homeotic transformation towards more posterior identities. These results suggest that corto participates in maintenance of Abd-B repression in embryos.
Figure 6. Expression of Hox genes in corto mutant embryos.
A. Anti-Ubx antibody staining of w1118 embryos, or embryos depleted of maternal and zygotic Corto. Upper row, lateral view and lower row, dorsal view. (B) Dorsal view. In wild-type embryos (left), Ubx is expressed from parasegment PS5 to PS12 with a higher expression in PS6. In corto embryos (right), the anterior boundary of Ubx is not modified but Ubx is strongly downregulated in PS11 and PS12 (arrowheads) . Parasegment numbers are indicated in white and the anterior border of Ubx expression by arrows. B. Anti-AbdB antibody staining of w1118 embryos, or embryos depleted of maternal and zygotic Corto. Upper row, close-ups of the ventral nerve chord. In wild-type embryos (left), Abd-B is expressed in a decreasing gradient from PS13 to PS10. In corto embryos (right), this gradient extends anteriorly to PS9 and PS8. Moreover, expression of Abd-B seems to be higher in PS13 to PS10. Lower row, dorsal view of whole mount embryos showing ectodermal Abd-B expression; note ectopic expression of Abd-B in anterior parasegments in corto mutant embryos (arrows).
Corto and Cyclin G bind the iab-7 PRE and the promoter of Abd-B
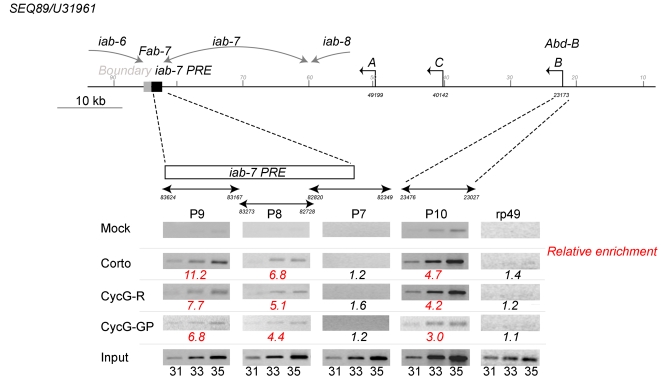
Co-localization of Corto and CycG proteins on polytene chromosomes raises the possibility that CycG and Corto belong to a complex that regulates Abd-B expression. To address this possibility, we investigated whether both proteins bind to Abd-B cis-regulatory sequences in embryos. We performed immunoprecipitation on formaldehyde cross-linked chromatin (XChIP) from 0–14h embryos. The co-immunoprecipitated DNA was amplified using primer pairs corresponding either to the promoter region of Abd-B (generating fragment p10), to the iab-7 PRE (generating fragments p9, p8 and p7) or to rp49 as a negative control. We found that Corto and CycG were both present on the promoter and on the iab-7 PRE (figure 7). As previously described for Polycomb (Pc), we observed prominent binding for the p9 fragment that contains the minimal PRE [42], [43]. These results strongly suggest that Corto and CycG directly regulate Abd-B expression.
Figure 7. Corto and CycG bind the iab-7 PRE and the promoter of Abd-B.
Schematic representation of the Abd-B locus (numbers and transcript names refer to SEQ89/U31961) [44]. Corto or CycG XChIP was performed using total chromatin from 0–14 h embryos. Fragments around 500 bp from the iab-7 PRE (p9, p8) and overlapping the iab-7 PRE (p7) or the Abd-B B promoter (p10) were amplified with specific primers using the immuno-precipitated DNA [42], [45]. A fragment from the rp49 gene was used as a negative control. The 31st, 33rd and 35th PCR cycle samples are shown. Relative enrichment was estimated for the 33rd PCR cycle sample from the ratio between Corto or CycG immunoprecipitations and mock signals from three independent experiments. The input track shows amplification of DNA from total chromatin with the same primers (Mock: rabbit preimmune serum, Corto: rabbit anti-Corto, CycG-R: rabbit anti-CycG, CycG-GP: guinea-pig anti-CycG). The iab-6, iab-7 and iab-8 cis-regulatory domains are indicated.
