Abstract
Epidermal desquamation accounts for 20% of the body's iron loss each day. Yet, little is known about how iron content in epidermis is regulated. To test the importance of the transferrin receptor in regulating iron content in epidermis, we created transgenic mice that have stratum-specific expression of the human transferrin receptor. The keratin 14 promoter targeted the receptor primarily to basal, proliferating keratinocytes; the involucrin (Inv) promoter targeted the receptor to suprabasal, differentiating keratinocytes. There were age- and site-dependent differences in iron content in the epidermis and hair. In both types of transgenic mice, epidermal iron content increased with age and at 8 weeks was 2−3-fold greater in transgenic mice compared to littermate controls. Iron was increased up to 2-fold in hair of keratin 14-human transferrin receptor (hTfR) transgenics and 30% in Inv-hTfR transgenics. No gross or histological changes were seen in transgenic animals with increased iron in the epidermis. Ferritin expression, which was low in normal epidermis, was greatly increased in both transgenic lines, indicating that it is the likely depot for the extra iron in these animals. These data show that control of transferrin receptor expression is sufficient to regulate iron content in proliferating or differentiating keratinocytes in the epidermis.
INTRODUCTION
Iron normally does not accumulate in the body, even though there is limited capacity to rid the body of excess iron. The 20−25% of absorbed iron that is lost through skin is eliminated through desquamation of keratinocytes (Weintraub et al., 1965; Jacob et al., 1981); very little iron is lost through sweat (Brune et al., 1986). We have proposed that if a systemic toxin could be made to accumulate in the epidermis, then epidermal desquamation might be harnessed to reduce the body burden of that toxin. We have chosen iron as a model toxin. One way to test the hypothesis is to create transgenic mice that accumulate iron in the epidermis so that they could be bred to mice with hereditary hemochromatosis.
Transferrin receptor endocytosis is the major pathway for iron uptake into cultured cells (Klausner et al., 1993; Hentze and Kuhn, 1996). Intracellular iron regulates surface expression of the transferrin receptor through post-transcriptional mechanisms (Owen and Kuhn, 1987) and through endosomal recycling (Callus et al., 1996). Transferrin receptor expression is reduced on differentiated cells in tissue sections from a variety of organs (Gatter et al., 1983), and its synthesis is reduced by transcriptional regulation following experimental induction of differentiation in hematopoietic cells (Ho et al., 1986). In cells transfected with transferrin receptor cDNA, there is a correlation between transferrin receptor expression and iron uptake (Callus et al., 1996; Luttropp et al., 1998).
Expression of the transferrin receptor is limited to the basal layer of the epidermis (Gatter et al., 1983). Its absence on differentiated cells and on stem cells (Tani et al., 2000) suggests that the transferrin receptor is an important determinant of the amount and location of iron in the epidermis.
We report here the production and analysis of transgenic mice that express the human transferrin receptor in a stratum-specific manner in the epidermis and in sufficient amount to overcome homeostatic signals that control iron stores in the epidermis.
RESULTS
Analysis of K14-hTfR transgenic mice
Two independent lines of mice were established using the keratin 14 (K14)-human transferrin receptor (hTfR) construct (Figure 1a), after demonstrating that it could direct synthesis of the human transferrin receptor in transfected murine keratinocytes (Supplementary Figure 1a). Each line had litters with the expected number of males and females, and the expected number of transgenic and wild-type littermates. Both lines have been bred for at least six generations; both demonstrate quantitatively and qualitatively similar expression of hTfR and effects on epidermal iron. Descriptions will therefore be limited to line A.
