The solution structure of A. thaliana protein At5g39720.1 reported here is the first for a member of the AIG2-like family (PF06094). The three-dimensional structure shows similarity to those determined for members of the uncharacterized Pfam family UPF0131.
Keywords: AIG2, NMR, structural genomics
Abstract
The three-dimensional structure of Arabidopsis thaliana protein At5g39720.1 was determined by NMR spectroscopy. It is the first representative structure of Pfam family PF06094, which contains protein sequences similar to that of AIG2, an A. thaliana protein of unknown function induced upon infection by the bacterial pathogen Pseudomonas syringae. The At5g39720.1 structure consists of a five-stranded β-barrel surrounded by two α-helices and a small β-sheet. A long flexible α-helix protrudes from the structure at the C-terminal end. A structural homology search revealed similarity to three members of Pfam family UPF0131. Conservation of residues in a hydrophilic cavity able to bind small ligands in UPF0131 proteins suggests that this may also serve as an active site in AIG2-like proteins.
1. Introduction
Arabidopsis thaliana protein At5g39720.1 was selected as a target by the Center for Eukaryotic Structural Genomics (CESG) because it had less than 30% sequence identity to any protein of known three-dimensional structure. It is a member of the AIG2-like protein family (PF06094), which thus far comprises seven A. thaliana homologs with sequence identities ranging from 47 to 75%, as well as proteins from Oryza sativa, fungi, bacteria and archaebacteria. The AIG2 (avrRpt2-induced gene) gene product is a functionally uncharacterized protein that is induced in A. thaliana as part of its resistance response upon infection by the bacterial pathogen Pseudomonas syringae (Reuber & Ausubel, 1996 ▶). Plant pathogens such as P. syringae secrete a diverse group of effector proteins into the host plant cell that are indirectly detected by corresponding resistance (R) proteins, triggering a defense response. This response includes rapid ion fluxes, generation of reactive oxygen species (ROS) and production of antimicrobial compounds and is often accompanied by localized programmed cell death, the hypersensitive response (HR), at the site of pathogen invasion (Hammond-Kosack et al., 1996 ▶). Numerous R genes and their corresponding effectors have been cloned and characterized in an effort to understand the complex surveillance mechanisms that control the so-called ‘gene-for-gene’ resistance response in Arabidopsis. The Arabidopsis AIG1 and AIG2 genes were identified and cloned in a study designed to isolate defense-related genes specifically induced by single effector genes and their corresponding R genes. These two genes were shown to be induced as part of the signal transduction pathway initiated by P. syringae effector protein avrRpt2 and the corresponding R protein, RPS2 (Reuber & Ausubel, 1996 ▶). It was suggested that AIG2 is a member of a small gene family because the clone hybridized to multiple bands of the Arabidopsis Col-0 DNA digest. Thus, At5g39720.1 is likely to represent one of these AIG2 homologs. Although much progress has been made in the last decade in understanding the mechanisms underlying effector-triggered immune responses in plants, as recently reviewed by Chisholm et al. (2006 ▶) and Mudgett (2005 ▶), the function of AIG2 is unknown. Interestingly, an AIG2-like domain is also contained in the two-domain tellurite-resistance protein from Rhodobacteracea. The three-dimensional structure of the At5g39720.1 protein reported here represents the first structure of an AIG2-like family member. We also show that it is structurally similar to members of the larger uncharacterized protein family UPF0131.
2. Materials and methods
2.1. Protein expression and purification
Samples of the gene product of A. thaliana gene At5g39720.1 were prepared in (U-13C,15N)-labeled form according to CESG wheat germ cell-free protocols as described previously (Vinarov et al., 2004 ▶). Briefly, the protein was expressed with an N-terminal His6 fusion tag in wheat germ extract supplemented with (U-13C,15N) amino acids (Cambridge Isotope Labs) and purified by His-Trap HP chelating affinity chromatography followed by size-exclusion chromatography. The sample used for NMR structure determination contained approximately 0.9 mM protein, 10 mM deuterated bis-tris pH 7.0, 100 mM sodium chloride and 5 mM dithiothreitol.
