Abstract
Although an excitotoxic mechanism of neuronal injury has been proposed to play a role in chronic neurodegenerative disorders such as Alzheimer’s disease, and neurotrophic factors have been put forward as potential therapeutic agents, direct evidence is lacking. Taking advantage of the fact that mutations in the presenilin-1 (PS1) gene are causally linked to many cases of early-onset inherited Alzheimer’s disease, we generated PS1 mutant knock-in mice and directly tested the excitotoxic and neurotrophic hypotheses of Alzheimer’s disease. Primary hippocampal neurons from PS1 mutant knock-in mice exhibited increased production of amyloid β-peptide 42/43 and increased vulnerability to excitotoxicity, which occurred in a gene dosage-dependent manner. Neurons expressing mutant PS1 exhibited enhanced calcium responses to glutamate and increased oxyradical production and mitochondrial dysfunction. Pretreatment with either basic fibroblast growth factor or activity-dependent neurotrophic factor protected neurons expressing mutant PS1 against excitotoxicity. Both basic fibroblast growth factor and activity-dependent neurotrophic factor stabilized intracellular calcium levels and abrogated the increased oxyradical production and mitochondrial dysfunction otherwise caused by the PS1 mutation. Our data indicate that neurotrophic factors can interrupt excitotoxic neurodegenerative cascades promoted by PS1 mutations.
Excessive activation of excitatory amino acid receptors, resulting in cytoplasmic calcium overload and oxyradical production, has been implicated in the neuronal death process that occurs in a variety of pathological settings, including Alzheimer’s disease (AD; refs. 1–3). Age-related factors that may predispose neurons to excitotoxicity in AD include reduced energy availability (4, 5), increased accumulation of neurotoxic forms of amyloid β-peptide (Aβ; refs. 6 and 7), and increased levels of oxidative stress (8–10). Some cases of AD are characterized by an early age of onset and a dominant inheritance pattern (11). Mutations in the gene encoding presenilin-1 (PS1) on chromosome 14 are responsible for many cases of early-onset autosomal dominant inherited AD, and mutations in a homologous gene on chromosome 1 (presenilin-2) account for a smaller fraction of familial AD cases (11–14). Presenilins are integral membrane proteins that are widely expressed in neurons throughout the brain, wherein they are localized mainly in the endoplasmic reticulum (15–17). Overexpression of mutant PS1 in cultured cells and transgenic mice causes altered proteolytic processing of the amyloid precursor protein (APP), resulting in increased production of a neurotoxic form of Aβ (Aβ 1–42; refs. 18–20), and decreased levels of a neuroprotective secreted form of APP (21, 22). Overexpression of presenilin mutations in cultured cells results in increased vulnerability of the cells to apoptosis induced by trophic factor withdrawal, Aβ, and metabolic insults (22–28). It is not known whether presenilin mutations similarly endanger primary neurons, nor whether the mutations sensitize neurons to excitotoxicity.
Because of their ability to promote neuronal survival and to protect neurons against various insults, neurotrophic factors have been put forward as potential therapeutic agents in neurodegenerative disorders (29–33). Recent studies have identified basic fibroblast growth factor (bFGF) and activity-dependent neurotrophic factor (ADNF) as two factors that can protect a wide range of neuronal populations against insults relevant to the pathogenesis of AD, including exposures to excitotoxins (34–37), Aβ (38–40), and oxidative insults (41, 42). The mechanisms whereby bFGF protects neurons include stabilization of calcium homeostasis and suppression of oxidative stress, and may involve modulation of expression of calcium-regulating proteins and antioxidant enzymes (43, 44). The neuroprotective action of ADNF appears to involve activation of NF-κB (42), a transcription factor previously shown to mediate anti-apoptotic and anti-excitotoxic actions in cultured hippocampal neurons (45, 46). Although neurotrophic factors can protect neurons against insults relevant to AD, it is not known whether they can counteract the adverse effects of genetic mutations that cause AD. We now report that bFGF and ADNF can protect primary hippocampal neurons against the excitotoxicity-enhancing action of a PS1 mutation.
MATERIALS AND METHODS
Generation and Characterization of PS1 Mutant Knock-in Mice.
The targeting strategy and methods for generating knock-in mice expressing the human AD-linked M146V mutation are detailed elsewhere (47). The assembled vector was linearized with PvuI and electroporated into 129/Sv-derived R1 embryonic stem cells (48). Genomic DNA was isolated from 250 clones surviving double selection, digested with HindIII and BglI, and analyzed by Southern blot using a 600-bp HindIII-HpaI 5′-probe. Nineteen of the clones produced the expected HindIII and BglI polymorphisms, and 4 of the 19 targeted cell lines (19, 106, 157, 179) were injected into recipient blastocysts and transferred to foster mothers to produce male chimeras which were then mated with C57BL/6 females to produce heterozygous PS1 mv(±) mice (129/Sv × C57BL/6 F1s). Our studies employed mice derived from either cell line 106 or 179. Tail DNA samples were used for genotyping in a PCR assay that employed two PCR primers (AGGCAGGAAGATCACGTGTTCAAGTAC-3′ and 5′-CACACGCACACTCTGACATGCACAGGC-3′) to amplify genomic DNA sequences flanking exon 5 prior to digestion of the amplified DNA with the restriction enzyme BstEII. The full-length PCR product is 530 bp, and the product sizes for the wild-type and targeted allele (PS1 mv) following BstEII enzyme digestion are 530 and 350/180 bp, respectively.
Quantification of Levels of Aβ1–40 and Aβ1–42.
Aβ1–40 and Aβ1–42 levels in brain tissue from wild-type and PS1M146VKI heterozygous and homozygous mice were measured using a fluorescence-based sandwich ELISA using antibodies specific for Aβ1–40 and Aβ1–42 (QCB, Hopkinton, MA). Individual brains were homogenized in 2 ml of 70% formic aid. The homogenates were centrifuged for 1 h at 100,000 × g and the supernatant was collected and neutralized with 1 M Tris base (20-fold dilution). A 100-μl neutralized sample was then mixed with 50 μl of phosphate-buffered Tween (PBT; 0.02% KCl, 0.02% KH2PO4, 0.8% NaCl, 0.216% Na2HPO4⋅7H2O, 5% BSA, 0.03% Tween-20; pH 7.4) and analyzed by ELISA. Fluorescence was quantified with a fluorescence plate reader with excitation at 460 nm and emission at 560 nm. Standard curves were generated using synthetic Aβ1–40 and Aβ1–42.