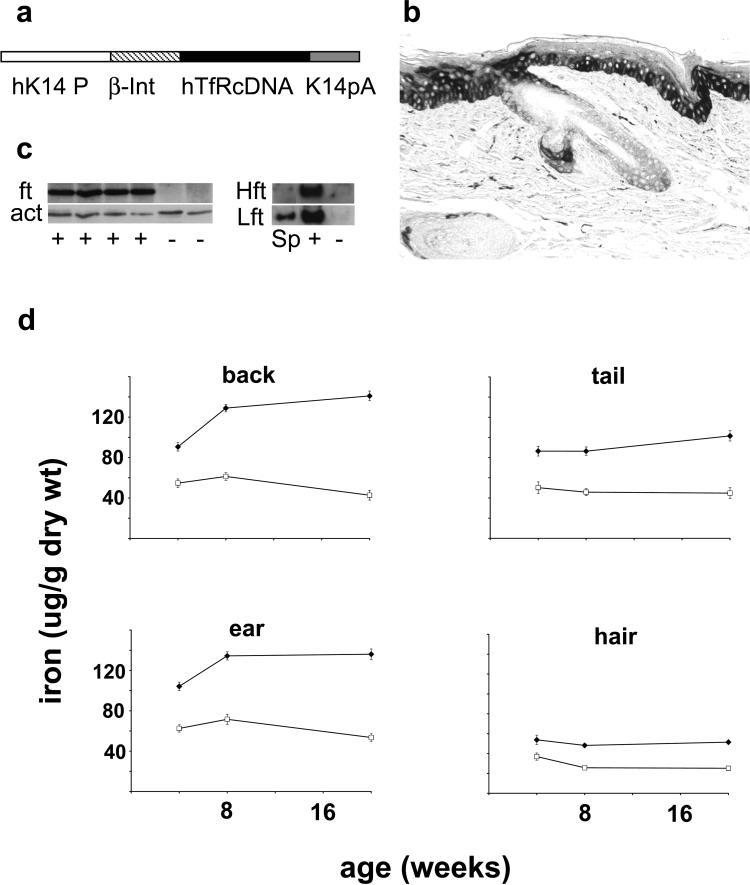
Figure 1. Basal layer expression of human transferrin receptor in K14-hTfR transgenic mice increases iron in epidermis.
(a) The transgene consists of the human transferrin receptor coding sequence (hTfRcDNA) in an expression cassette that contains a human keratin promoter (hK14 P), a β-globin Intron (β-Int), and human K14 polyA (K14pA). (b) Immunohistochemistry of tail skin using antibody that reacts with human transferrin receptor but not normal mouse epidermis (see Figure S1d). (c) Western blots of epidermal extracts from K14-hTfR transgenic (+) or control littermates (−) were probed (left panel) for total ferritin (ft) and reprobed for actin (act) or were probed (right panel) for heavy (Hft) and light (Lft) chains of ferritin using spleen (Sp) as control. (d) Iron in the epidermis and hair of K14-hTfR transgenic mice. Iron was measured in epidermis from various locations and in hair. Each point is the mean±SD from eight to 13 animals. Iron content in samples from transgenic mice (◆) was significantly greater (at least P≤0.008) than wild-type littermates (□) at all locations and at all times.
Expression of the human transferrin receptor was highest in the basal layer of the epidermis and the outer root sheath of the hair follicle (Figure 1b); expression showed localized extension into the spinous layer of the epidermis. Human transferrin receptor was undetectable in the epidermis of wild-type littermates (Figures S1d). Newly synthesized hTfR was identified in cultured keratinocytes from transgenic but not wild-type mice (Figures S1b). Reverse transcriptase-PCR revealed the expected amplicon in the RNA from transgenic but not wild-type epidermis (Figures S1e).
There were no gross phenotypic or histological differences between skin from transgenic and wild-type littermates. In particular, there were no differences in thickness, organization, or differentiation of the epidermis in specimens harvested from different anatomic locations of animals up to 40 weeks of age (data not shown).
Epidermal iron content was considerably greater in the K14-hTfR transgenic mice than in wild-type littermates (Figure 1d). Relative differences in epidermal iron between transgenic and control mice were dependent on age and anatomic location. Iron content in tail epidermis was generally lower than iron content in epidermis from other locations in both transgenic mice and controls. Epidermal iron content in the K14-hTfR transgenic mice increased at most body sites between 4 and 20 weeks of age. By contrast, epidermal iron content in wild-type littermates remained the same or decreased slightly. As a result, iron content in the dorsal epidermis of K14-hTfR transgenics was 1.7-fold greater than in controls at 4 weeks, but 3.3-fold greater than controls at 20 weeks. Iron content in hair of the K14-hTfR transgenic mice was as much as 2-fold greater than that in controls at all times and both locations sampled (dorsal and ventral hair). Iron content of the liver was equal in transgenic and wild-type littermates, was greater in females than males, and slowly increased over time in the females (data not shown).
Total ferritin, ferritin heavy chain, and ferritin light chain were greatly increased in epidermal extracts from K14-hTfR transgenic mice compared to wild-type littermates (Figure 1c).
Analysis Inv-hTfR transgenic mice
Two independent lines of mice were established using the involucrin (Inv)-hTfR construct (Figure 2a), after demonstrating that it could direct synthesis of the human transferrin receptor in transfected murine keratinocytes (Figure S2a). Each line had litters with the expected number of males and females, and the expected number of transgenic and wild-type littermates. Both lines have been bred for at least six generations; both demonstrate quantitatively and qualitatively similar expression of hTfR and effects on epidermal iron. Line B bred poorly, so descriptions will be limited to line A.