2.2. NMR spectroscopy
All NMR data were acquired at 298 K on a Bruker 600 MHz spectrometer equipped with a triple-resonance CryoProbe and processed with NMRPipe software (Delaglio et al., 1995 ▶). The total acquisition time for all NMR spectra was ∼260 h and no protein degradation or unfolding was detected in comparisons of one-dimensional and two-dimensional spectra collected at the start and end of data collection. Heteronuclear 15N–1H NOE values were determined from an interleaved pair of two-dimensional gradient sensitivity-enhanced correlation spectra of (U-13C,15N) At5g39720.1 acquired with and without a 5 s proton saturation period (Farrow et al., 1994 ▶) and analyzed using CARA, freely available at http://www.nmr.ch/ (Keller, 2004 ▶). The translational self-diffusion coefficient of At5g39720.1 was measured as described previously (Veldkamp et al., 2005 ▶). Over 90% of the backbone 1H, 15N and 13C resonance assignments were obtained in an automated manner using the program Garant (Bartels et al., 1996 ▶), with peaklists from three-dimensional HNCO, HNCACO, HNCA, HNCOCA, HNCACB, HCCONH and CCONH spectra that were generated automatically with SPSCAN. Side-chain assignments were completed manually from three-dimensional HBHACONH, HCCONH, HCCH-TOCSY and 13C(aromatic)-edited NOESY-HSQC spectra analyzed using XEASY (Bartels et al., 1995 ▶).
2.3. Structure determination
A total of 1989 unique non-trivial NOE distance constraints were obtained from three-dimensional 15N-edited NOESY-HSQC, 13C(aromatic)-edited NOESY-HSQC and 13C-edited NOESY-HSQC spectra (τmix = 80 ms). A total of 189 backbone ϕ and ψ dihedral angle constraints were generated from secondary shifts of the 1Hα, 13Cα, 13Cβ, 13C′ and 15N nuclei shifts using the program TALOS (Cornilescu et al., 1999 ▶). Structures were generated in an automated manner using the NOEASSIGN module of the torsion-angle dynamics program CYANA 2.1 (Herrmann et al., 2002 ▶), which produced an ensemble with high precision and low residual constraint violations that required minimal manual refinement. The 20 CYANA conformers with the lowest target function were subjected to a molecular-dynamics protocol in explicit solvent (Linge et al., 2003 ▶) using XPLOR-NIH (Schwieters et al., 2003 ▶).
3. Results and discussion
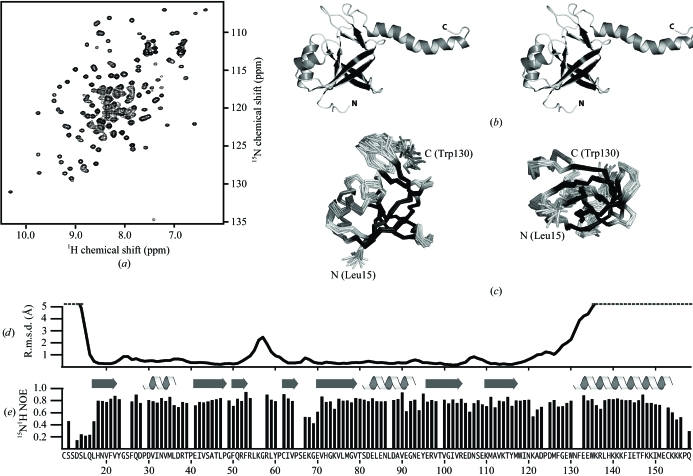
The At5g39720.1 construct consists of 173 residues, including nine residues from the His tag, with molecular weight of 21.2 kDa. The two-dimensional 15N–1H HSQC spectrum (Fig. 1 ▶ a) displayed a pattern of well dispersed peaks with uniform intensity, consistent with a stable folded domain. The structure (Fig. 1 ▶ b) was solved by an automatic iterative NOE refinement method (Herrmann et al., 2002 ▶). Chemical shift assignments and NMR data were deposited in the BioMagResBank (entry 7007). The final ensemble of 20 conformers (PDB code 2g0q) is shown in Fig. 1 ▶(c) and a summary of the experimental restraints and structural statistics is provided in Table 1 ▶. The structure revealed a five-stranded antiparallel β-barrel (β1-β5-β2-β6-β7) flanked by two α-helices and a β-sheet (β3-β4). The two β-sheets interact via a hydrogen bond between the Trp118 amide and the Ile63 carbonyl. A highly positively charged flexible 26-residue α-helix protrudes from the molecule at the C-terminal end. Consistent patterns of 13C and 1H secondary shifts and d NN(i, i + 1), d αN(i, i + 3) and d αβ(i, i + 3) NOEs provide dihedral and distance constraints that clearly define the helical conformation of residues 132–154. However, high backbone atomic r.m.s.d. values (Fig. 1 ▶ d) illustrate that these residues are unconstrained relative to the rest of the protein, because long-range NOEs between protons on the helix and protons in the rest of the protein were not observed in any of the NMR spectra.
