Abstract
Identification of a biomarker of prognosis and response to therapy that can be assessed preoperatively would significantly improve overall outcomes for patients with pancreatic cancer. In this study, patients whose tumours exhibited high LMO4 expression had a significant survival advantage following operative resection, whereas the survival of those patients whose tumours had low or no LMO4 expression was not significantly different when resection was compared with operative biopsy alone.
Keywords: LMO4, prognosis, outcome, pancreatic cancer, surgical resection, therapeutic response
Pancreatic cancer (PC) remains the fourth leading cause of cancer death in western societies with a 5-year survival rate of less than 5%. Pancreatectomy is the only therapeutic intervention that can increase long-term survival. However, only 10–20% of patients who undergo pancreatectomy survive for more than 3 years, and there is no method to predict preoperatively as to which patients will benefit from resection (Yeo et al, 1997). For the majority of patients who are unsuitable for operative resection, non-operative approaches, including chemotherapy and chemoradiotherapy, have met with limited success. To date, there are no molecular markers of clinical utility, or rationally designed, molecularly targeted therapies, for PC. Thus, there is a crucial need for the identification of novel molecules important in PC that may also have diagnostic, therapeutic and prognostic utility.
LMO4 is a member of the LIM-only (LMO) family of transcription regulators that act as molecular adaptors, providing a scaffold for multiprotein complexes of DNA-binding factors and transcriptional regulatory proteins (Rabbitts, 1998; Visvader et al, 2001; Sum et al, 2002, 2005b). Here, we show that aberrant expression of LMO4 occurs in a significant proportion of PCs and that low/no LMO4 expression is associated with a poor outcome in PC. Multivariate analyses identified LMO4 expression as an independent predictor of survival in this cohort and also in a subgroup of patients who underwent pancreatic resection. LMO4 expression cosegregated with resectability and was associated with a significant survival advantage following operative resection. Thus, LMO4 expression may have potential clinical utility in estimating prognosis and response to operative resection.
MATERIALS AND METHODS
Patient cohort
We identified a cohort of 120 patients with the diagnosis of pancreatic ductal adenocarcinoma who underwent pancreatic resection or biopsy from Westmead Hospital, Concord Hospital, Royal Prince Alfred Hospital and St Vincent's Hospital in Sydney, Australia (Table 1). This cohort represents a subset of a previously described group of 348 patients (Biankin et al, 2002). Multicentre ethical approval for data collection and tissue use was granted by the Human Research Ethics Committees of the above hospitals.
Table 1. Clinicopathological and outcome data for all patients in the cohort.
| Whole cohort | n=120 | Resected cohort | n=75 | |||
|---|---|---|---|---|---|---|
| Parameter | No. (%) | Median survival (months) | P-value (log-rank) | No. (%) | Median survival (months) | P-value (log-rank) |
| Sex | ||||||
| Female | 50 (41.7) | 30 (40) | ||||
| Male | 70 (58.3) | 45 (60) | ||||
| Age at diagnosis (years) | ||||||
| Mean | 64.3 | 62.3 | ||||
| Median | 66.5 | 65 | ||||