Hippocampal Cell Cultures and Quantification of Neuronal Survival.
Dissociated hippocampal cell cultures were prepared from postnatal day 1 mice and were maintained using methods similar to those described previously (9, 34, 47). Experiments were performed in 8-day-old cultures. More than 90% of the cells in these cultures were neurons. Immediately prior to experimental treatment, the medium was replaced with Locke’s buffer (NaCl, 154 mM; KCl, 5.6 mM; CaCl2, 2.3 mM; MgCl2, 1.0 mM; NaHCO3, 3.6 mM; glucose, 5 mM; Hepes, 5 mM; pH 7.2). bFGF (Boehringer Mannheim) was prepared as a 1000× stock in Locke’s buffer, and was stored at −80°C. ADNF9 (the nonapeptide SALLRSIPA; Molecular Genetics) was prepared as a 100× stock in Locke’s buffer and stored at room temperature. Glutamate was prepared as a 10 mM stock in Locke’s solution (pH 7.2). The method for quantification of neuron survival in hippocampal cell cultures was performed as described previously (34, 46, 47).
Measurement of Intracellular Calcium Levels.
Cytoplasmic free calcium levels were quantified by fluorescence ratio imaging of the calcium indicator dye fura-2 using methods described previously (25, 38). Briefly, cells were loaded with the acetoxymethyl ester form of fura-2 (30-min incubation in the presence of 10 μM fura-2) and imaged using a Zeiss AttoFluor system with a 40× oil-immersion objective. The average [Ca2+]i in individual neuronal cell bodies was determined from the ratio of the fluorescence emissions obtained using two different excitation wavelengths (334 nm and 380 nm). The system was calibrated using solutions containing either no Ca2+ or a saturating level of Ca2+ (1 mM) using the formula: [Ca2+]i = Kd[(R − Rmin)/(Rmax − R)](Fo/Fs).
Measurements of Cellular Peroxides, Mitochondrial Reactive Oxygen Species (ROS), and Membrane Lipid Peroxidation.
Peroxide levels were measured by confocal microscope analysis of cellular dichlorofluorescein (DCF) fluorescence as described previously (44). The dye dihydrorhodamine 123, which enters mitochondria and fluoresces when oxidized by ROS (principally peroxynitrite) to the positively charged rhodamine 123 derivative, was used to quantify levels of mitochondrial ROS using methods described previously (46). The thiobarbituric acid reactive substances fluorescence method was used to measure levels of membrane lipid peroxidation as described previously (49).
Assessments of Mitochondrial Functional Parameters.
Levels of 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction to formazan dye crystals in cells, a measure of mitochondrial respiratory chain activity and mitochondrial redox state were quantified as described previously (50). Briefly, MTT solution (5 mg/ml) was added to cultures and incubated for 1 h. Cells were then washed three times in Locke’s solution and solublized in dimethyl sulfoxide, and absorbance (592 nm) was quantified by using a plate reader. The dye rhodamine 123 (Molecular Probes) was employed as a measure of mitochondrial transmembrane potential using methods described previously (47, 50).
Western Blot Analysis.
Relative levels of PS1 protein in brain tissue from neonatal mice were characterized by Western blot analysis using methods similar to those described previously (25, 51). Briefly, proteins were separated by electrophoresis through an SDS/polyacrylamide gel and then transferred to a sheet of Immobilon-P. The membrane was then probed with αPS1 loop antiserum (51).
RESULTS
Characterization of PS1 M146V Knock-in Mice and Assessment of Aβ Production.
PS1 mutant knock-in mice were generated by targeting the M146V mutation to the third coding exon (exon 5) of PS1 (47). The distribution of genotypes among the progeny of intercrossed PS1M146V/wild-type heterozygotes appears to be normal and we have not observed any lethality in homozygous PS1M146VKI mice. RT-PCR, Northern blot, and Western blot analyses indicated that the targeted allele produces a properly spliced cDNA transcript and that relative expression levels of PS1 mRNA and protein are similar in wild-type and PS1M146VKI mice (47). Homozygous PS1M146VKI mice (up to 16 months of age) have not exhibited any signs of an overt phenotype, demonstrating that the targeted M146V mutation does not dramatically impair the normal developmental and physiological functions of PS1 (52, 53). These findings are consistent with data showing that ectopic expression of human PS1 protein (with familial AD mutations) in mice can fully suppress the PS1 null phenotype (54, 55).
In light of prior studies demonstrating increased levels of Aβ1–42 in brain tissue from PS1 mutant transgenic mice (19, 20), we quantified levels of Aβ1–40 and Aβ1–42 in brain tissue from young (1- to 4-month-old) wild-type mice and heterozygous and homozygous PS1M146VKI mice. Aβ1–40 levels were (pmol/g wt/wt): wild-type, 2.63 ± 0.10; PS1M146VKI heterozygous, 2.68 ± 0.13; and PS1M146VKI homozygous, 2.88 ± 0.16. Aβ1–42 levels were (pmol/g wt/wt; n = 5–8 mice): wild-type, 0.76 ± 0.06; PS1M146VKI heterozygous, 1.14 ± 0.08 (P < 0.05); and PS1M146VKI homozygous, 1.38 ± 0.07 (P < 0.01). Aβ1–42/Aβ1–40 ratios were: wild-type, 0.289; PS1M146VKI heterozygous, 0.396 (P < 0.05); and PS1M146VKI homozygous, 0.515 (P < 0.01).
Mutant PS1 Increases Neuronal Vulnerability to Excitotoxicity: Protection by bFGF and ADNF.
