The N-terminal, cytoplasmic domain of the Na+-coupled HCO3 cotransporter NBCe1-A crystallizes in the trigonal space group P3121 or P31 with pseudo P3121 symmetry and diffracts X-rays to 3.0 Å resolution. The crystal packing demonstrates a domain-swap mechanism for dimerization.
Keywords: SLC4 family, Na+-coupled HCO3− transport, crystal annealing, Laue symmetry, crystal packing
Abstract
The N-terminal cytoplasmic domain of the Na+-coupled HCO cotransporter NBCe1-A (NtNBCe1) has been linked with proximal renal tubular acidosis. In a previous purification study of recombinant NtNBCe1, crystal growth at a suboptimal protein concentration (<1 mg ml−1) yielded small single diamond-shaped crystals that diffracted poorly. In the present study, by increasing the protein concentration 50-fold, the crystal size was doubled and robustness was also improved. Crystal annealing made the crystals suitable for X-ray diffraction. The crystals either belong to space group P3121 or P31 with pseudo P3121 symmetry, with unit-cell parameters a = 51.7, b = 51.7, c = 200.6 Å, α = β = 90, γ = 120°, and diffract X-rays to 3.0 Å resolution. The calculated Matthews number is 1.9 Å3 Da−1, with two monomers of molecular weight ∼83 kDa in the asymmetric unit. The molecular- replacement packing solution shows that the molecules form dimers by a domain-swapping mechanism.
cotransporter NBCe1-A (NtNBCe1) has been linked with proximal renal tubular acidosis. In a previous purification study of recombinant NtNBCe1, crystal growth at a suboptimal protein concentration (<1 mg ml−1) yielded small single diamond-shaped crystals that diffracted poorly. In the present study, by increasing the protein concentration 50-fold, the crystal size was doubled and robustness was also improved. Crystal annealing made the crystals suitable for X-ray diffraction. The crystals either belong to space group P3121 or P31 with pseudo P3121 symmetry, with unit-cell parameters a = 51.7, b = 51.7, c = 200.6 Å, α = β = 90, γ = 120°, and diffract X-rays to 3.0 Å resolution. The calculated Matthews number is 1.9 Å3 Da−1, with two monomers of molecular weight ∼83 kDa in the asymmetric unit. The molecular- replacement packing solution shows that the molecules form dimers by a domain-swapping mechanism.
1. Introduction
The five known Na+-coupled HCO transporters are integral membrane proteins that play a vital role in transporting Na+ and HCO
transporters are integral membrane proteins that play a vital role in transporting Na+ and HCO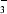 across plasma membranes in tissues throughout the body (Boron, 2004 ▶; Boron & Boulpaep, 1983 ▶; Romero et al., 1997 ▶). They represent at least half of the SLC4 family of HCO
across plasma membranes in tissues throughout the body (Boron, 2004 ▶; Boron & Boulpaep, 1983 ▶; Romero et al., 1997 ▶). They represent at least half of the SLC4 family of HCO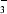 transporters. In humans, mutations in the electrogenic Na/HCO3 cotransporter NBCe1 (SLC4A4) can cause autosomal recessive disorders that may include severe proximal renal tubular acidosis (pRTA), glaucoma and mental retardation (Igarashi et al., 2002 ▶; Laing et al., 2005 ▶). NBCe1 has three known splice variants. In the present study, we focus on the renal splice variant NBCe1-A, which is localized to the basolateral membrane of the proximal tubule. Renal NBCe1-A is central to the reabsorption of HCO
transporters. In humans, mutations in the electrogenic Na/HCO3 cotransporter NBCe1 (SLC4A4) can cause autosomal recessive disorders that may include severe proximal renal tubular acidosis (pRTA), glaucoma and mental retardation (Igarashi et al., 2002 ▶; Laing et al., 2005 ▶). NBCe1 has three known splice variants. In the present study, we focus on the renal splice variant NBCe1-A, which is localized to the basolateral membrane of the proximal tubule. Renal NBCe1-A is central to the reabsorption of HCO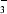 from lumen to blood and thus to the control of blood pH.
from lumen to blood and thus to the control of blood pH.