Figure 1.
NMR data and structure of At5g39720.1. (a) Two-dimensional 1H–15N HSQC spectrum for (U-13C,15N) At5g39720.1 acquired at 298 K and 600 MHz. (b) Ribbon diagram of a representative conformer in stereoview. Residues of the N-terminal tag (1–9) and the disordered C-terminus (159–173) are omitted for clarity and were not included in the coordinates deposited in the PDB. (c) Ensemble of 20 conformers shown as a Cα trace in the same orientation as in (b) (left) and rotated 90° about the horizontal axis (right). (d) Backbone (N, Cα, C′) r.m.s.d. values and (e) 1H–15N heteronuclear NOEs plotted for each residue of At5g39720.1.
Table 1. Structural statistics for the 20 At5g397201 conformers.
| Non-redundant distance constraints | |
| Long | 636 |
| Medium [1 < (i − j) ≤ 5] | 267 |
| Sequential [(i − j) = 1] | 564 |
| Intraresidue (i = j) | 522 |
| Total | 1989 |
| Dihedral angle constraints (ϕ and ψ) | 189 |
| Average atomic r.m.s.d. to the mean structure (Å) | |
| Residues 17–53, 61–121 | |
| Backbone (N, Cα, C′) | 0.47 ± 0.09 |
| Heavy atoms | 0.92 ± 0.08 |
| Deviations from idealized covalent geometry† | |
| Bond length r.m.s.d. (Å) | 0.015 |
| Torsion angle r.m.s.d. (°) | 1.2 |
| WHATCHECK quality indicators | |
| Z score | −2.16 ± 0.17 |
| R.m.s. Z score | |
| Bond lengths | 0.77 ± 0.02 |
| Bond angles | 0.76 ± 0.02 |
| Bumps | 0 ± 0 |
| Lennard–Jones energy‡ (kJ mol−1) | −3608 ± 125 |
| Constraint violations | |
| NOE distance violations > 0.5 ŧ | 0 ± 0 |
| NOE distance r.m.s.d. (Å) | 0.021 ± 0.001 |
| Torsion-angle violations > 5°¶ | 0.2 ± 0.62 |
| Torsion-angle r.m.s.d. (°) | 0.766 ± 0.131 |
| Ramachandran statistics (% of residues 10–158) | |
| Most favored | 87.7 ± 1.5 |
| Additionally allowed | 9.8 ± 1.5 |
| Generously allowed | 1.9 ± 0.6 |
| Disallowed | 0.6 ± 0.7 |
Final X-PLOR force constants were 250 (bonds), 250 (angles), 300 (impropers), 100 (chirality), 100 (ω), 50 (NOE constraints) and 200 (torsion-angle constraints).
Nonbonded energy was calculated using XPLOR-NIH.
The largest NOE violation in the ensemble of structures was 0.35 Å.
The largest torsion-angle violation in the ensemble of structures was 6.2°.
Heteronuclear NOE values (Fig. 1 ▶ e) reflect the relative mobility of each 1H–15N bond vector in the polypeptide chain. NOE values of ∼0.8 throughout most of the domain indicate a highly ordered backbone with a few flexible loops (e.g. β4–β5). Values below 0.4 for the C-terminal 15 residues are diagnostic of dynamic disorder on the picosecond–nanosecond timescale and these residues, along with the N-terminal affinity tag, were excluded from the models deposited in the Protein Data Bank. Interestingly, the untethered C-terminal helix (residues 132–154) displays 15N–1H NOE values similar to those of the folded domain, suggesting that the helix may be mobile on slower (microsecond–millisecond) timescales, but probably as a relatively rigid structural unit. With NOE values of 0.5–0.7, residues 121–123 appear to be more flexible and may serve as a hinge allowing the helix to sample multiple orientations.
