The C-terminal protease domain of Venezuelan equine encephalitis virus (VEEV) nsP2 has been overexpressed in E. coli, purified and successfully crystallized. Native crystals diffract to beyond 2.5 Å resolution and isomorphous heavy-atom derivatives suitable for phase analysis have been identified.
Keywords: alphaviruses, Venezuelan equine encephalitis virus, cysteine proteases, nsP2
Abstract
The C-terminal region of Venezuelan equine encephalitis virus (VEEV) nsP2 is responsible for proteolytic processing of the VEEV polyprotein replication complex. This action regulates the activity of the replication complex and is essential for viral replication, thus making nsP2 a very attractive target for development of VEEV therapeutics. The 338-amino-acid C-terminal region of VEEV nsP2 has been overexpressed in Escherichia coli, purified and crystallized. Crystals diffract to beyond 2.5 Å resolution and belong to the orthorhombic space group P212121. Isomorphous heavy-atom derivatives suitable for phase analysis have been obtained and work on building a complete structural model is under way.
1. Introduction
Venezuelan equine encephalitis virus (VEEV) is an alphavirus responsible for significant human and livestock disease in Central and South America. Epidemics reported as recently as 1995 in Venezuela and Colombia caused widespread illness and had mortality rates approaching 1% (Weaver et al., 1996 ▶). In addition, VEEV is a recognized weapon of biological warfare and terrorism, with several nations successfully developing and stockpiling weapons-grade VEEV (Bronze et al., 2002 ▶). There are no therapeutics to treat VEEV infection; the VEEV TC-83 vaccine strain only provides partial protection against infection and is not approved for general human immunizations (Weaver et al., 1999 ▶).
The nsP2 protein of alphaviruses has multiple enzymatic activities. The N-terminal domain, consisting of 456 amino acids (Gly1–Ile456; VEEV numbering), has been shown to possess ATPase and GTPase activity (Rikkonen et al., 1994 ▶), RNA helicase activity (Gomez de Cedron et al., 1999 ▶) and RNA 5′-triphosphatase activity (Vasiljeva et al., 2000 ▶). The C-terminal domain of nsP2, consisting of 338 amino acids (Met457–Cys794; VEEV numbering), is responsible for regulation of 26S subgenomic RNA synthesis (Suopanki et al., 1998 ▶), switching between minus- and plus-strand RNA synthesis (Sawicki et al., 2006 ▶; Sawicki & Sawicki, 1993 ▶), targeting nsP2 for nuclear transport (Peranen et al., 1990 ▶) and proteolytic processing of the alphavirus nonstructural polyprotein replication complex through a cysteine protease mechanism of action (Merits et al., 2001 ▶; Vasiljeva et al., 2001 ▶, 2003 ▶).
No structural information is available for nsP2 and there are no protein structures with sufficient sequence identity to nsP2 to allow construction of homology models. Sequence analysis places alphavirus nsP2 proteins in peptidase family C9 of clan CA (Rawlings & Barrett, 1993 ▶; Rawlings et al., 2006 ▶). The cysteine protease activity of nsP2 is specific for substrates with certain target sequences (AGA or AGC in VEEV; Strauss & Strauss, 1994 ▶) and is essential for alphavirus replication, making nsP2 an attractive target for the development of therapeutics for VEEV infection. Structural information on VEEV nsP2 would greatly facilitate drug-discovery and development efforts for VEEV and related alphaviruses. Here, we report the purification, crystallization and preliminary X-ray diffraction studies of the C-terminal protease domain of VEEV nsP2 (nsP2pro).
2. Materials and methods
2.1. Materials
Tryptone, yeast extract, K2HPO4, Na2HPO4, imidazole, NaCl, Triton X-100, MgCl2, zinc acetate and Tris buffers were purchased from Fisher. Carbenicillin was from Invitrogen. Chloramphenicol, EDTA, l-arginine–HCl, l-glutamic acid monosodium salt, hen egg-white lysozyme and DNAse I were purchased from Sigma–Aldrich. Isopropyl β-d-thiogalactopyranoside (IPTG) was obtained from Gold Biotechnologies. All materials were reagent grade or better.