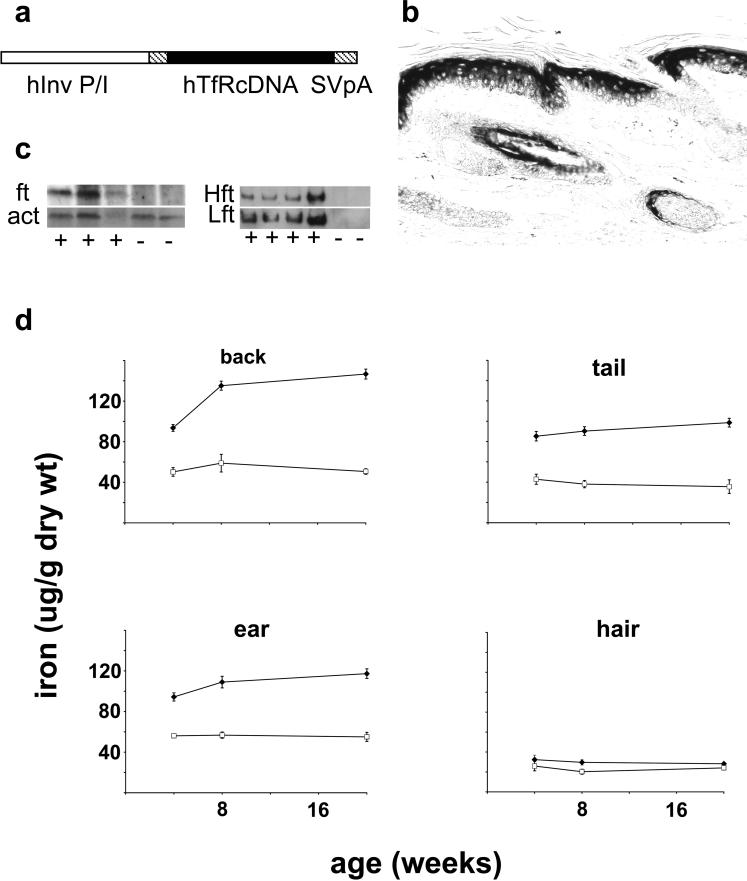
Figure 2. Suprabasal expression of human transferrin receptor in Inv-hTfR transgenic mice increases iron in the epidermis.
(a) The transgene consists of the human transferrin receptor coding sequence (hTfRcDNA) in an expression cassette that contains a human involucrin promoter/intron (hInv P/I), and an SV40 polyA (SVpA). (b) Immunohistochemistry of tail skin using antibody that reacts with hTfR but not normal mouse epidermis (see Figures S2d). (c) Western blots of epidermal extracts from Inv-hTfR transgenic (+) or control littermates (−) were probed (left panel) for ferritin (ft) and reprobed for actin (act) or were probed (right panel) for heavy (Hft) and light (Lft) chains of ferritin using spleen (Sp) as control. (d) Iron in the epidermis and hair of Inv-hTfR transgenic mice. Iron was measured in the epidermis from various locations and in hair. Each point is the mean±SD from six to 10 animals. Iron content in samples from transgenic mice (◆) was significantly greater (at least P≤0.04) than wild-type littermates (□) at all locations and at all times.
Expression of the human transferrin receptor was highest in suprabasal cells in the epidermis from the first layer above the basal layer extending to the stratum corneum (Figure 2b). There was intense staining of the inner root sheath and in some matrix cells in the hair follicle. Human transferrin receptor was undetectable in the epidermis of wild-type littermates (Figure S2d). Newly synthesized hTfR was identified in cultured keratinocytes from transgenic but not wild-type mice, and was greatly increased by subjecting the cells to calcium-induced differentiation (Figure S2b). Reverse transcriptase-PCR revealed the expected amplicon in the RNA from transgenic but not wild-type epidermis (Figure S2e).
There were no gross phenotypic or histological differences between the transgenic animals and their wild-type littermates. In particular, there were no discernable differences in thickness, organization, or differentiation of the epidermis in specimens harvested from different anatomic locations of animals up to 40 weeks of age (data not shown).
Epidermal iron content was considerably greater in the Inv-hTfR transgenic mice compared to wild-type littermates (Figure 2d). Relative differences in epidermal iron between transgenic and control mice were dependent on age and anatomic location. Iron content in tail epidermis was generally lower than iron content in the epidermis from other locations in both transgenic mice and controls. Epidermal iron content in the Inv-hTfR transgenic mice increased at most body sites between 4 and 20 weeks of age. By contrast, epidermal iron content in wild-type littermates remained the same. As a result, iron content in the dorsal epidermis of transgenics was 1.9-fold greater than in controls at 4 weeks, but 2.9-fold greater than controls at 20 weeks. Iron content in hair of the Inv-hTfR transgenic mice was as much as 50% greater than that in controls at all times and from both locations sampled (dorsal and ventral hair). Iron content of liver was the same in transgenic animals and wild-type littermates (data not shown).