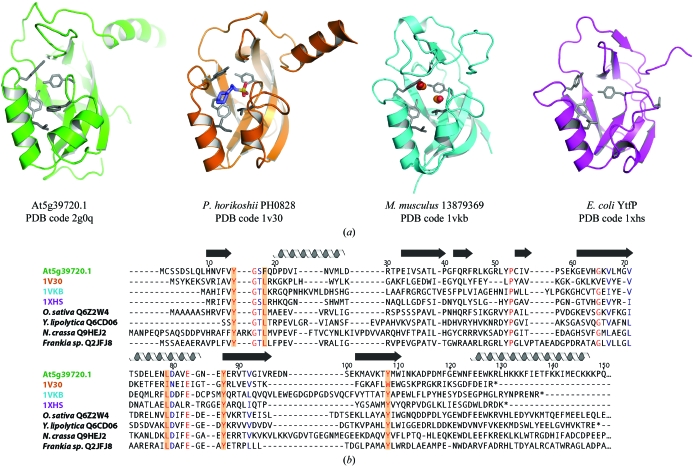
Searches using VAST (Gibbs et al., 1998 ▶) and FATCAT (Ye & Godzik, 2003 ▶) revealed significant structural similarity to three members of the uncharacterized Pfam family UPF0131 (Fig. 2 ▶ a). The hypothetical protein YtfP from E. coli (PDB code 1xhs) has an r.m.s.d. value of 2.31 Å over 105 aligned Cα atoms and a sequence identity of 21%, hypothetical protein PH0828 from Pyrococcus horikoshii (PDB code 1v30; Tajika et al., 2004 ▶) has an r.m.s.d. value of 2.58 Å over 115 aligned Cα atoms and a sequence identity of 17% and mouse protein 13879369 (PDB code 1vkb; Klock et al., 2005 ▶) has an r.m.s.d. value of 2.81 Å over 118 aligned Cα atoms and a sequence identity of 15%. The structures of PH0828 and 13879369 are more similar to that of At5g39720.1 in that they both have the same β-barrel topology, whereas in YtfP the β-barrel is open. Of the three structures, PH0828 is the only one that, like At5g39720.1, contains a C-terminal α-helix. However, the helix is only five residues long, compared with 26 residues in At5g39720.1. Another difference in the At5g39720.1 structure is that the first α-helix is rotated 45° compared with the same helix in the other three structures.
Figure 2.
At5g39720.1 and its structural homologs. (a) Ribbon diagrams of At5g39720.1, E. coli YtfP (PDB code 1xhs), Pyrococcus horikoshii PH0828 (PDB code 1v30) and murine 13879369 (PDB code 1vkb). (b) Multiple sequence alignment of the At5g39720.1 homologs shown in (a) and representative members of the AIG2-like family (identified by species and SWISS-PROT ID) from a plant (Oryza sativa Q6Z2W4), fungi (Yarrowia lipolytica Q6CD06 and Neuropora crassa Q9HEJ2) and a bacterium (Frankia sp. CcI3 Q2JFJ8). The residue numbering corresponds to the actual At5g39720.1 sequence without N-terminal His tag and thus differs by eight from the PDB numbering shown in Fig. 1 ▶(a). Residues identical in at least seven of eight sequences are shaded red and highly conserved residues are colored blue. Conserved residues lining the cavity are highlighted in orange.