Discussion
We have identified Cyclin G as a new binding partner of the ETP Corto in Drosophila melanogaster. CycG inactivation leads to lethality showing that this gene is essential in flies. Mammalian genomes encode two G-type cyclins, CycG1 and CycG2, the first one being mainly nuclear whereas the second is mainly cytoplasmic [46]. Drosophila has a single homologue, however, it produces at least two different protein isoforms, only the larger being associated with chromatin. These isoforms could combine CycG1 and CycG2 functions. In Drosophila, large scale two-hybrid screens suggested binding of CycG to various Cyclin-Dependent Kinases (CDK) (Cdc2 and Cdk4) [47], [48]. Corto and CycG interact in vitro as well as in vivo and form a complex in embryos and presumably also on chromatin. Moreover, Corto interacts with the amino-terminal domain of CycG, which is compatible with the simultaneous binding of CDK and cell-cycle control function of CycG.
Requirement of PcG, trxG and ETP genes in cell-cycle control has already been shown in Drosophila [49], [50]. Interestingly, PcG and trxG genes are also involved in self-renewal and proliferation of hematopoietic stem cells in vertebrates [51], [52]. One way they might control cell proliferation is by an epigenetic regulation of genes involved in cell cycle and cell proliferation. Indeed, homologues of Drosophila E(z) and Brm participate in the transcriptional regulation of Cyclin A and E in vertebrates, and in Drosophila, Cyclin A is a PcG target [53]–[55]. Alternatively, PcG, TrxG or ETP proteins may interact directly with cell cycle regulatory proteins. Indeed, it has been shown that Brm interacts with Cyclin E, that Mel-18, a human homologue of Posterior Sex Combs, interacts with Cyclin D2 possibly blocking its interaction with Cdks [56], [57] and we show here that the ETP Corto interacts with CycG. These interactions reveal a potential role for these maintenance proteins in regulating the cell cycle independently of transcriptional regulation. This could be a widespread mechanism by which PcG, TrxG and ETP coordinate the chromatin activity status.
CycG and Corto co-localize on many sites on polytene chromosomes suggesting that they may have regulated associations. Our data show that Corto represses Abd-B in embryos and although we were not able to test the role of CycG in regulating Abd-B expression in embryos, we observed that both Corto and CycG bind the iab-7 PRE and the promoter of Abd-B suggesting that they could cooperate in this function. Nevertheless, neither Corto nor CycG were detected on the BX-C locus in salivary glands suggesting that they regulate Abd-B in a tissue-specific manner. The role of the CycG-Corto interaction needs to be further investigated. CycG could regulate Corto activity directly on chromatin by recruiting other factors like kinases or phosphatases thus modifying the phosphorylation status of Corto itself, of histones or other proteins at PRE/TRE and promoters. It has been shown that binding of the PcG protein Bmi1 to chromatin correlates with its phosphorylation status [58], [59]. It will be interesting to investigate whether Corto and CycG bind the iab-7 PRE and promoter of Abd-B simultaneously, to examine their phosphorylation status when bound to chromatin, and to determine if their presence correlates with Abd-B transcriptional activity. One interesting possibility would be that CycG is involved in changing Corto from an Enhancer of TrxG into an Enhancer of PcG.
Materials and Methods
Drosophila strains and genetics
Details on Drosophila strains can be found in Flybase [60]. corto420 and corto07128 are strong hypomorphic alleles; corto420/TM6B was a gift from Roland Rosset [27]. corto-deficient germline clones were obtained as previously described [15]. Other strains were obtained from either Bloomington or Kyoto Drosophila stock centers. CycG transgenic lines were established by standard P-element mediated transformation.
Plasmid constructs
Corto-GST fusions and pBS-Corto were previously described [16]. The pJG-CycG plasmid served for PCR amplification of full-length or truncated forms of CycG that were subsequently cloned into pGEX4T-1. Full length cDNAs were subcloned into pENTR/D-TOPO® and transferred into Gateway™ vectors: corto into pAWF to obtain pAct::Corto-Flag, and CycG into pAMW to obtain pAct::Myc-CycG [61].