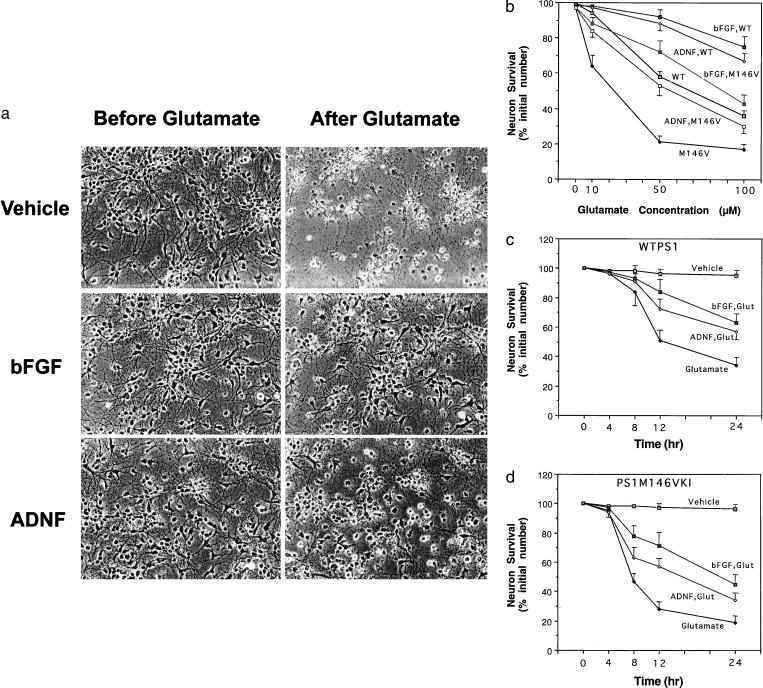
Primary neurons in hippocampal cultures established from postnatal day 1 wild-type and PS1M146VKI mice maintained viability for at least 2 wk, with no apparent adverse effect of the PS1 mutation on long-term survival during this time period in culture (data not shown). Exposure of hippocampal cultures from wild-type mice to 50 μM glutamate resulted in death of approximately 40% of the neurons during a 24-h exposure period (Fig. 1). In contrast, exposure of hippocampal cultures from homozygous PS1M146VKI mice to the same concentration of glutamate resulted in massive neuronal death, such that approximately 80% of the neurons were dead within 24 h (Fig. 1). Neurons from heterozygous PS1M146VKI mice also exhibited increased vulnerability to glutamate toxicity (values for survival 24 h following exposure to 50 μM glutamate were: cultures from wild-type mice, 57 ± 4%; cultures from heterozygous PS1M146VKI mice, 31 ± 3%; P < 0.01, n = 4). The morphological manifestations of glutamate neurotoxicity were essentially the same as those described previously (34, 43), and included neurite fragmentation and cell-body swelling and vacuolation (Fig. 1a). There was a leftward shift in the concentration-effect curve for PS1M146VKI neurons compared with wild-type neurons (Fig. 1b) such that approximately 40% of PS1M146VKI neurons were killed by a concentration of glutamate (10 μM) that was essentially nontoxic to wild-type neurons (Fig. 1b). The time course of neuronal death was also shifted to the left in cultures of PS1M146V hippocampal cells compared with cultures of wild-type hippocampal cells, such that over 50% of PS1M146VKI neurons were killed within 8 h of exposure to 100 μM glutamate, compared with less than 20% neuronal death in wild-type cultures (Fig. 1 c and d).
Figure 1.
PS1 mutation renders hippocampal neurons vulnerable to excitotoxicity: protection by bFGF and ADNF. (a) Phase-contrast micrographs of cultured hippocampal neurons from PS1M146VKI mice shown before and 24 h following exposure to 50 μM glutamate. (Top) Cells pretreated with vehicle (saline). (Middle) Cells pretreated for 24 h with 100 ng/ml bFGF. (Bottom) cells pretreated for 24 h with 0.1 pM ADNF9. Note extensive neuronal loss induced by glutamate in the vehicle-treated culture, and reduced neuronal loss in cultures pretreated with bFGF or ADNF9. (b) Hippocampal cultures from wild-type (WT) and PS1M146VKI mice were pretreated for 24 h with vehicle, 100 ng/ml bFGF, or 0.1 pM ADNF9. Cultures were then exposed for 24 h to the indicated concentrations of glutamate, and neuron survival was quantified. Values for cultures from PS1M146VKI were significantly less than each of the other values at each glutamate concentration (P < 0.01–0.001). (c and d) Hippocampal cultures from wild-type mice (WTPS1; c) or PS1M146VKI mice (d) were pretreated for 24 h with vehicle, 100 ng/ml bFGF, or 0.1 pM ADNF9. Cultures were then exposed for the indicated time periods to 100 μM glutamate, and neuron survival was quantified. Values for glutamate-treated cultures pretreated with bFGF or ADNF were significantly greater than corresponding values for cultures exposed to glutamate alone at the 12- and 24-h time points (P < 0.01). Probability values were determined by ANOVA with Scheffe’s post hoc tests. In each graph, values are the mean and SE of determinations made in four separate cultures.
Pretreatment of wild-type and PS1M146V mouse hippocampal cultures with bFGF (10 ng/ml) or ADNF9 (0.1 pM) for 24 h prior to exposure to glutamate resulted in significant decreases in neuronal death in both wild-type and PS1M146VKI cultures (Fig. 1). Concentration-effect and time-course curves revealed that both bFGF and ADNF9 shifted the curves to the right. However, the level of glutamate-induced neuronal death in bFGF- and ADNF9-treated cultures was greater in PS1M146VKI cultures than in wild-type cultures (Fig. 1 b–d), indicating that the neurotrophic factors did not completely prevent the death-enhancing effect of mutant PS1.
Enhanced Elevation of [Ca2+]i and Oxidative Stress Following Exposure to Glutamate in Neurons Expressing Mutant PS1: Protection by bFGF and ADNF.
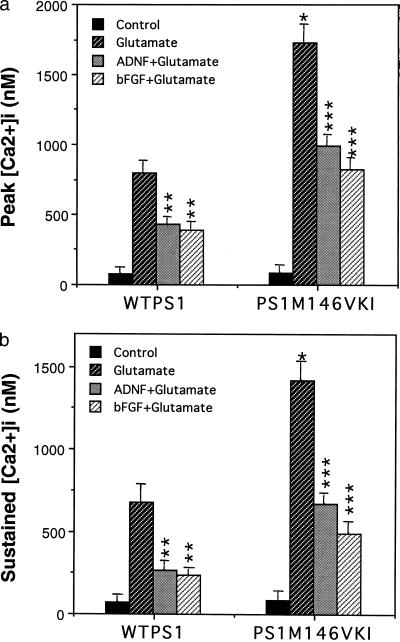
Because excessive calcium influx plays a central role in excitotoxic injury to neurons, we performed imaging analyses of [Ca2+]i responses to glutamate in cultured hippocampal neurons from wild-type and PS1M146VKI mice. Basal [Ca2+]i was similar in neurons from wild-type and PS1M146VKI mice. In contrast, both the peak and sustained components of the glutamate-induced [Ca2+]i increase were significantly greater in neurons from the PS1M146VKI mice compared with the wild-type mice (Fig. 2). In cultures of wild-type or PS1M146VKI neurons pretreated with either bFGF or ADNF9, both the peak and sustained increases of [Ca2+]i were significantly reduced compared with cultures not pretreated with a trophic factor (Fig. 2).