| Range | 34.4–83.8 | 34.4–82.6 | ||||
| Specimen | ||||||
| Resection | 75 (62.5) | 12.2 | ||||
| Biopsy | 39 (32.5) | 4.6 | <0.0001 | |||
| Post-mortem | 6 (5) | |||||
| Outcome | ||||||
| Follow-up | 0–117.4 | 0.2–117.4 | ||||
| Median follow-up | 7.8 | 10.8 | ||||
| 30-day mortality | 2 (2.7) | |||||
| Death from PC | 106 (88.3) | 62 (82.6) | ||||
| Death from other cause | 2 (1.7) | 2 (2.7) | ||||
| Alive | 8 (6.7) | 8 (10.7) | ||||
| Lost to follow-up | 4 (3.3) | 3 (4) | ||||
| Stage a | 119 | |||||
| I | 27 (22.7) | |||||
| II | 10 (8.4) | 14.8 | ||||
| III | 67 (56.3) | |||||
| IV | 15 (12.6) | 7.2 | 0.0001 | |||
| Differentiation b | 119 | |||||
| Well | 9 (7.6) | 7 (9.3) | ||||
| Moderate | 65 (54.6) | 9.5 | 44 (58.7) | 14.5 | ||
| Poor | 45 (37.8) | 6.2 | 0.0050 | 24 (32) | 9.7 | 0.0398 |
| Tumour size | ||||||
| ⩽20 mm | 14 (18.7) | 17.1 | ||||
| >20 mm | 61 (81.3) | 10.5 | 0.0455 | |||
| Margins | ||||||
| Clear | 40 (53.3) | 16.2 | ||||
| Involved | 35 (46.7) | 8.6 | 0.0008 | |||
| Lymph node status c | 73 | |||||
| Negative | 35 (47.9) | 16.2 | ||||
| Positive | 38 (52.1) | 9.7 | 0.0182 | |||
| LMO4 status | ||||||
| Negative | 20 (16.7) | 3.7 | 9 (12) | 6.2 | ||
| Positive | 100 (83.3) | 9.2 | 0.0092 | 66 (88) | 14.2 | 0.0514 |
PC=pancreatic cancer.
Stage I and II vs stage III and IV tumours for survival analysis.
Well- and moderately differentiated tumours grouped together for survival analysis.
Lymph node status was only available in 73 patients in the resected cohort.
Immunohistochemistry
Pancreatic tissue microarrays were dewaxed and rehydrated before antigen unmasking, using target retrieval solution (DAKO Corporation, Carpenteria, CA, USA), in a water bath for 30 min. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide in methanol, followed by avidin/biotin and serum-free protein blocks (DAKO Corporation). Rat anti-LMO4 monoclonal antibody was generated as described previously (Sum et al, 2005a). Sections were incubated for 30 min in anti-LMO4 monoclonal antibody (20F8) followed by biotinylated rabbit anti-rat IgG (DAKO Corporation). A streptavidin–biotin–peroxidase system was used with 3,3′-diaminobenzidine as a substrate (DAKO Corporation). Counterstaining was performed with Mayer's hematoxylin (DAKO Corporation).
Immunohistochemical scoring
Staining was assessed by two separate observers for each case (DS and JGK), one of whom is a pathologist. Both observers were blinded to patient identification, clinicopathological variables and outcome. Standardisation of scoring was achieved by comparison of scores between observers, and by conferencing, where any discrepancies were resolved by consensus. For resected specimens, an average of 3 × 1.6 mm cores were assessed on tissue microarrays; however, due to limitations in tissue availability, only one or two cores were assessed for biopsy specimens. Scores were given as percentage of cells with positive nuclear staining within the representative area of the tissue microarray core and the absolute intensity of nuclear staining on a scale of 0–3 (0 representing no staining, 1 representing mild nuclear staining, 2 representing moderate nuclear staining and 3 representing strong nuclear staining) (Figure 1A–D). The following criterion was used to achieve a positive score for LMO4 overexpression: nuclear intensity ⩾2 in >50% of nuclei (Sum et al, 2005b).
Figure 1.