The crystal structures of PH0828 and 13879369 each revealed ligands (CHES and two formate molecules, respectively) bound in a hydrophilic cavity surrounded by the β-barrel and adjacent helices. The cavity is lined by strictly conserved or conservatively substituted residues that are also conserved in At5g39720.1, as indicated in the sequence alignment shown in Fig. 2 ▶(b). It was proposed that this cavity serves as an active site for the protein (Klock et al., 2005 ▶; Tajika et al., 2004 ▶). Further comparison of the UPF0131 family sequences to At5g39720.1 and other AIG2-like proteins revealed the substitution of a strictly conserved solvent-accessible glutamate residue by lysine (Lys74) in all of the AIG2-like plant sequences, indicating a possible functional role for this residue. In addition, the C-terminal α-helix, which contains a number of conserved residues, may serve as a protein-binding site. It is noteworthy that an AIG2-like domain is one of two domains comprising the tellurite-resistance proteins from Rhodobacteraceae. The other domain is a NUDIX domain, named as such because substrates of this enzyme family are all nucleoside diphosphates linked to some other moiety X (Bessman et al., 1996 ▶). The tellurite-resistance protein trgB from Rhodobacter sphaeroides is a predicted ADP-ribose pyrophosphatase that increases resistance to tellurite, a toxic oxyanion (Dunn et al., 1999 ▶). Although it is unknown whether the AIG2-like domain is necessary for trgB function, its presence suggests a possible role for AIG2-like domains in recognition of signaling molecules common to both tellurite- and RPS2-induced resistance pathways. The structure of At5g39720.1 is a first step toward understanding the functional role of plant AIG2-like proteins in host defense.
Supplementary Material
PDB reference: At5g39720.1, 2g0q
Acknowledgments
This work was supported by the NIGMS Protein Structure Initiative (GM074901, John L. Markley, PI).
References
- Bartels, C., Billeter, M., Güntert, P. & Wüthrich, K. (1996). J. Biomol. NMR, 7, 207–213. [DOI] [PubMed]
- Bartels, C., Xia, T.-H., Billeter, M., Güntert, P. & Wüthrich, K. (1995). J. Biomol. NMR, 5, 1–10. [DOI] [PubMed] [Google Scholar]
- Bessman, M. J., Frick, D. N. & O’Handley, S. F. (1996). J. Biol. Chem.271, 25059–25062. [DOI] [PubMed] [Google Scholar]
- Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. (2006). Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Cornilescu, G., Delaglio, F. & Bax, A. (1999). J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995). J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Dunn, C. A., O’Handley, S. F., Frick, D. N. & Bessman, M. J. (1999). J. Biol. Chem.274, 32318–32324. [DOI] [PubMed] [Google Scholar]
- Farrow, N. A., Muhandiram, R., Singer, A. U., Pascal, S. M., Kay, C. M., Gish, G., Shoelson, S. E., Pawson, T., Forman-Kay, J. D. & Kay, L. E. (1994). Biochemistry, 33, 5984–6003. [DOI] [PubMed] [Google Scholar]
- Gibbs, A. C., Kondejewski, L. H., Gronwald, W., Nip, A. M., Hodges, R. S., Sykes, B. D. & Wishart, D. S. (1998). Nature Struct. Biol.5, 284–288. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K. E., Silverman, P., Raskin, I. & Jones, J. (1996). Plant Physiol.110, 1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, T., Güntert, P. & Wüthrich, K. (2002). J. Mol. Biol.319, 209–227. [DOI] [PubMed] [Google Scholar]
- Keller, R. (2004). The Computer-Aided Resonance Assignment Tutorial. Goldau: Cantina Verlag.
- Klock, H. E. et al. (2005). Proteins, 61, 1132–1136. [DOI] [PubMed] [Google Scholar]
- Linge, J. P., Williams, M. A., Spronk, C. A., Bonvin, A. M. & Nilges, M. (2003). Proteins, 50, 496–506. [DOI] [PubMed] [Google Scholar]
- Mudgett, M. B. (2005). Annu. Rev. Plant Biol.56, 509–531. [DOI] [PubMed] [Google Scholar]
- Reuber, T. L. & Ausubel, F. M. (1996). Plant Cell, 8, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Clore, G. M. (2003). J. Magn. Reson.160, 65–73. [DOI] [PubMed] [Google Scholar]
- Tajika, Y., Sakai, N., Tanaka, Y., Yao, M., Watanabe, N. & Tanaka, I. (2004). Proteins, 55, 210–213. [DOI] [PubMed] [Google Scholar]
- Veldkamp, C. T., Peterson, F. C., Pelzek, A. J. & Volkman, B. F. (2005). Protein Sci.14, 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinarov, D. A., Lytle, B. L., Peterson, F. C., Tyler, E. M., Volkman, B. F. & Markley, J. L. (2004). Nature Methods, 1, 149–153. [DOI] [PubMed] [Google Scholar]
- Ye, Y. & Godzik, A. (2003). Bioinformatics, 19, ii246–ii255. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: At5g39720.1, 2g0q