Figure 2.
Hippocampal neurons from PS1M146VKI mice exhibit enhanced calcium responses to glutamate: suppression by bFGF and ADNF. Hippocampal cultures from wild-type mice WTPS1 and PS1M146VKI mice were pretreated for 24 h with saline (control), 100 ng/ml bFGF, or 0.1 pM ADNF9. The [Ca2+]i was then monitored before and after exposure to 50 μM glutamate. Cumulative data for the peak [Ca2+]i (a) and the sustained [Ca2+]i (5 min after glutamate; b) following exposure to 50 μM glutamate. Values are the mean and SE of determinations made in 4–6 cultures (10–18 neurons analyzed per culture). ∗, P < 0.01 compared with the value for WTPS1 cells exposed to glutamate. ∗∗, P < 0.01 compared with the value for WTPS1 cells exposed to glutamate alone. ∗∗∗, P < 0.01 compared with the corresponding value for WTPS1 cells and to the value for PS1M146VKI cells exposed to glutamate alone. Probability values were determined by ANOVA with Scheffe’s post hoc tests.
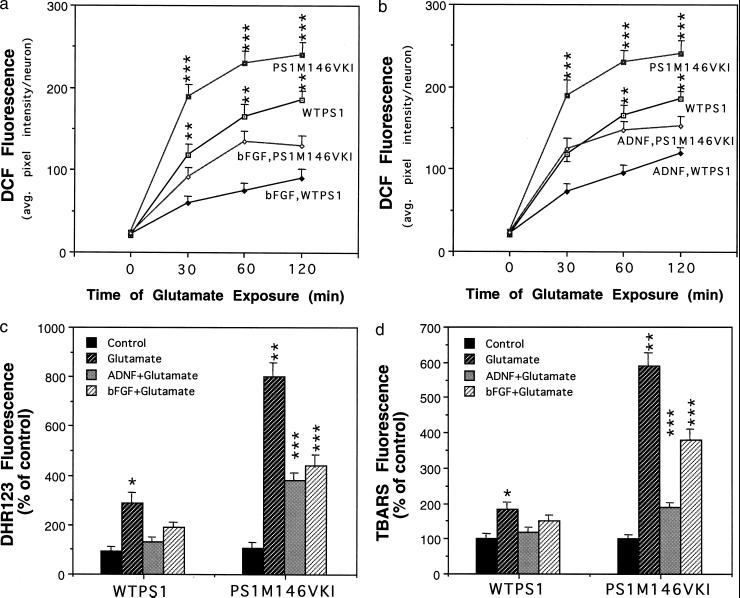
The excitotoxic mechanism involves excessive calcium influx, which leads to increased levels of oxidative stress and subsequent cell damage and death (44). Glutamate induced a progressive increase in levels of DCF fluorescence (a measure of peroxide levels), the magnitude of which was significantly greater in PS1M146VKI neurons than in wild-type neurons (Fig. 3 a and b). The glutamate-induced increase of DCF fluorescence occurred in neurites as well as in the cell body. Pretreatment with either bFGF or ADNF9 significantly attenuated the glutamate-induced increase of DCF fluorescence in both wild-type and PS1M146VKI neurons (Fig. 3 a and b). Glutamate induced increases in levels of mitochondrial ROS (dihydrorhodamine 123 fluorescence; Fig. 3c) and membrane lipid peroxidation (thiobarbituric acid reactive substances fluorescence; Fig. 3d) which were significantly greater in PS1M146VKI neurons than in wild-type neurons. Pretreatment with either bFGF or ADNF9 significantly attenuated the glutamate-induced increases of mitochondrial ROS and lipid peroxidation in neurons from both wild-type and PS1M146VKI mice (Fig. 3 c and d).
Figure 3.
Hippocampal neurons from PS1M146VKI mice exhibit enhanced oxidative stress following exposure to glutamate: suppression by bFGF and ADNF. (a and b) Hippocampal cultures from wild-type mice and PS1M146VKI mice were pretreated for 24 h with vehicle, 100 ng/ml bFGF, or 0.1 pM ADNF9. Cultures were then exposed for the indicated time periods to 100 μM glutamate, and levels of DCF were quantified. Values are the mean and SE of determinations made in four separate cultures (12–20 neurons analyzed per culture). ∗∗, P < 0.01 compared with the corresponding value for PS1M146VKI cells and to WTPS1 cells pretreated with bFGF or ADNF. ∗∗∗, P < 0.001 compared with the corresponding value for PS1M146VKI cells pretreated with bFGF or ADNF. (c and d) Hippocampal cultures from wild-type mice WTPS1 and PS1M146VKI mice were left untreated, or were pretreated for 24 h with vehicle (control), 100 ng/ml bFGF, or 0.1 pM ADNF9. Cultures were then exposed for 4 h to 100 μM glutamate, and levels of dihydrorhodamine 123 fluorescence and thiobarbituric acid reactive substances fluorescence were quantified. Values are the mean and SE of determinations made in 4–6 cultures (10–18 neurons analyzed per culture). ∗, P < 0.01 compared with value for control WTPS1 cells and WTPS1 cells pretreated with bFGF or ADNF9. ∗∗, P < 0.01 compared with the corresponding value for WTPS1 cells. ∗∗∗, P < 0.01 compared with the corresponding value for WTPS1 cells and to the value for PS1M146VKI cells exposed to glutamate alone. Probability values were determined by ANOVA with Scheffe’s post hoc tests.
Enhanced Mitochondrial Dysfunction Following Exposure to Glutamate in Neurons Expressing Mutant PS1: Protection by bFGF and ADNF.