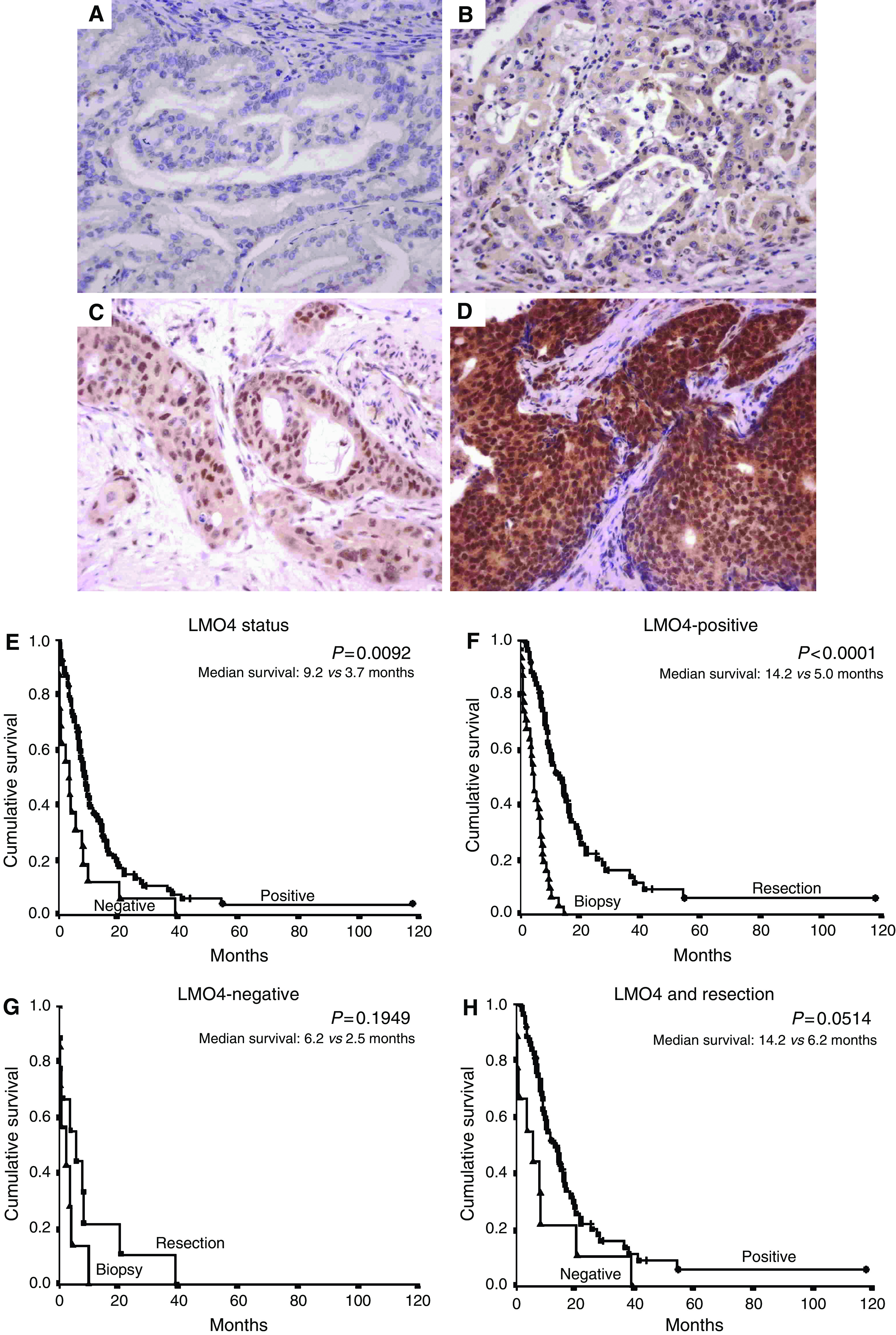
Pancreatic ductal adenocarcinoma: LMO4 nuclear expression score 0–3. (A) 0 representing no staining, (B) 1 representing mild nuclear staining, (C) 2 representing moderate nuclear staining and (D) 3 representing strong nuclear staining. Kaplan–Meier survival curves for whole cohort: (E) low/no vs high (>50% of nuclei with intensity ⩾2) LMO4 nuclear expression. Effect of resection on prognosis in the following subgroups: (F) LMO4-positive, (G) LMO4-negative; resected cohort: (H) low/no vs high (>50% of nuclei with intensity ⩾2) LMO4 nuclear expression.
Statistical evaluation
Statistical evaluation was performed using the Kaplan–Meier survival for univariate analysis and the Cox proportional hazards model for multivariate analysis using the Statview 5.0 software (Abacus Systems, Berkeley, CA, USA).