P{UAS::CycG} was constructed by cloning the entire cDNA as EcoRI/XhoI fragment into pUAST [62]. UAS::dsCycG was constructed as outlined before [39]. About 600 nucleotide coding sequence (codons 72 to 268) was chosen to prepare the RNAi-construct. This segment shows only limited identity to other Drosophila genes and none of them conforms to an optimal siRNA. Two possible “Off-Targets” were found, LvpL encoding a larval protein with a predicted role in glucose metabolism and CG15639 encoding an unknown product. In both cases, 21 nucleotides are identical with a GC content of 55–58% instead of the optimal 43–53% [63]. Cloning details are available upon request.
Antibodies
Corto and Polyhomeotic (PH) antibodies were used as described previously [17]. Antibodies against Abd-B (clone 1A2E9) were obtained from the Developmental Studies Hybridoma Bank. Polyclonal antibodies against CycG were raised against the N-terminal 276 amino-acids of CycG fused to maltose binding protein in rabbit, rat and guinea pig. Their specificity was checked on CycG protein generated by in vitro transcription/translation. Monoclonal anti-Flag M2 and anti-HA was from Sigma (F-3165, H-3663) and anti-Myc clone 9E10 from Santa-Cruz Biotechnology.
Histology
Antibody staining of embryos and larvae was performed using rabbit anti-CycG (1∶40) or mouse monoclonal anti-Abd-B (1∶10) [64]. Co-immunostaining of polytene chromosomes was performed with rabbit anti-Corto (1∶40) and guinea-pig anti-CycG (1∶40) according to [16]. Secondary antibodies (Alexa Fluor® 594 goat anti-rabbit IgG and Alexa Fluor® 488 goat anti-guinea pig; Molecular Probes) were used at a 1∶1000 dilution.
Protein-protein interactions
A two-hybrid screen was performed using pEG-C1/324 that encodes amino-acids 1 to 324 of Corto as bait [16]. The embryonic RFLY1 library [65] was transformed into EGY48SHΔSpe [MATα his3, trp1, ura3, LexAop(x6)-LEU2] containing the bait. In vitro transcription/translation and GST pull-down assays were performed as described [16].
For co-immunoprecipitations, 1 g of 0–14 h w1118 embryos were homogenized in RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP40, 0.1% SDS, 1 mM PMSF) with protease inhibitors (Roche Diagnostics). 500 µl of total extract (about 1 mg) were pre-cleared with protein A plus protein G agarose beads (for polyclonal antibody IP) or protein G beads (for monoclonal anti-Myc, anti-Flag or anti-HA). Input was 20 µl of this mixture. Incubation was with 10 µl of either rabbit pre-immune or Corto antiserum, mouse anti-Flag, anti-Myc or anti-HA overnight at 4°C. The appropriate beads were added and further incubated for 2 h at 4°C. The supernatant was kept; the beads were washed five times with RIPA buffer and finally resuspended in 40 µl of Laemmli buffer. 20 µl of input (4%), 20 µl of supernatant and half of the beads (20 µl) were loaded. Immunoprecipitates were detected with respective antisera developed in rat. Drosophila S2 cells were cultivated at 25°C in Schneider medium supplemented with 10% fetal calf serum and antibiotics. Cells were transfected using Effecten® transfection reagent according to the manufacturer (Qiagen). Commonly, 2×106 cells were transfected with 1 µg of each DNA. Cells were collected after 48 h of incubation and homogenized in 500 µl of RIPA buffer.
Immunoprecipitation of crosslinked chromatin (XChIP)
Chromatin from 0–14 h embryos was formaldehyde cross-linked and immunoprecipitated as described [66] using rabbit anti-Corto (1∶20), rabbit anti-CycG (1∶20), guinea-pig anti-CycG (1:20) or rabbit pre-immune sera (mock) (1∶20). Three independent immunoprecipitations were performed and further analysed. The precipitated DNA was dissolved in 100 µl of TE [10 mM Tris (pH 8.0), 1 mM EDTA] and 1 µl was used per PCR reaction. Three primer pairs spanning the iab-7 PRE (p7, p8, p9) and one primer pair from the promoter region of Abd-B (p10) were used [42], [45]. rp49 was used as negative control (primers 5′ CCC AAG ATC GTG AAG AAG CG 3′ and 5′ AGA TAC TGT CCC TTG AAG CG 3′). PCR schemes were as follows: 94°C for 3 minutes; 94°C for one minute, 45°C (p7, p9), 50°C (p8, p10, rp49) for one minute, 72°C for one minute, 36 cycles; 72°C for 10 minutes. 5 µl samples were taken every 2 cycles from the 29th to the 35th cycle to determine the linear range of amplification. PCR products were quantified using ImageJ and results of three independent experiments were normalized against the mock immunoprecipitation.