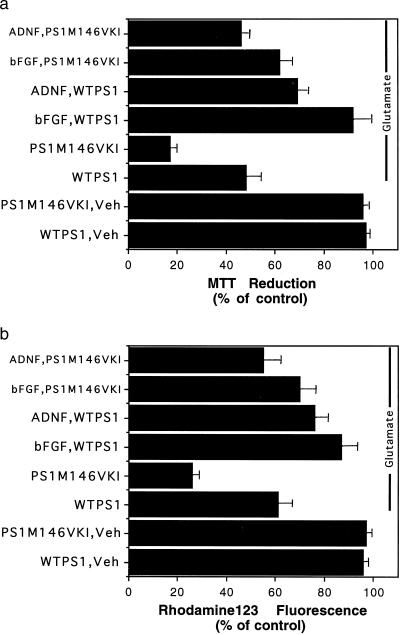
Mitochondrial dysfunction may contribute to the neurodegenerative process in AD (4), and occurs during the process of excitotoxic neuronal injury (56). Therefore, we quantified levels of MTT reduction (a measure of mitochondrial energy charge/redox state) and rhodamine 123 fluorescence (a measure of mitochondrial transmembrane potential) in hippocampal neurons from wild-type and PS1M146VKI mice following exposure to glutamate. Glutamate caused a 50% decrease in the level of MTT reduction in wild-type neurons, and a significantly greater (80%) decrease in the level of MTT reduction in PS1M146VKI neurons (Fig. 4a). Both bFGF and ADNF9 significantly attenuated the decreases in MTT reduction in both wild-type and PS1M146VKI neurons (Fig. 4a). Glutamate caused a 40% decrease in the level of rhodamine 123 fluorescence in wild-type neurons, and a significantly greater (75%) decrease in the level of rhodamine 123 fluorescence in PS1M146VKI neurons (Fig. 4b). Both bFGF and ADNF9 significantly attenuated the decreases in rhodamine 123 fluorescence in both wild-type and PS1M146VKI neurons (Fig. 4b).
Figure 4.
Glutamate-induced mitochondrial dysfunction is exacerbated in hippocampal neurons from PS1M146VKI mice: protection by bFGF and ADNF. Hippocampal cultures from wild-type mice and PS1M146VKI mice were pretreated for 24 h with vehicle (control), 100 ng/ml bFGF, or 0.1 pM ADNF9. Cultures were then exposed for 12 h to 50 μM glutamate, and levels of MTT reduction (a) or rhodamine 123 fluorescence (b) were quantified. Values are the mean and SE of determinations made in four separate cultures (15–20 neurons analyzed per culture). The control values were the values in untreated cultures and were 237 ± 7 absorbance units per culture for MTT reduction and 29 ± 3 average fluorescence pixel intensity per neuron for rhodamine 123 fluorescence. The value for PS1M146VKI cultures exposed to glutamate was significantly less than the value for WTPS1 cultures exposed to glutamate (P < 0.01). The values for cultures pretreated with bFGF or ADNF were significantly greater than the corresponding values for cultures not pretreated with a trophic factor (P < 0.05–0.01). Probability values were determined by ANOVA with Scheffe’s post hoc tests.
DISCUSSION
Previous studies have shown that bFGF (34, 35, 38) and transforming growth factor-β (57, 58) can protect cultured hippocampal neurons against death induced by Aβ and metabolic and oxidative insults. Our data provide evidence that neurotrophic factors can interrupt the neurodegenerative cascade promoted by PS1 mutations. Both bFGF and ADNF9 greatly reduced the level of neuronal death following exposure of cultures from PS1M146VKI mice to glutamate. bFGF and ADNF9 also suppressed both the increased oxyradical production, and the mitochondrial dysfunction caused by glutamate and exacerbated by mutant PS1. The neuroprotective mechanisms of bFGF and ADNF9 appear to involve stabilization of cellular calcium homeostasis and enhancement of antioxidant defense systems. As evidence, we found that both the peak and sustained [Ca2+]i responses to glutamate were suppressed in neurons pretreated with bFGF and ADNF9, and that levels of glutamate-induced ROS were also suppressed in neurons pretreated with each trophic factor. It was previously shown that bFGF pretreatment can suppress calcium responses to glutamate, possibly by altering the expression of glutamate receptor proteins and calcium binding proteins (34, 43, 59). In rat hippocampal cultures, bFGF treatment increased levels of Cu/Zn-SOD and glutathione reductase (44), whereas ADNF9 induced activation of the transcription factor NF-κB (42), which has been linked to increased expression of Mn-SOD (46). The improved mitochondrial function of trophic-factor-treated hippocampal neurons expressing mutant PS1 following exposure to glutamate is likely to contribute to enhanced cell survival because mitochondrial alterations are now recognized as playing pivotal roles in the cell death process. The potential for neurotrophic factor therapy to retard the neurodegenerative process in AD patients is currently being pursued in patients with sporadic AD (60, 61). The present data suggest the possibility that neurotrophic factors may also reduce neuronal degeneration in early-onset familial AD resulting from PS1 mutations.
The increased vulnerability to excitotoxicity of hippocampal neurons expressing a PS1 mutation provides direct evidence that this mutation can endanger neurons, and suggests a role for excitotoxicity in the pathogenesis of a familial form of AD. Previous indirect evidence for a role for excitotoxicity in AD includes: insults believed relevant to AD (e.g., exposure to Aβ, or metabolic and oxidative insults) can increase neuronal vulnerability to excitotoxicity (6–10, 35, 41); neuronal populations vulnerable to AD express high levels of glutamate receptors (62); exposure of hippocampal neurons to excitotoxic and metabolic insults induces several alterations in cytoskeleton similar to those seen in neurofibrillary tangles in AD (7, 35, 63–67); and primary hippocampal neurons are more vulnerable than tumor cell lines to Aβ and oxidative insults, and expression of glutamate receptors appears to be a major factor in this increased vulnerability (5–10, 68). Our data provide direct support for the hypothesis that activation of glutamate receptors contributes to the neurodegenerative process in AD. We recently found that hippocampal neurons from PS1M146VKI mice are more vulnerable to apoptosis induced by Aβ (69). When considered together with the compelling evidence that PS1 mutations alter APP processing so as to increase Aβ1–42 production (18–20), our data suggest that PS1 mutations promote a cascade of events involving increased vulnerability of neurons to Aβ and excitotoxicity. The collective findings suggest a scenario in which PS1 mutations exert two adverse effects that promote a degenerative cascade involving both Aβ and activation of glutamate receptors. The increased Aβ1–42 that results from PS1 mutations may disrupt calcium regulation and render neurons vulnerable to excitotoxicity. Alternatively, PS1 mutations may more directly perturb calcium homeostasis (25, 27) and thereby render neurons vulnerable to Aβ toxicity and excitotoxicity. Based upon the available data, it seems most likely that the adverse effects of PS1 mutations on Aβ production and calcium homeostasis work in concert in a cyclic cascade in which Aβ perturbs calcium homeostasis and perturbed calcium homeostasis alters APP processing. Therefore, our data argue for an important role for activation of glutamate receptors in the pathogenic mechanism of PS1 mutations, which may be mechanistically linked to the amyloid hypothesis of AD.