RESULTS
Clinicopathological parameters
Clinicopathological parameters for the cohort are presented in Table 1.
LMO4 protein expression
Immunohistochemical staining data were analysed in two groups; initially all patients were analysed, then subsequently only the subgroup of patients who underwent pancreatic resections. Significant nuclear LMO4 expression, as defined by the presence of nuclear staining in >50% of tumour cell nuclei with intensity ⩾2, was identified in 100 (83.3%) out of 120 tumours (Table 1).
Survival analysis
Kaplan–Meier analysis identified that operative resection of the tumours, stage 1 and 2 tumours (lymph node negative) and non-poorly differentiated tumours, was associated with longer survival (Table 1). Overall disease-specific 1-year survival was 32%, with a 3-year survival of 6% and a 5-year survival of 1%.
Low or no LMO4 expression was associated with a poor outcome (Figure 1E). Multivariate analysis using Cox proportional hazards modelling for those factors that were prognostic on univariate analysis identified LMO4 expression status as an independent prognostic factor when modelled together with resection, differentiation and Union Internationale Centre le Cancer (UICC) stage (Table 2, a). This model was refined by stepwise removal of the redundant variable of differentiation (Table 2, b). Operative resection was beneficial to patients who expressed LMO4 (median survival=14.2 vs 5.0 months, respectively; log-rank: P<0.0001; Figure 1F); in contrast, those patients whose tumours did not express LMO4 had no survival advantage over those who underwent biopsy alone (median survival=6.2 vs 5.0 months, respectively; Figure 1G). Hence, in this cohort, LMO4 expression cosegregated with response to operative resection, with patients whose tumours were LMO4-negative having no detectable survival advantage from operative resection.
Table 2. Multivariate analysis for clinicopathological parameters and LMO4 expression in the whole and resected cohorts of PC.
| Variable | Hazard ratio (95% confidence interval) | P-value |
|---|---|---|
| (a) Whole cohort (n=120) | ||
| LMO4 expression | 0.535 (0.297–0.961) | 0.0364 |
| Operative resection | 0.315 (0.191–0.519) | <0.0001 |
| Stage I/II vs stage III/IV | 1.894 (1.088–3.298) | 0.0239 |
| Differentiation | 1.252 (0.799–1.962) | 0.3275 |
| (b) Whole cohort (n=120) | ||
| LMO4 expression | 0.486 (0.279–0.847) | 0.0108 |
| Operative resection | 0.313 (0.190–0.515) | <0.0001 |
| Stage I/II vs stage III/IV | 2.024 (1.182–3.466) | 0.0102 |
| (c) Resected subgroup (n=75) | ||
| LMO4 expression | 0.504 (0.229–1.110) | 0.0888 |
| Tumour size >20 mm | 1.422 (0.697–2.901) | 0.3330 |
| Margin involvement | 1.575 (0.832–2.981) | 0.1629 |
| Lymph node involvement | 1.699 (0.912–3.167) | 0.0950 |
| Differentiation | 1.238 (0.694–2.210) | 0.4698 |
| (d) Resected subgroup (n=75) | ||
| LMO4 expression | 0.487 (0.223–1.063) | 0.0708 |
| Tumour size >20 mm | 1.439 (0.708–2.933) | 0.3144 |
| Margin involvement | 1.663 (0.892–3.101) | 0.1098 |
| Lymph node involvement | 1.721 (0.921–3.215) | 0.0887 |
| (e) Resected subgroup (n=75) | ||
| LMO4 expression | 0.460 (0.212–0.997) | 0.0492 |
| Margin involvement | 1.816 (0.990–3.332) | 0.0539 |
| Lymph node involvement | 1.734 (0.927–3.243) | 0.0848 |
| (f) Resected subgroup (n=75) | ||
| LMO4 expression | 0.382 (0.180–0.810) | 0.0121 |
| Lymph node involvement | 2.215 (1.251–3.922) | 0.0063 |
| (g) Resected subgroup (n=75) | ||
| LMO4 expression | 0.579 (0.282–1.191) | 0.1376 |
| Margin involvement | 0.426 (0.246–0.737) | 0.0023 |
PC=pancreatic cancer.