The Cl–HCO3 exchanger AE1 (SLC4A1) is 37% identical at the amino-acid level to NBCe1. The large cytoplasmic N-terminal domain of AE1 is important for the binding of ankyrin and other cytoskeletal proteins, as well as several glycolytic enzymes (for a review, see Alper, 2006 ▶). Zhang and coworkers solved the crystal structure to 2.6 Å resolution of most of the N-terminus of AE1 (NtAE1; Zhang et al., 2000 ▶). They found that two NtAE1 monomers form a dimer by domain swapping, each contributing an arm made up of three α-helices and one β-strand.
The cytoplasmic N-terminal domain of NBCe1 (NtNBCe1) appears to be essential for the transmembrane domain (TMD) to function as a transporter and additionally modulates the rate of HCO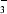 transport (Liu et al., 2005 ▶). In fact, a missense mutation mapped to the N-terminus of NBCe1 is associated with pRTA (Igarashi et al., 2002 ▶). NtNBCe1 is the first N-terminus of an Na+-coupled HCO
transport (Liu et al., 2005 ▶). In fact, a missense mutation mapped to the N-terminus of NBCe1 is associated with pRTA (Igarashi et al., 2002 ▶). NtNBCe1 is the first N-terminus of an Na+-coupled HCO transporter to be crystallized (Gill & Boron, 2006 ▶) and only the second N-terminus of the entire SLC4 family. Here, we report (i) the crystallization conditions that yield crystals of sufficient size and robustness for X-ray diffraction studies, (ii) the annealing techniques that allow the crystals to diffract X-rays, (iii) the crystal properties of NtNBCe1 and (iv) the molecular-replacement crystal-packing solution.
transporter to be crystallized (Gill & Boron, 2006 ▶) and only the second N-terminus of the entire SLC4 family. Here, we report (i) the crystallization conditions that yield crystals of sufficient size and robustness for X-ray diffraction studies, (ii) the annealing techniques that allow the crystals to diffract X-rays, (iii) the crystal properties of NtNBCe1 and (iv) the molecular-replacement crystal-packing solution.
2. Materials and methods
2.1. Engineering, expression and purification of NtNBCe1
We engineered an expression construct containing residues 1–365 of NBCe1-A based on protein-sequence analysis and the known proteolytic sensitivity of the related AE1 protein. Hydropathy plots were performed using the program TMPRED (Hofmann & Stoffel, 1993 ▶) to identify the start of a long hydrophobic rich region, which we assume to be the first membrane-spanning segment of the TMD. Secondary-structure plots were predicted using the program PHD (Przybylski & Rost, 2002 ▶).
NtNBCe1 was then recombinantly expressed in Escherichia coli and purified using the method described elsewhere (Gill & Boron, 2006 ▶), except that the optional DEAE-filtration step was omitted. Briefly, the corresponding DNA fragment of NBCe1-A, preceded by six repeats of the CAT nucleotide sequence that encodes a hexahistidine motif, was PCR-amplified and inserted at the NcoI and XhoI sites of the E. coli expression vector pET15B (Novagen, San Diego, CA, USA). The construct was transformed into the Rossetta2 cell line (Novagen, San Diego, CA, USA) and grown in Luria Broth at 310 K to an OD600 of 1, at which point 0.5 mM IPTG was added and the temperature lowered to 288 K. The cells were harvested after 18 h, lysed by sonication and spun down at 10 000g for 20 min. A 1% streptomycin sulfate cut was then performed on the supernatant before applying it to Ni2+ beads. After the Ni2+ beads had been washed with 20 mM imidazole, the supernatant was eluted with 200 mM imidazole, concentrated and applied onto a size-exclusion column. The buffer of the protein solution contained 20 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM TCEP and 0.02% NaN3 (Gill & Boron, 2006 ▶).
SDS–PAGE was performed using the method of Laemmli (1970 ▶), staining the gels with Coomassie Brilliant Blue R-250. Purity was checked by overloading 40 µg NtNBCe1 on SDS–polyacrylamide gel and visual inspection of contaminating proteins after staining. Protein concentration was determined spectrophotometrically by absorbance at 280 nm using a calculated extinction coefficient for NtNBCe1 of 3 × 104 M −1 cm−1, and was cross-checked by comparing against BSA standards on SDS–PAGE stained with Coomassie dye.