We found that the Aβ1–42 to Aβ1–40 ratio in brain tissue from heterozygous PS1M146VKI mice was greater than in wild-type mice but less than in homozygous PS1M146VKI mice, and that hippocampal neurons from heterozygous PS1M146VKI exhibit a level of vulnerability to glutamate toxicity that is intermediate to those from wild-type and heterozygous PS1M146VKI mice. Our data on altered APP processing suggest a gene-dosage-dependent gain-of-function of mutated PS1, and are consistent with a previous study showing that the magnitude of increase in Aβ1–42 levels is correlated with the magnitude of mutant PS1 transgene overexpression in transgenic mice (20). The gene-dosage effect on neuronal vulnerability to excitotoxicity further suggests a gain-of-function action of the PS1 mutations in promoting neuronal degeneration. Importantly, the present findings demonstrate adverse effects of PS1 mutations when expressed at normal physiological levels and in the heterozygous state, indicating that prior data in studies of transfected cell lines and transgenic mice were not the result of nonspecific effects of PS1 overexpression.
The mechanism whereby PS1 mutations increase neuronal vulnerability to excitotoxicity appears to involve enhanced calcium responses to glutamate and increased oxidative stress and mitochondrial dysfunction. Whereas basal [Ca2+]i was not different in wild-type and PS1M146VKI neurons, both the peak and sustained levels of [Ca2+]i were greatly increased in the neurons expressing mutant PS1. It has been reported that PC12 cells overexpressing PS1 mutations exhibit altered calcium regulation in the endoplasmic reticulum, which appears to contribute to their increased vulnerability to apoptosis induced by Aβ (23, 25, 27). We have found that hippocampal neurons from PS1M146VKI mice are more vulnerable to disruption of calcium homeostasis and apoptosis induced by Aβ (69). Because calcium influx through N-methyl-d-aspartate receptors and voltage-dependent calcium channels is a trigger that initiates the excitotoxic process, an exacerbation of the disruption of endoplasmic reticulum calcium homeostasis may play a role in the increased sensitivity of neurons expressing mutant PS1 to excitotoxicity.
Levels of peroxides, mitochondrial ROS, and membrane lipid peroxidation were increased following exposure to glutamate in hippocampal neurons expressing mutant PS1 compared with wild-type neurons. These findings suggest a widespread exacerbation of oxidative stress in neurons expressing mutant PS1, which would be expected to damage proteins, nucleic acids, and membrane lipids. Greater oxidative stress would be expected to promote excitotoxic cascades by impairing the function of ion-motive ATPases, which results in membrane depolarization and increased calcium influx through the N-methyl-d-aspartate receptor (9, 10). In addition, membrane lipid peroxidation has been shown to impair glucose transport and reduce ATP levels in cultured hippocampal neurons (5), which would further impair ATP-dependent calcium-regulating systems and promote excitotoxicity. Hippocampal neurons expressing the M146V PS1 mutation exhibited greater decreases in MTT reduction and mitochondrial transmembrane potential following glutamate exposure compared with wild-type neurons, which is consistent with a role for impaired mitochondrial function in the endangering action of PS1 mutations (28).
PS1M146VKI mice lack endogenous wild-type PS1 and yet show no overt phenotype, strongly suggesting that the mutant PS1 protein can substitute for wild-type PS1 in regulating developmental processes such as determination of cell fate (52–55). Hippocampal neurons from the PS1M146VKI mice did not undergo spontaneous cell death under the culture conditions employed (i.e., during the first 8 days in culture), but nevertheless exhibited a dramatically increased vulnerability to excitotoxicity. The collective data therefore indicate that PS1 mutations, though not affecting brain development or function under normal conditions, may render neurons vulnerable to age-related stressors such as reduced energy availability and increased levels of oxidative stress.
Acknowledgments
We thank J. Partin and L. Yan for technical assistance, and S. Sisodia for the generous gift of PS1 loop antibody. This work was supported by grants to M.P.M. from the National Institutes of Health (AG14554, AG05144, and NS35253), to C.B.W. from the National Institute on Aging (AG05136), to B.L.S. from the University of Washington Nathan Shock Center for Excellence in the Basic Biology of Aging, and to G.M.M. from the National Institute on Aging (ADRC AG05136).