Acknowledgments
We thank Dr S. Celniker, R. Finley, T.D. Murphy, the Developmental Studies Hybridoma Bank and the Bloomington stock center for reagents, Dr J. Deutsch, J-M. Gibert, N. Randsholt and J. Szawinski for critically reading of the manuscript, and V. Ribeiro and N. Salmon for excellent technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by CNRS and UPMC and by grant 4492 from ARC to F.P. J.S. was partly supported by a scholarship from ARC.
References
- 1.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 2.Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- 3.Ingham PW. A clonal analysis of the requirement for the trithorax gene in the diversification of segments in Drosophila. J Embryol Exp Morphol. 1985;89:349–365. [PubMed] [Google Scholar]
- 4.Moehrle A, Paro R. Spreading the silence: epigenetic transcriptional regulation during Drosophila development. Dev Genet. 1994;15:478–484. doi: 10.1002/dvg.1020150606. [DOI] [PubMed] [Google Scholar]
- 5.Brock HW, Fisher CL. Maintenance of gene expression patterns. Dev Dyn. 2005;232:633–655. doi: 10.1002/dvdy.20298. [DOI] [PubMed] [Google Scholar]
- 6.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 7.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. Embo J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gildea JJ, Lopez R, Shearn A. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 11.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, et al. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 12.Poux S, Melfi R, Pirrotta V. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 2001;15:2509–2514. doi: 10.1101/gad.208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decoville M, Giacomello E, Leng M, Locker D. DSP1, an HMG-like protein, is involved in the regulation of homeotic genes. Genetics. 2001;157:237–244. doi: 10.1093/genetics/157.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dejardin J, Rappailles A, Cuvier O, Grimaud C, Decoville M, et al. Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature. 2005;434:533–538. doi: 10.1038/nature03386. [DOI] [PubMed] [Google Scholar]
- 15.Lopez A, Higuet D, Rosset R, Deutsch J, Peronnet F. corto genetically interacts with Pc-G and trx-G genes and maintains the anterior boundary of Ultrabithorax expression in Drosophila larvae. Mol Genet Genomics. 2001;266:572–583. doi: 10.1007/s004380100572. [DOI] [PubMed] [Google Scholar]
- 16.Salvaing J, Lopez A, Boivin A, Deutsch JS, Peronnet F. The Drosophila Corto protein interacts with Polycomb-group proteins and the GAGA factor. Nucleic Acids Res. 2003;31:2873–2882. doi: 10.1093/nar/gkg381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salvaing J, Decoville M, Mouchel-Vielh E, Bussiere M, Daulny A, et al. Corto and DSP1 interact and bind to a maintenance element of the Scr Hox gene: understanding the role of Enhancers of trithorax and Polycomb. BMC Biol. 2006;4:9. doi: 10.1186/1741-7007-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 19.Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 20.Maurange C, Paro R. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev. 2002;16:2672–2683. doi: 10.1101/gad.242702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 23.Tolhuis B, Muijrers I, de Wit E, Teunissen H, Talhout W, et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 24.Gatti M, Baker BS. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- 25.Rastelli L, Chan CS, Pirrotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. Embo J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, et al. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- 27.Kodjabachian L, Delaage M, Maurel C, Miassod R, Jacq B, et al. Mutations in ccf, a novel Drosophila gene encoding a chromosomal factor, affect progression through mitosis and interact with Pc-G mutations. Embo J. 1998;17:1063–1075. doi: 10.1093/emboj/17.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Dor E, Beck SA, Brock HW. Polycomb group mutants exhibit mitotic defects in syncytial cell cycles of Drosophila embryos. Dev Biol. 2006;290:312–322. doi: 10.1016/j.ydbio.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Kanaoka Y, Jinno S, Nagata A, Ogiso Y, et al. Cyclin G: a new mammalian cyclin with homology to fission yeast Cig1. Oncogene. 1993;8:2113–2118. [PubMed] [Google Scholar]