2.2. Crystallizing NtNBCe1
Crystals were grown by the hanging-drop vapor-diffusion method (McPherson et al., 1995 ▶). Crystal conditions were screened using (i) the commercially available kits Crystal Screens I and II (Hampton Research, Irvine, CA, USA) and (ii) solutions comprising ammonium sulfate as a precipitant, buffered with sodium citrate around the calculated isoelectric point of NtNBCe1 (pI 6.4). Each drop had a total volume of 4 µl and consisted of equal parts of well solution and a 47 mg ml−1 NtNBCe1 stock solution. The volume of the well was 0.5 ml. The well solution that yielded the best quality crystals consisted of a 38%(v/v) saturated ammonium sulfate solution in 100 mM sodium citrate pH 6.5.
2.3. X-ray data collection and processing
The crystals were flash-frozen using the well solution as a cryoprotectant, which was remade by replacing some water with glycerol to create a 20%(v/v) solution. Data were collected on the X29A beamline using a Quantum 3×3 detector (Area Systems Detector Corporation, San Diego, CA, USA) at the National Synchrotron Light Source (NSLS), Brookhaven, Long Island, NY, USA. At the synchrotron, the crystal was oscillated through 120° with an oscillation angle of 1.0° and each frame was exposed for 3 s. The crystal-to-detector distance was set to 400 mm. Data were processed with the program HKL2000 (Otwinowski et al., 2003 ▶).
3. Results and discussion
3.1. Agreement of the N-terminus–TMD boundary with SLC4 family member AE1
The programs TMPRED and PHD independently predicted the same beginning for the first TMD helix of NBCe1 (i.e. residue ∼422). Because PHD predicted a large coiled or non-structured region (∼58 residues) just before the N-terminus–TMD boundary, we defined NtNBCe1 as residues 1–365 of full-length NBCe1-A. These 365 amino acids of NBCe1-A are 37% identical to the corresponding region of AE1. The predicted non-structured region of NBCe1 is homologous to a region in AE1 that is susceptible to proteolytic cleavage (Kiyatkin et al., 1995 ▶; Low, 1986 ▶). Indeed, treating inside-out erythrocyte membranes with proteases cleaves AE1 just after an amino acid that corresponds to residue ∼409 in NBCe1, releasing nearly the entire cytoplasmic N-terminal domain of AE1 and leaving behind 13 amino acids plus the entire TMD of AE1.
3.2. Improved and new crystallization of NtNBCe1
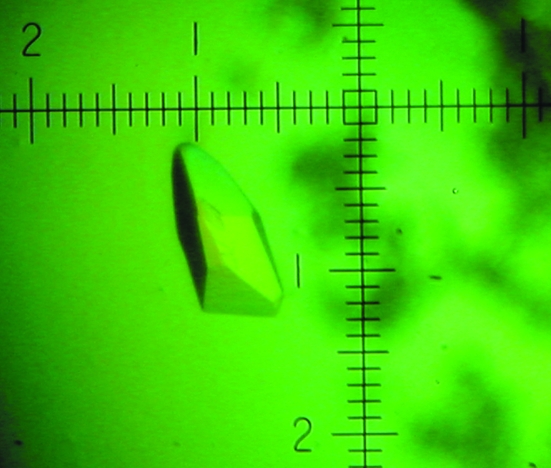
In our earlier study, the best crystals of NtNBCe1, grown at <1 mg ml−1 protein concentration, were small single and diamond-shaped and diffracted to only 10 Å (Gill & Boron, 2006 ▶). In the present study, we screened for crystallization conditions at protein concentrations of ∼50 mg ml−1, obtaining crystals that were twice as large and more robust than before (see §2). Fig. 1 ▶ shows a crystal (condition 1) that grew over a period of three weeks at room temperature and is that used for data collection here. Another condition, which yielded a 1.0 × 0.3 × 0.3 mm crystal (condition 2) after ∼4 months at room temperature, was an equal mixture of NtNBCe1 and 1.4 M trisodium citrate dihydrate in 0.1 M HEPES pH 7.5. Because of its size and fragility, this single crystal could only be mounted with difficulty and diffracted X-rays poorly.
Figure 1.
Crystal of NtNBCe1. The crystal is shown colored by a polarization filter. Every ten small divisions of the scale is equivalent to 0.25 mm. The NtNBCe1 crystal forms in a rock shape with dimensions of 0.125 × 0.125 × 0.25 mm.
3.3. Crystal annealing
Although the condition 1 NtNBCe1 crystals grew to sufficient size for mounting, they initially diffracted X-rays poorly or not at all. We attributed this problem to a slightly exacerbated mosaicity of the crystal lattice and overcame the problem by thawing the flash-frozen crystal in mother liquor at room temperature for ∼3 min and then flash-freezing it again. We repeated the process two more times, after which the crystals diffracted X-rays reasonably well.