ABBREVIATIONS
- AD
Alzheimer’s disease
- ADNF
activity-dependent neurotrophic factor
- APP
amyloid precursor protein
- bFGF
basic fibroblast growth factor
- DCF
dichlorofluorescein
- MTT
3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PS1
presenilin-1
- ROS
reactive oxygen species
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Choi D W. Neuron. 1988;1:623–634. doi: 10.1016/0896-6273(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 2.Coyle J, Bird S, Evans R, Gulley R, Nadler J, Nicklas W, Olney J. Neurosci Res Prog Bull. 1980;19:330–425. [PubMed] [Google Scholar]
- 3.Mattson M P. Physiol Rev. 1997;77:1081–1132. doi: 10.1152/physrev.1997.77.4.1081. [DOI] [PubMed] [Google Scholar]
- 4.Bowling A C, Beal M F. Life Sci. 1995;56:1151–1171. doi: 10.1016/0024-3205(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 5.Mark R J, Pang Z, Geddes J W, Uchida K, Mattson M P. J Neurosci. 1997;17:1046–1054. doi: 10.1523/JNEUROSCI.17-03-01046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh J, Yang L L, Cotman C W. Brain Res. 1990;533:315–320. doi: 10.1016/0006-8993(90)91355-k. [DOI] [PubMed] [Google Scholar]
- 7.Mattson M P, Cheng B, Davis D, Bryant K, Lieberburg I, Rydel R E. J Neurosci. 1992;12:376–389. doi: 10.1523/JNEUROSCI.12-02-00376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattson M P, Mark R J, Furukawa K. Chem Res Toxicol. 1997;10:507–517. doi: 10.1021/tx9601317. [DOI] [PubMed] [Google Scholar]
- 9.Mark R J, Hensley K, Butterfield D A, Mattson M P. J Neurosci. 1995;15:6239–6249. doi: 10.1523/JNEUROSCI.15-09-06239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mark R J, Lovell M A, Markesbery W R, Uchida K, Mattson M P. J Neurochem. 1997;68:255–264. doi: 10.1046/j.1471-4159.1997.68010255.x. [DOI] [PubMed] [Google Scholar]
- 11.Tanzi R E, Kovacs D M, Kim T W, Moir R D, Guenette S Y, Wasco W. Neurobiol Dis. 1996;3:159–168. doi: 10.1006/nbdi.1996.0016. [DOI] [PubMed] [Google Scholar]
- 12.Hardy J. Trends Neurosci. 1997;20:154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- 13.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 14.Levy-Lahad E, Wasco W, Poorkaj P, Romano D M, Oshima J, Pettingell W H, Yu C-E, Jondro P D, Schmidt S D, Wang K, et al. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 15.Kovacs D M, Fausett H J, Page K J, Kim T-W, Moir R D, Merriam D E, Hollister R D, Hallmark O G, Mancini R, Felsenstein K M, et al. Nat Med. 1996;2:224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- 16.Cook D G, Sung J C, Golde T E, Felsenstein K M, Wojczyk B S, Tanzi R E, Trojanowski J Q, Lee V M, Doms R W. Proc Natl Acad Sci USA. 1996;93:9223–9228. doi: 10.1073/pnas.93.17.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doan A, Thinakaran G, Borchelt D R, Slunt H H, Ratovistsky T, Podlisny M, Selkoe D J, Seeger M, Gandy S E, Price D L, Sisodia S S. Neuron. 1996;17:1023–1030. doi: 10.1016/s0896-6273(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 18.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird T D, Hardy J, Hutton M, Kukull W, et al. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 19.Borchelt D R, Thinakaran G, Eckman C B, Lee M K, Davenport F, Ratovitsky T, Prada C-M, Kim G, Seekins S, Yager D, et al. Neuron. 1996;17:1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 20.Duff K, Eckman C, Zehr C, Yu X, Prada C-M, Perez-Tur J, Hutton M, Buee L, Harigaya Y, Yager D, Morgan D, Gordon M N, Holcomb L, Refolo L, Zenk B, Hardy J, Younkin S. Nature (London) 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 21.Ancolio K, Marambaud P, Dauch P, Checler F. J Neurochem. 1997;69:2494–2499. doi: 10.1046/j.1471-4159.1997.69062494.x. [DOI] [PubMed] [Google Scholar]
- 22.Guo Q, Robinson N, Mattson M P. J Biol Chem. 1998;273:12341–12351. doi: 10.1074/jbc.273.20.12341. [DOI] [PubMed] [Google Scholar]
- 23.Guo Q, Furukawa K, Sopher B L, Pham D G, Xie J, Robinson N, Martin G M, Mattson M P. NeuroReport. 1996;8:379–383. doi: 10.1097/00001756-199612200-00074. [DOI] [PubMed] [Google Scholar]
- 24.Wolozin B, Iwasaki K, Vito P, Ganjei J K, Lacana E, Sunderland T, Zhao B, Kusiak J W, Wasco W, D’Adamio L. Science. 1996;274:1710–1713. doi: 10.1126/science.274.5293.1710. [DOI] [PubMed] [Google Scholar]
- 25.Guo Q, Sopher B L, Pham D G, Furukawa K, Robinson N, Martin G M, Mattson M P. J Neurosci. 1997;17:4212–4222. doi: 10.1523/JNEUROSCI.17-11-04212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janicki S, Monteiro M J. J Cell Biol. 1997;139:485–495. doi: 10.1083/jcb.139.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Q, Christakos S, Robinson N, Mattson M P. Proc Natl Acad Sci USA. 1998;95:3227–3232. doi: 10.1073/pnas.95.6.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller J N, Guo Q, Holtsberg F W, Bruce-Keller A J, Mattson M P. J Neurosci. 1998;18:4439–4450. doi: 10.1523/JNEUROSCI.18-12-04439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oppenheim R W. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 30.Raff M C, Barres B A, Burne J F, Coles H S, Ishizaki Y, Jacobson M D. Science. 1993;262:695–700. doi: 10.1126/science.8235590. [DOI] [PubMed] [Google Scholar]
- 31.Mattson M P, Cheng B, Smith-Swintosky V L. Exp Neurol. 1993;124:89–95. doi: 10.1006/exnr.1993.1178. [DOI] [PubMed] [Google Scholar]
- 32.Williams L R. Cerebrovasc Brain Metab Rev. 1995;7:55–73. [PubMed] [Google Scholar]
- 33.Mattson M P, Lindvall O. Adv Cell Aging Gerontol. 1997;2:299–345. [Google Scholar]