2.2. Cloning, expression and purification of the C-terminal protease domain of VEEV nsP2 (nsP2pro)
The DNA coding for VEEV nsP2pro (residues 457–794) was amplified by PCR from full-length VEEV TC83 infectious cDNA maintained in pUC19 cloning vector (generously provided by Dr S. Weaver, UTMB). PCR products were blunt-end cloned into pETBlue1 cloning and expression vector using the Perfectly Blunt cloning kit (Novagen). Resulting constructs (pETBlue/nsP2pro) were propagated in NovaBlue Escherichia coli. The nucleic acid sequence of the cloned gene was verified and constructs with correct sequence were used to transform E. coli expression strain Tuner(DE3)pLacI (Novagen).
Tuner(DE3)pLacI cells transformed with pETBlue/nsP2pro were grown at 310 K in phosphate-buffered 2×YT (16 g tryptone, 10 g yeast extract, 10 g K2HPO4 per litre) supplemented with 50 µg ml−1 carbenicillin, 34 µg ml−1 chloramphenicol and 1 mM MgCl2. Cultures were induced by addition of 0.2 mM IPTG at an OD600 of 1.5 and were grown for 3 h before collection by centrifugation.
E. coli pellets were lysed in SP column buffer (20 mM Tris, 14 mM 2-mercaptoethanol pH 8.5) by addition of 5 mM EDTA, 0.2 mg ml−1 hen egg-white lysozyme and 1.0% Triton X-100 while stirring at room temperature. Lysis was monitored by observing the viscosity of the suspension. When no further increase in the viscosity was noticed, 10 µg ml−1 DNAse I and 20 mM MgSO4 were added with stirring to digest the DNA. When the viscosity was sufficiently reduced, the lysate was transferred to polycarbonate round-bottom tubes and centrifuged at 34 000 rev min−1 for 15 min in a Sorvall SS-34 rotor at 277 K. The supernatant was applied onto an SP high-performance Sepharose column (Amersham Pharmacia) equilibrated in SP column buffer. After thorough washing, the column was eluted with 300 mM NaCl in SP column buffer. Since nsP2pro shows intrinsic metal affinity, eluted fractions containing nsP2pro were applied onto a nickel high-performance Sepharose column (Amersham Pharmacia) equilibrated in Ni-IMAC buffer (50 mM Na2HPO4, 500 mM NaCl, 14 mM 2-mercaptoethanol pH 7.0). NsP2pro was eluted with 25 mM imidazole in Ni-IMAC buffer. The identity of the purified material was verified by N-terminal sequencing and MALDI–TOF. After addition of 50 mM arginine and 50 mM glutamic acid, this material was concentrated to >15 mg ml−1 total protein and applied onto a Superdex 200 gel-filtration column (Amersham Pharmacia) in 50 mM arginine, 50 mM glutamic acid pH 8.0 (GF buffer). Protein concentration was determined by absorbance at 280 nm using an extinction coefficient of 0.92 ml mg−1 cm−1 calculated from the amino-acid composition using the Lasergene software package (DNAStar Inc.). Fractions from this column showing >99% purity by SDS–PAGE were used in crystallization trials. Fractions were stored at 277 K until needed.
2.3. Crystallization of nsP2pro
NsP2pro from gel filtration was concentrated to 6.0 mg ml−1 in GF buffer using Millipore Ultrafree centrifugal concentrators with 10 000 NMWL Biomax filters. Concentrated nsP2pro was filtered using 0.22 µm Amicon Ultrafree MC centrifugal filters (Millipore) prior to setup of crystallization drops. Crystallization conditions were screened by hanging-drop vapour diffusion using Hampton Research Crystal Screens I and II, Salt Rx and Index Screens. All commercial screens were used at twofold dilution in Milli-Q grade dH2O. Promising conditions were optimized by varying the pH and precipitant concentration. Conditions were further improved using the Hampton Research Additive Screens 1, 2 and 3 and Detergent Screens 1, 2 and 3. Crystals were obtained at 290 K using hanging-drop vapour diffusion over 0.5 ml 3.0 M ammonium formate (pH unadjusted), 2% 2-methyl-2,4-pentanediol (MPD), 1% glycerol and 0.2 mM zinc acetate with drops made by mixing 2 µl nsP2pro with 2 µl reservoir solution. Suitable crystals typically grew in less than 7 d, although some drops required two to three weeks to produce useable crystals.