- 30.Okamoto K, Beach D. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. Embo J. 1994;13:4816–4822. doi: 10.1002/j.1460-2075.1994.tb06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reimer CL, Borras AM, Kurdistani SK, Garreau JR, Chung M, et al. Altered regulation of cyclin G in human breast cancer and its specific localization at replication foci in response to DNA damage in p53+/+ cells. J Biol Chem. 1999;274:11022–11029. doi: 10.1074/jbc.274.16.11022. [DOI] [PubMed] [Google Scholar]
- 32.Baek WK, Kim D, Jung N, Yi YW, Kim JM, et al. Increased expression of cyclin G1 in leiomyoma compared with normal myometrium. Am J Obstet Gynecol. 2003;188:634–639. doi: 10.1067/mob.2003.140. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto K, Prives C. A role of cyclin G in the process of apoptosis. Oncogene. 1999;18:4606–4615. doi: 10.1038/sj.onc.1202821. [DOI] [PubMed] [Google Scholar]
- 34.Kimura SH, Ikawa M, Ito A, Okabe M, Nojima H. Cyclin G1 is involved in G2/M arrest in response to DNA damage and in growth control after damage recovery. Oncogene. 2001;20:3290–3300. doi: 10.1038/sj.onc.1204270. [DOI] [PubMed] [Google Scholar]
- 35.Seo HR, Lee DH, Lee HJ, Baek M, Bae S, et al. Cyclin G1 overcomes radiation-induced G2 arrest and increases cell death through transcriptional activation of cyclin B1. Cell Death Differ. 2006;13:1475–1484. doi: 10.1038/sj.cdd.4401822. [DOI] [PubMed] [Google Scholar]
- 36.Bates S, Rowan S, Vousden KH. Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene. 1996;13:1103–1109. [PubMed] [Google Scholar]
- 37.Bennin DA, Don AS, Brake T, McKenzie JL, Rosenbaum H, et al. Cyclin G2 associates with protein phosphatase 2A catalytic and regulatory B' subunits in active complexes and induces nuclear aberrations and a G1/S phase cell cycle arrest. J Biol Chem. 2002;277:27449–27467. doi: 10.1074/jbc.M111693200. [DOI] [PubMed] [Google Scholar]
- 38.Salamov AA, Nishikawa T, Swindells MB. Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics. 1998;14:384–390. doi: 10.1093/bioinformatics/14.5.384. [DOI] [PubMed] [Google Scholar]
- 39.Nagel AC, Maier D, Preiss A. Green fluorescent protein as a convenient and versatile marker for studies on functional genomics in Drosophila. Dev Genes Evol. 2002;212:93–98. doi: 10.1007/s00427-002-0210-y. [DOI] [PubMed] [Google Scholar]
- 40.Celniker SE, Keelan DJ, Lewis EB. The molecular genetics of the bithorax complex of Drosophila: characterization of the products of the Abdominal-B domain. Genes Dev. 1989;3:1424–1436. doi: 10.1101/gad.3.9.1424. [DOI] [PubMed] [Google Scholar]
- 41.Delorenzi M, Bienz M. Expression of Abdominal-B homeoproteins in Drosophila embryos. Development. 1990;108:323–329. doi: 10.1242/dev.108.2.323. [DOI] [PubMed] [Google Scholar]
- 42.Breiling A, O'Neill LP, D'Eliseo D, Turner BM, Orlando V. Epigenome changes in active and inactive polycomb-group-controlled regions. EMBO Rep. 2004;5:976–982. doi: 10.1038/sj.embor.7400260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dejardin J, Cavalli G. Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module. Embo J. 2004;23:857–868. doi: 10.1038/sj.emboj.7600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin CH, Mayeda CA, Davis CA, Ericsson CL, Knafels JD, et al. Complete sequence of the bithorax complex of Drosophila. Proc Natl Acad Sci U S A. 1995;92:8398–8402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- 46.Horne MC, Goolsby GL, Donaldson KL, Tran D, Neubauer M, et al. Cyclin G1 and cyclin G2 comprise a new family of cyclins with contrasting tissue-specific and cell cycle-regulated expression. J Biol Chem. 1996;271:6050–6061. doi: 10.1074/jbc.271.11.6050. [DOI] [PubMed] [Google Scholar]