- 34.Furukawa K, Fu W, Li Y, Witke W, Kwiatkowski D J, Mattson M P. J Neurosci. 1997;17:8178–8186. doi: 10.1523/JNEUROSCI.17-21-08178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng B, Mattson M P. Exp Neurol. 1992;117:114–123. doi: 10.1016/0014-4886(92)90120-f. [DOI] [PubMed] [Google Scholar]
- 36.Nozaki K, Finklestein S P, Beal M F. J Cereb Blood Flow Metab. 1993;13:221–228. doi: 10.1038/jcbfm.1993.27. [DOI] [PubMed] [Google Scholar]
- 37.Brenneman D E, Gozes I. J Clin Invest. 1996;97:2299–2307. doi: 10.1172/JCI118672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattson M P, Tomaselli K, Rydel R E. Brain Res. 1993;621:35–49. doi: 10.1016/0006-8993(93)90295-x. [DOI] [PubMed] [Google Scholar]
- 39.Mark R J, Keller J N, Kruman I, Mattson M P. Brain Res. 1997;756:205–214. doi: 10.1016/s0006-8993(97)00196-0. [DOI] [PubMed] [Google Scholar]
- 40.Brenneman D E, Hauser J, Neale E, Rubinraut S, Fridkin M, Davidson A, Gozes I. J Pharmacol Exp Ther. 1998;285:619–627. [PubMed] [Google Scholar]
- 41.Zhang Y, Tatsuno T, Carney J, Mattson M P. J Cerebral Blood Flow Metab. 1993;13:378–388. doi: 10.1038/jcbfm.1993.51. [DOI] [PubMed] [Google Scholar]
- 42.Glazner G W, Boland A, Brenneman D E, Mattson M P. J Cell Biol. 1998;9:353a. [Google Scholar]
- 43.Mattson M P, Kumar K, Cheng B, Wang H, Michaelis E K. J Neurosci. 1993;13:4575–4588. doi: 10.1523/JNEUROSCI.13-11-04575.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattson M P, Lovell M A, Furukawa K, Markesbery W R. J Neurochem. 1995;65:1740–1751. doi: 10.1046/j.1471-4159.1995.65041740.x. [DOI] [PubMed] [Google Scholar]
- 45.Barger S W, Horster D, Furukawa K, Goodman Y, Krieglstein J, Mattson M P. Proc Natl Acad Sci USA. 1995;92:9328–9332. doi: 10.1073/pnas.92.20.9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattson M P, Goodman Y, Luo H, Fu W, Furukawa K. J Neurosci Res. 1997;49:681–697. doi: 10.1002/(SICI)1097-4547(19970915)49:6<681::AID-JNR3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 47.Guo Q, Fu W, Sopher B L, Miller M W, Ware C B, Martin G M, Mattson M P. Nat Med. 1999;5:101–107. doi: 10.1038/4789. [DOI] [PubMed] [Google Scholar]
- 48.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodman Y, Mattson M P. J Neurochem. 1996;66:869–872. doi: 10.1046/j.1471-4159.1996.66020869.x. [DOI] [PubMed] [Google Scholar]
- 50.Mattson M P, Robinson N, Guo Q. NeuroReport. 1997;8:3817–3821. doi: 10.1097/00001756-199712010-00031. [DOI] [PubMed] [Google Scholar]
- 51.Thinakaran G, Borchelt D R, Lee M K, Slunt H H, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, et al. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 52.Wong P C, Zheng H, Chen H, Becher M W, Sirinathsinghji D J S, Trumbauer M E, Chen H Y, Price D L, Van der Ploeg L H T, Sisodia S S. Nature (London) 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 53.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D J, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 54.Qian S, Jiang P, Guan X M, Singh G, Trumbauer M E, Yu H, Chen H Y, Van de Ploeg L H, Zheng H. Neuron. 1998;20:611–617. doi: 10.1016/s0896-6273(00)80999-x. [DOI] [PubMed] [Google Scholar]
- 55.Davis J A, Naruse S, Chen H, Eckman C, Younkin S, Price D L, Borchelt D R, Sisodia S S, Wong P C. Neuron. 1998;20:603–609. doi: 10.1016/s0896-6273(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 56.Schinder A F, Olson E C, Spitzer N C, Montal M. J Neurosci. 1996;16:6125–6133. doi: 10.1523/JNEUROSCI.16-19-06125.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prehn J H, Bindokas V P, Jordan J, Galindo M F, Ghadge G D, Roos R P, Boise L H, Thompson C B, Krajewski S, Reed J C, Miller R J. Mol Pharmacol. 1996;49:319–328. [PubMed] [Google Scholar]
- 58.Prehn J H, Bindokas V P, Marcuccilli C J, Krajewski S, Reed J C, Miller R J. Proc Natl Acad Sci USA. 1994;91:12599–12603. doi: 10.1073/pnas.91.26.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson D A, Lucidi-Phillipi C A, Murphy D P, Ray J, Gage F H. J Neurosci. 1996;16:886–898. doi: 10.1523/JNEUROSCI.16-03-00886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koliatsos V E. Crit Rev Neurobiol. 1996;10:205–238. doi: 10.1615/critrevneurobiol.v10.i2.40. [DOI] [PubMed] [Google Scholar]
- 61.Hoffer B, Olson L. J Neural Transm Suppl. 1997;49:1–10. doi: 10.1007/978-3-7091-6844-8_1. [DOI] [PubMed] [Google Scholar]
- 62.Martin L J, Al-Abdulla N A, Brambrink A M, Kirsch J R, Sieber F E, Portera-Cailliau C. Brain Res Bull. 1998;46:281–309. doi: 10.1016/s0361-9230(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 63.Mattson M P. Neuron. 1990;4:105–117. doi: 10.1016/0896-6273(90)90447-n. [DOI] [PubMed] [Google Scholar]
- 64.Busciglio J, Lorenzo A, Yeh J, Yankner B A. Neuron. 1995;14:879–888. doi: 10.1016/0896-6273(95)90232-5. [DOI] [PubMed] [Google Scholar]
- 65.Stein-Behrens B, Mattson M P, Chang I, Yeh M, Sapolsky R M. J Neurosci. 1994;14:5373–5380. doi: 10.1523/JNEUROSCI.14-09-05373.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith-Swintosky V L, Pettigrew L C, Sapolsky R M, Phares C, Craddock S D, Brooke S M, Mattson M P. J Cerebr Blood Flow Metab. 1996;16:585–598. doi: 10.1097/00004647-199607000-00008. [DOI] [PubMed] [Google Scholar]
- 67.Geula C, Wu C K, Saroff D, Lorenzo A, Yuan M, Yankner B A. Nat Med. 1998;4:827–831. doi: 10.1038/nm0798-827. [DOI] [PubMed] [Google Scholar]
- 68.Kruman I, Bruce-Keller A J, Bredesen D E, Waeg G, Mattson M P. J Neurosci. 1997;17:5089–5100. doi: 10.1523/JNEUROSCI.17-13-05089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo Q, Sebastian L, Sopher B L, Miller M W, Ware C B, Martin G M, Mattson M P. J Neurochem. 1999;72:1019–1029. doi: 10.1046/j.1471-4159.1999.0721019.x. [DOI] [PubMed] [Google Scholar]