2.4. Preparation of isomorphous heavy-atom derivatives
Identification of heavy-atom derivatives for phase determination was accomplished by soaking crystals of nsP2pro in a stabilizing solution containing various heavy-atom compounds in sitting-drop format. Crystals were transferred from crystallization drops and washed in stabilizing solution containing 3.0 M lithium formate, 2% MPD, 1% glycerol. Crystals were then transferred to stabilizing solution containing between 1 and 20 mM concentration of various heavy-atom compounds. Crystals were soaked for 2–7 d prior to diffraction analysis to identify suitable derivatives.
2.5. Data collection and analysis
Crystals were soaked in a cryoprotectant solution containing 2.5 M lithium formate, 2% MPD and 40% glycerol for 5–10 min prior to mounting in a 100 K nitrogen stream. Diffraction data were collected over 0.25–1° oscillation ranges at the Cu Kα wavelength (1.5418 Å) using a MacScience M06HF rotating-anode X-ray generator with a DIP2030 imaging-plate detector. Data were also collected at The Gulf Coast Consortium Protein Crystallography PX1 beamline at the Center for Advanced Microstructures and Devices (CAMD, Baton Rouge, LA, USA). Diffraction data were indexed, integrated and scaled using the HKL2000 package (HKL Research Inc). Identification of heavy-atom sites, determination of phases and initial phase refinement were performed using the SHARP software package (Buster Development Group). Initial protein model building was performed using TEXTAL (Holton et al., 2000 ▶; Ioerger & Sacchettini, 2003 ▶; Romo et al., 2005 ▶). Further model building is currently in progress using the Xtalview software package (McRee, 1999 ▶).
3. Results and discussion
3.1. Expression and purification of VEEV nsP2pro
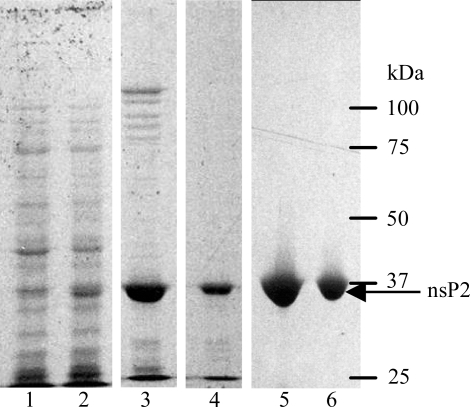
Upon induction of E. coli containing the pETBlue1/nsP2pro construct, a protein band of about 35 kDa is observed in Coomassie Blue-stained polyacrylamide gels following SDS–PAGE (Fig. 1 ▶). The majority of this material is expressed in inclusion bodies. After adsorption of the soluble fraction on SP-Sepharose and elution in 300 mM NaCl, soluble nsP2pro is observed as an enriched band at around 35 kDa. Further purification by Ni-affinity and gel-filtration chromatography results in >99% pure nsP2pro (Fig. 1 ▶). Approximately 2 mg of pure material was obtained per litre of culture. Interestingly, in order to obtain soluble nsP2pro it was necessary to add K2HPO4 prior to autoclaving the media. In the absence of K2HPO4 or if phosphate buffer is sterilized separately and then added to the media, soluble nsP2pro was produced at barely detectable levels.
Figure 1.
9% SDS–PAGE of nsP2pro expression and purification. Lane 1, uninduced E. coli Tuner(DE3)pLacI; lane 2, 3 h post-induction; lane 3, 300 mM NaCl fraction from SP-Sepharose; lane 4, 25 mM imidazole fraction from Ni-Sepharose HP; lanes 5 and 6, peak fractions from Superdex-200 gel-filtration column.