A and B, multivariate analysis of the whole cohort, with the final model (B) following removal of redundant variables. C–G, multivariate analysis of the resected subgroup. The initial model (C) with stepwise removal of redundant variables through D to E, where LMO4 expression is the only independent prognostic factor. Removal of margin status (F) shows that LMO4 expression is independent of lymph node involvement, but not margin involvement by tumour (G).
Resected cohort
Overall disease-specific 1-year survival following resection was 50%, with a 3-year survival of 16% and a 5-year survival of 6%. Patients in the cohort did not receive chemotherapy as either primary treatment, or in a neoadjuvant or adjuvant setting. Chemotherapy of any type was only given to 10 patients for the palliation of symptoms, and was not associated with a survival advantage.
Survival analysis of patients who underwent operative resection identified a trend toward decreased survival with negative LMO4 expression (borderline statistical significance; Figure 1H). Table 2 (c)–(f) shows the multivariate models for resected cancers. The initial model (Table 2, c) was subjected to stepwise removal of redundant variables to Table 2 (e), where LMO4 expression became the only independent prognostic factor. Removal of margin status demonstrated that LMO4 expression was independent of lymph node involvement (Table 2, f); however, LMO4 expression was not independent of margin involvement by the tumour (Table 2, g).
DISCUSSION
Low or no LMO4 expression was an independent poor prognostic factor for all patients with PC as well as in the subgroup of patients who underwent pancreatic resection. Importantly, high LMO4 expression was associated with a significant survival advantage following operative resection. In contrast, the survival of those patients whose tumours did not express LMO4 was not significantly different with resection compared with operative biopsy alone. Although operative resection is currently the best method available to treat PC, it is a procedure that carries significant morbidity and mortality. LMO4 status can potentially be assessed using preoperative biopsy material without resection using techniques such as PCR, which overcome the problems of low tissue yield associated with fine-needle aspirate biopsies. Other factors such as tumour size, resection margins, perineural invasion and lymph node status are determined after resection. The ability to reliably predict which patients will or will not benefit from surgery would be a significant advance in the treatment of PC. LMO4 may have a potential role as a marker for determining the suitability for resection and the prognosis of patients with PC.
Deregulation of LMO4 has been previously described in several tumour types including breast, prostate and squamous cell carcinoma of the oral cavity (Visvader et al, 2001; Mousses et al, 2002; Mizunuma et al, 2003). Like the other LMO family members, LMO4 is a transcriptional cofactor that functions as a scaffold for the generation of multiprotein complexes (Visvader et al, 2001). Several LMO4-interacting proteins have been identified, including the ubiquitous nuclear adaptor protein Lbd1 (Grutz et al, 1998), the transcription factor deformed epidermal autoregulatory factor 1 (Sugihara et al, 1998), the basic helix-loop-helix protein HEN1 (Manetopoulos et al, 2003) and the grainyhead-like epithelial transactivator (Kudryavtseva et al, 2003), as well as the cofactor CtIP and BRCA1.
In conclusion, current prognostic markers in PCs remain poorly defined and cannot be determined preoperatively. Assessment of LMO4 status may provide an additional method for determining the suitability for resection and the prognosis of patients with PC. Although this study demonstrates the potential utility of LMO4 as a prognostic marker for PC, validation of these data is required in large independent cohorts. Further studies to elucidate the pathways regulated by LMO4 and mechanisms by which LMO4 contributes to the development and progression of PC will be an essential step toward further assessing the potential clinical utility of LMO4 expression levels in PC.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia, The Cancer Council New South Wales, the St Vincent's Clinic Foundation, the Royal Australasian College of Surgeons, the Prostate Cancer Foundation of Australia, the R. T. Hall Trust and the Victorian Breast Cancer Research Consortium. AVB, CJS, EAM and SMH are Cancer Institute New South Wales Fellows. We thank Dr Catherine Langusch and Ms Tristan Barnes for their help with the maintenance of the Garvan Institute of Medical Research Pancreatic Cancer Tissue Bank and clinical database.