3.4. Space-group determination and molecular-replacement solution
The crystals diffract X-rays to 3.0 Å resolution. The unit-cell parameters and data-reduction statistics are summarized in Table 1 ▶. We initially processed the data in space group P3, but inspection of systematic absences indicates the presence of a threefold screw axis, suggesting a P31 or P32 space group. Calculation of the Matthews coefficient (Kantardjieff & Rupp, 2003 ▶; Matthews, 1968 ▶) results in 3.7 Å3 Da−1 for a monomer in the asymmetric unit and 1.9 Å3 Da−1 for a dimer in the asymmetric unit. The latter corresponds more closely to the usual range of 1.6–3.6 Å3 Da−1 (Kantardjieff & Rupp, 2003 ▶; Matthews, 1968 ▶) and the presence of a dimer would be consistent with our previous molecular-weight calculation of 81 kDa by gel-filtration chromatography (Gill & Boron, 2006 ▶).
Table 1. NtNBCe1 crystal parameters.
Values in parentheses are for the highest resolution shell.
| Radiation wavelength (Å) | 1.1 | |
| Resolution range (Å) | 50.0–2.94 | |
| Space group | P31 | P3121 |
| Unit-cell parameters | ||
| a (Å) | 51.7 | 51.7 |
| b (Å) | 51.7 | 51.7 |
| c (Å) | 200.6 | 200.6 |
| β (°) | 120 | 120 |
| Mosaicity | 1.1 | 1.1 |
| Completeness (%) | 91.4 (86.0) | 95.7 (93.4) |
| Observed reflections | 28904 | 28617 |
| Unique reflections | 11833 | 6985 |
| Redundancy | 2.4 (2.0) | 4.1 (3.3) |
| Rsym† (%) | 7.5 (85.0) | 8.2 (98.2) |
| Overall I/σ(I) | 26.1 (2.2) | 34.0 (2.6) |
R
sym = 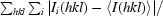
 , where Ii(hkl) is the ith measurement and 〈I(hkl)〉 is the mean of all measurements of I for Miller indices hkl.
, where Ii(hkl) is the ith measurement and 〈I(hkl)〉 is the mean of all measurements of I for Miller indices hkl.
To confirm the presence of non-crystallographic symmetry, we calculated a self-rotation function in the κ = 180° section. The self-rotation showed three dyads in the bc plane spaced approximately every 60° along ψ starting at (30, 0°); that is, at (ψ, ϕ) = (30, 0), (90, 0), (150, 0°). This confirms the presence of a dimer in our trigonal crystals; however, the location of the dyads also corresponds to that expected for P321 space groups. Rescaling and merging in this space group gave similar R sym values (Table 1 ▶).
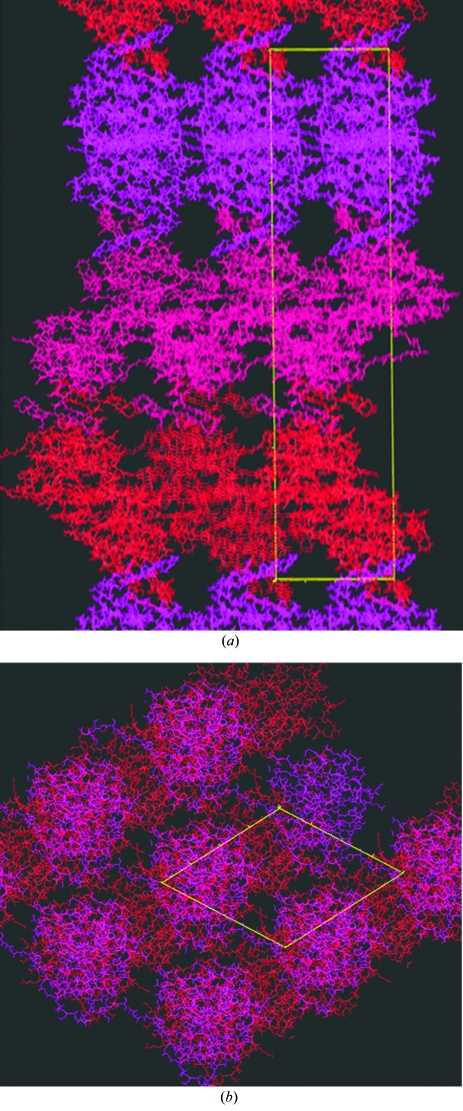
To address the remaining space-group ambiguities, we carried out a molecular-replacement search with Phaser (Collaborative Computational Project, Number 4, 1994 ▶; McCoy et al., 2005 ▶) using the N-terminus of an AE1 monomer (Zhang et al., 2000 ▶) as a molecular probe. We obtain the highest peak (Z = 12 with the second highest at Z = 4) in space group P31. For space group P32 the highest peak is only Z = 3.5. Interestingly, the molecular-packing solution (Fig. 2 ▶) demonstrates that the NtNBCe1 molecules indeed have two monomers related by a twofold axis. The dimer architecture is very similar to the domain-swapped dimers observed for the NtAE1 crystals (Zhang et al., 2000 ▶), suggesting that NtNBCe1 forms a similar domain-swapped dimer. Further refinement will resolve those structural details and reveal whether our crystals have space group P3121 or P31 with pseudo-P3121 symmetry.