3.2. Crystallization of nsP2pro
Of the initial crystallization conditions examined, crystals (short needles) were only obtained from 2.0 M sodium formate. After several rounds of optimization, crystals suitable for X-ray diffraction studies were obtained from 3.0 M ammonium formate (pH unadjusted), MPD, 1% glycerol and 200 µM zinc acetate (Fig. 2 ▶). Crystals of similar appearance were also obtained if CdCl2 was substituted for zinc, but these crystals have not been used in diffraction experiments. The most useful crystals grew as clusters of thin blades measuring ∼300 µm in length, ∼50 µm in width and ∼5–10 µm in thickness.
Figure 2.
nsP2pro crystals. Long blades of dimensions 500 × 30 × 5 µm obtained from unbuffered 3.0 M ammonium formate, 2% MPD, 1% glycerol and 0.2 mM zinc acetate. Crystals such as these were used in all data collections.
3.3. X-ray diffraction of nsP2pro
Data were collected on the PX beamline at CAMD to 2.5 Å using a single crystal. Analysis of diffraction data indicated that the crystals belonged to space group P212121. Based on the Matthews coefficient (V M = 2.4 Å3 Da−1; Matthews, 1968 ▶), there was only a single monomer of nsP2pro per asymmetric unit and the solvent content was calculated as ∼48.7%. Data-collection statistics are presented in Table 1 ▶.
Table 1. Diffraction data-collection and processing statistics.
Values in parentheses are for the highest resolution shell. All reflections were processed.
| Heavy-atom derivative data | ||||
|---|---|---|---|---|
| Native data | KAu(CN)2 | Thimerosal | Terbium | |
| X-ray source | CAMD PX beamline | MacScience M06HF | MacScience M06HF | MacScience M06HF |
| Wavelength (Å) | 1.38079 | 1.5418 | 1.5418 | 1.5418 |
| Oscillation angle (°) | 0.25 | 1.0 | 1.0 | 1.0 |
| No. of images | 600 | 67 | 63 | 158 |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit-cell parameters | ||||
| a (Å) | 47.5 | 47.3 | 47.5 | 47.3 |
| b (Å) | 72.6 | 72.7 | 72.9 | 72.8 |
| c (Å) | 106.7 | 105.6 | 105.6 | 105.9 |
| Resolution limits (Å) | 40–2.50 (2.59–2.50) | 40.0–3.0 (3.11–3.00) | 40.0–3.0 (3.11–3.00) | 40.0–3.0 (3.11–3.00) |
| Mosaicity (°) | 0.44 | 0.92 | 0.71 | 0.48 |
| Total reflections | 74982 | 19245 | 18632 | 45962 |
| Unique reflections | 13340 | 7529 | 7699 | 7593 |
| Redundancy | 5.6 (5.7) | 3.3 (3.2) | 2.6 (2.5) | 6.1 (6.1) |
| Completeness (%) | 99.7 (100.0) | 76.7 (81.0) | 91.8 (95.7) | 100.0 (100.0) |
| 〈I/σ(I)〉 | 14.2 (3.1) | 7.8 (2.8) | 6.1 (2.2) | 10.8 (4.4) |
| Rsym† (%) | 9.4 (34.0) | 18.0 (44.4) | 16.1 (44.4) | 16.8 (41.4) |
R
sym = 
 , where I
i(hkl) is the ith measured diffraction intensity and 〈I(hkl)〉 is the mean of the intensity for the Miller index hkl.
, where I
i(hkl) is the ith measured diffraction intensity and 〈I(hkl)〉 is the mean of the intensity for the Miller index hkl.
3.4. Isomorphous heavy-atom derivatives of nsP2pro
After screening crystals against multiple heavy-metal compounds, isomorphous derivatives were obtained for thimerosal [ethyl-2-mercaptobenzoato(2-)-O,S-mercurate sodium], potassium dicyanoaurate(I) and terbium(III) chloride. Diffraction results for these crystals are presented in Table 1 ▶. Phase determination using these data is currently under way.