References
- Biankin AV, Morey AL, Lee C-S, Kench JG, Biankin SA, Head DR, Hugh TB, Sutherland RL, Henshall SM (2002) DPC4/Smad4 expression and outcome in pancreatic ductal adenocarcinoma. J Clin Oncol 20: 4531–4542 [DOI] [PubMed] [Google Scholar]
- Grutz G, Forster A, Rabbitts TH (1998) Identification of the LMO4 gene encoding an interaction partner of the LIM-binding protein LDB1/NLI1: a candidate for displacement by LMO proteins in T cell acute leukaemia. Oncogene 17: 2799–2803 [DOI] [PubMed] [Google Scholar]
- Kudryavtseva EI, Sugihara TM, Wang N, Lasso RJ, Gudnason JF, Lipkin SM, Andersen B (2003) Identification and characterization of Grainyhead-like epithelial transactivator (GET-1), a novel mammalian Grainyhead-like factor. Dev Dyn 226: 604–617 [DOI] [PubMed] [Google Scholar]
- Manetopoulos C, Hansson A, Karlsson J, Jonsson JI, Axelson H (2003) The LIM-only protein LMO4 modulates the transcriptional activity of HEN1. Biochem Biophys Res Commun 307: 891–899 [DOI] [PubMed] [Google Scholar]
- Mizunuma H, Miyazawa J, Sanada K, Imai K (2003) The LIM-only protein, LMO4, and the LIM domain-binding protein, LDB1, expression in squamous cell carcinomas of the oral cavity. Br J Cancer 88: 1543–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousses S, Bubendorf L, Wagner U, Hostetter G, Kononen J, Cornelison R, Goldberger N, Elkahloun AG, Willi N, Koivisto P, Ferhle W, Raffeld M, Sauter G, Kallioniemi OP (2002) Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res 62: 1256–1260 [PubMed] [Google Scholar]
- Rabbitts TH (1998) LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev 12: 2651–2657 [DOI] [PubMed] [Google Scholar]
- Sugihara TM, Bach I, Kioussi C, Rosenfeld MG, Andersen B (1998) Mouse deformed epidermal autoregulatory factor 1 recruits a LIM domain factor, LMO-4, and CLIM coregulators. Proc Natl Acad Sci USA 95: 15418–15423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sum EY, O'Reilly LA, Jonas N, Lindeman GJ, Visvader JE (2005a) The LIM domain protein LMO4 is highly expressed in proliferating mouse epithelial tissues. J Histochem Cytochem 53: 475–486 [DOI] [PubMed] [Google Scholar]
- Sum EY, Peng B, Yu X, Chen J, Byrne J, Lindeman GJ, Visvader JE (2002) The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J Biol Chem 277: 7849–7856 [DOI] [PubMed] [Google Scholar]
- Sum EY, Segara D, Duscio B, Bath ML, Field AS, Sutherland RL, Lindeman GJ, Visvader JE (2005b) Overexpression of LMO4 induces mammary hyperplasia, promotes cell invasion, and is a predictor of poor outcome in breast cancer. Proc Natl Acad Sci USA 102: 7659–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Venter D, Hahm K, Santamaria M, Sum EY, O'Reilly L, White D, Williams R, Armes J, Lindeman GJ (2001) The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci USA 98: 14452–14457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo CJ, Cameron JL, Sohn TA, Lillemoe KD, Pitt HA, Talamini MA, Hruban RH, Ord SE, Sauter PK, Coleman J, Zahurak ML, Grochow LB, Abrams RA (1997) Six hundred fifty consecutive pancreaticoduodenectomies in the 1990s: pathology, complications, and outcomes. Ann Surg 226: 248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]