Figure 2.
The crystal packing of NtNBCe1 molecules. The figure shows the crystal packing in the P31 space group. Each asymmetric unit contains two monomers, which are identically colored violet, red or pink. (a) The bc plane is shown. The c axis is vertical and the b axis is horizontal. (b) The ab plane is shown and rotated 45°. The molecular-replacement packing solution was found using the program Phaser (Collaborative Computational Project, Number 4, 1994 ▶; McCoy et al., 2005 ▶). The starting R cryst was 54%.
Acknowledgments
We thank Lauren Dutcher for help with crystal growth, Dr Michael Sawaya for insightful discussion and Dr Howard Robinson at the X29A beamline for rapid access. HG was supported by NIH Postdoctoral Training Grant 5T32 DK007259. This work was supported by NIH Grants R37 DK30344 and R01 NS18400. Data for this study were measured at the NSLS, which is supported by the Offices of Biological and Environmental Research and Basic Energy Sciences of the US-DOE and the National Center for Research Resources of the NIH.
References
- Alper, S. L. (2006). Exp. Physiol.91, 153–161. [DOI] [PubMed] [Google Scholar]
- Boron, W. F. (2004). Adv. Physiol. Educ.28, 160–179. [DOI] [PubMed] [Google Scholar]
- Boron, W. F. & Boulpaep, E. L. (1983). J. Gen. Physiol.81, 53–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Gill, H. & Boron, W. (2006). In the press.
- Hofmann, K. & Stoffel, W. (1993). Biol. Chem. Hoppe-Seyler, 374, 166. [DOI] [PubMed]
- Igarashi, T., Sekine, T., Inatomi, J. & Seki, G. (2002). J. Am. Soc. Nephrol.13, 2171–2177. [DOI] [PubMed] [Google Scholar]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyatkin, A. B., Natarajan, P., Munshi, S., Minor, W., Johnson, J. E. & Low, P. S. (1995). Proteins, 22, 293–297. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Nature (London), 227, 2680–2685.
- Laing, C. M., Toye, A. M., Capasso, G. & Unwin, R. J. (2005). Int. J. Biochem. Cell Biol.37, 1151–1161. [DOI] [PubMed] [Google Scholar]
- Liu, X. F., McAlear, S. D. & Bevensee, M. O. (2005). FASEB J.19, A142.
- Low, P. S. (1986). Biochim. Biophys. Acta, 864, 145–167. [DOI] [PubMed] [Google Scholar]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed] [Google Scholar]
- McPherson, A., Malkin, A. J. & Kuznetsov, Y. G. (1995). Structure, 3, 759–|768. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., Borek, D., Majewski, W. & Minor, W. (2003). Acta Cryst. A59, 228–234. [DOI] [PubMed] [Google Scholar]
- Przybylski, D. & Rost, B. (2002). Proteins, 46, 197–205. [DOI] [PubMed] [Google Scholar]
- Romero, M. F., Hediger, M. A., Boulpaep, E. L. & Boron, W. F. (1997). Nature (London), 387, 409–413. [DOI] [PubMed] [Google Scholar]
- Zhang, D., Kiyatkin, A., Bolin, J. T. & Low, P. S. (2000). Blood, 96, 2925–2933. [PubMed] [Google Scholar]