Acknowledgments
We thank Dr Henry Bellamy for assistance at the CAMD PX beamline and Dr Mark White, Dr Jebamony J. Robert and Dr Lokesh Rao for technical assistance and guidance in crystallization, sample preparation, data collection and analysis. Data used in this publication were collected at the Gulf Coast Protein Crystallography (GCPCC) Beamline at the Center for Advanced Microstructures and Devices (CAMD). This beamline is supported by National Science Foundation (NSF) grant DBI-9871464 with co-funding from the National Institute for General Medical Sciences (NIGMS).
References
- Bronze, M. S., Huycke, M. M., Machado, L. J., Voskuhl, G. W. & Greenfield, R. A. (2002). Am. J. Med. Sci.323, 316–325. [DOI] [PubMed] [Google Scholar]
- Gomez de Cedron, M., Ehsani, N., Mikkola, M. L., Garcia, J. A. & Kaariainen, L. (1999). FEBS Lett.448, 19–22. [DOI] [PubMed] [Google Scholar]
- Holton, T., Ioerger, T. R., Christopher, J. A. & Sacchettini, J. C. (2000). Acta Cryst. D56, 722–734. [DOI] [PubMed] [Google Scholar]
- Ioerger, T. R. & Sacchettini, J. C. (2003). Methods Enzymol.374, 244–270. [DOI] [PubMed] [Google Scholar]
- McRee, D. E. (1999). J. Struct. Biol.125, 156–165. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Merits, A., Vasiljeva, L., Ahola, T., Kaariainen, L. & Auvinen, P. (2001). J. Gen. Virol.82, 765–773. [DOI] [PubMed] [Google Scholar]
- Peranen, J., Rikkonen, M., Liljestrom, P. & Kaariainen, L. (1990). J. Virol.64, 1888–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N. D. & Barrett, A. J. (1993). Biochem. J.290, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N. D., Morton, F. R. & Barrett, A. J. (2006). Nucleic Acids Res.34, D270–D272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikkonen, M., Peranen, J. & Kaariainen, L. (1994). J. Virol.68, 5804–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romo, T., Gopal, K., McKee, E., Kanbi, L., Pai, R., Smith, J., Sacchettini, J. & Ioerger, T. (2005). IEEE Intell. Syst.20, 59–63.
- Sawicki, D. L., Perri, S., Polo, J. M. & Sawicki, S. G. (2006). J. Virol.80, 360–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki, D. L. & Sawicki, S. G. (1993). J. Virol.67, 3605–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, J. H. & Strauss, E. G. (1994). Microbiol. Rev.58, 491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suopanki, J., Sawicki, D. L., Sawicki, S. G. & Kaariainen, L. (1998). J. Gen. Virol.79, 309–319. [DOI] [PubMed] [Google Scholar]
- Vasiljeva, L., Merits, A., Auvinen, P. & Kaariainen, L. (2000). J. Biol. Chem.275, 17281–17287. [DOI] [PubMed] [Google Scholar]
- Vasiljeva, L., Merits, A., Golubtsov, A., Sizemskaja, V., Kaariainen, L. & Ahola, T. (2003). J. Biol. Chem.278, 41636–41645. [DOI] [PubMed] [Google Scholar]
- Vasiljeva, L., Valmu, L., Kaariainen, L. & Merits, A. (2001). J. Biol. Chem.276, 30786–30793. [DOI] [PubMed] [Google Scholar]
- Weaver, S. C., Salas, R., Rico-Hesse, R., Ludwig, G. V., Oberste, M. S., Boshell, J. & Tesh, R. B. (1996). Lancet, 348, 436–440. [DOI] [PubMed] [Google Scholar]
- Weaver, S. C., Tesh, R. B. & Shope, R. E. (1999). Tropical Infectious Diseases: Principles, Pathogens and Practice, edited by R. E. Guerrant, D. H. Walker & P. F. Weller, Vol. 2, pp. 1281–1287. New York: Churchill Livingstone.