- 47.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 48.Stanyon CA, Liu G, Mangiola BA, Patel N, Giot L, et al. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Remillieux-Leschelle N, Santamaria P, Randsholt NB. Regulation of larval hematopoiesis in Drosophila melanogaster: a role for the multi sex combs gene. Genetics. 2002;162:1259–1274. doi: 10.1093/genetics/162.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narbonne K, Besse F, Brissard-Zahraoui J, Pret AM, Busson D. polyhomeotic is required for somatic cell proliferation and differentiation during ovarian follicle formation in Drosophila. Development. 2004;131:1389–1400. doi: 10.1242/dev.01003. [DOI] [PubMed] [Google Scholar]
- 51.Lessard J, Sauvageau G. Polycomb group genes as epigenetic regulators of normal and leukemic hemopoiesis. Exp Hematol. 2003;31:567–585. doi: 10.1016/s0301-472x(03)00081-x. [DOI] [PubMed] [Google Scholar]
- 52.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coisy M, Roure V, Ribot M, Philips A, Muchardt C, et al. Cyclin A repression in quiescent cells is associated with chromatin remodeling of its promoter and requires Brahma/SNF2alpha. Mol Cell. 2004;15:43–56. doi: 10.1016/j.molcel.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 54.Tonini T, Bagella L, D'Andrilli G, Claudio PP, Giordano A. Ezh2 reduces the ability of HDAC1-dependent pRb2/p130 transcriptional repression of cyclin A. Oncogene. 2004;23:4930–4937. doi: 10.1038/sj.onc.1207608. [DOI] [PubMed] [Google Scholar]
- 55.Martinez AM, Colomb S, Dejardin J, Bantignies F, Cavalli G. Polycomb group-dependent Cyclin A repression in Drosophila. Genes Dev. 2006;20:501–513. doi: 10.1101/gad.357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brumby AM, Zraly CB, Horsfield JA, Secombe J, Saint R, et al. Drosophila cyclin E interacts with components of the Brahma complex. Embo J. 2002;21:3377–3389. doi: 10.1093/emboj/cdf334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chun T, Rho SB, Byun HJ, Lee JY, Kong G. The polycomb group gene product Mel-18 interacts with cyclin D2 and modulates its activity. FEBS Lett. 2005;579:5275–5280. doi: 10.1016/j.febslet.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 58.Voncken JW, Schweizer D, Aagaard L, Sattler L, Jantsch MF, et al. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J Cell Sci. 1999;112 ( Pt 24):4627–4639. doi: 10.1242/jcs.112.24.4627. [DOI] [PubMed] [Google Scholar]
- 59.Voncken JW, Niessen H, Neufeld B, Rennefahrt U, Dahlmans V, et al. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J Biol Chem. 2005;280:5178–5187. doi: 10.1074/jbc.M407155200. [DOI] [PubMed] [Google Scholar]
- 60.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huynh CQ, Zieler H. Construction of modular and versatile plasmid vectors for the high-level expression of single or multiple genes in insects and insect cell lines. J Mol Biol. 1999;288:13–20. doi: 10.1006/jmbi.1999.2674. [DOI] [PubMed] [Google Scholar]
- 62.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 63.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 64.LaJeunesse D, Shearn A. E(z): a polycomb group gene or a trithorax group gene? Development. 1996;122:2189–2197. doi: 10.1242/dev.122.7.2189. [DOI] [PubMed] [Google Scholar]
- 65.Finley RL, Jr., Thomas BJ, Zipursky SL, Brent R. Isolation of Drosophila cyclin D, a protein expressed in the morphogenetic furrow before entry into S phase. Proc Natl Acad Sci U S A. 1996;93:3011–3015. doi: 10.1073/pnas.93.7.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cavalli G, Orlando V, Paro R. Mapping DNA target sites of chromatin-associated proteins by formaldehyde cross-linking in Drosophila embryos. In: Bickmore WA, editor. Chromosome Structural Analysis: A Practical Approach: UK: Oxford University Press; 1999. pp. 20–37. [Google Scholar]