Total ferritin, ferritin heavy chain, and ferritin light chain were greatly increased in epidermal extracts from Inv-hTfR transgenic mice compared to wild-type littermates (Figure 2c).
DISCUSSION
Control of transferrin receptor expression is sufficient to regulate iron content in vivo, as shown by our analysis of transgenic mice that express human transferrin receptor 1 in the epidermis. This result was not a foregone conclusion: there have been no previous reports of transferrin receptor transgenic mice and there is increasing recognition that iron homeostasis involves multiple pathways for iron uptake, secretion, and hormonal regulation (Hentze et al., 2004).
The finding of increased iron in the epidermis of mice with stratum-specific expression of the transgene has several implications. Proteins needed for appropriate transferrin receptor trafficking or endosome recycling must be present in sufficient excess to allow increased iron uptake and storage. Moreover, proteins that traffic with transferrin receptor apparently are not coordinately downregulated by increased intracellular iron nor by the differentiation program that normally would downregulate transferrin receptor expression.
Epidermal iron has been measured in several diseases, and our results have implications for the interpretation of those data. As transferrin receptor-mediated transport of iron into keratinocytes appears to be a critical determinant of iron content in keratinocytes and as transferrin receptor expression normally is restricted to proliferating basal keratinocytes, any condition that increases the proportion of proliferating keratinocytes in the epidermis should be expected to increase iron content in the epidermis. Psoriasis and, to a lesser degree, dermatitis are two diseases in which the epidermis is hyperproliferative and may show evidence of proliferating, suprabasal keratinocytes. Data demonstrating increased iron in the epidermis in psoriasis and dermatitis (Forslind et al., 1999) may be a direct consequence of a change in the proportion of proliferating to differentiating cells, rather than an indication that iron has a role in the pathogenesis of those diseases.
Elevated ferritin in the epidermis of transgenic animals indicates that the epidermal response to increased intracellular iron is physiologically appropriate, even if the (transgenic) cause of its increase is not. In human skin, ferritin normally is detected only in the basal cell layer, but is increased and readily identified in all epidermal layers in response to UV light (Applegate et al., 1998; Seite et al., 2004). The increased ferritin in the K14-hTfR and Inv-hTfR transgenic animals proves that both proliferating and differentiating keratinocytes are capable of responding to an iron load by accumulating ferritin. The increase in both subunits of ferritin in the transgenic epidermis is consistent with current understanding that the H-chain is necessary to oxidize newly absorbed iron and that the L-chain is increased in tissues that store iron (Harrison and Arosio, 1996).
Iron-overloaded animals have increased susceptibility to inflammatory (Reelfs et al., 2004) and neoplastic skin disease (Bhasin et al., 2003). The K14-hTfR and Inv-hTfR transgenic mice that accumulate iron in the epidermis should be useful in distinguishing the effects of local from systemic iron overload. These animals should also be a useful tool in further defining the role of epidermal desquamation in iron homeostasis.
MATERIALS AND METHODS
Animals
The K14-hTfR mice were derived by the Yale Animal Research Core transgenic facility (Yale University School of Medicine, New Haven, CT) in C57Bl/6J × SJL/J mice. From 24 pups, six were positive for the transgene by dot blot. Inv-hTfR mice were derived by the Yale Immunobiology mouse transgenic facility in C57Bl/6J mice. From 100 pups, 25 were positive for the transgene by dot blot. All mouse production and experimentation was performed in compliance with guidelines of the Yale Institutional Animal Care and Use Committee. All animals reported here were fed standard mouse chow containing 0.226 g/kg iron (TD2018; Harlan Teklad, Madison, WI) and have been backcrossed into C57Bl/6J for at least six generations. Transgenic and non-transgenic littermates were housed together, but separated by sex.
Cells
The epidermis is separated from the dermis by overnight exposure to Dispase II (165−859; Roche, Indianapolis, IN) at 4°C (Kitano and Okada, 1983), and a single-cell suspension of keratinocytes obtained by brief exposure to trypsin:EDTA. Human foreskin keratinocytes were grown in low-calcium, serum-free medium (Zhou et al., 1999). Murine BALB/c keratinocytes and keratinocytes isolated from transgenic pups were grown in low-calcium, serum-containing medium (Bruegel Sanchez et al., 2004).
Transgene construction
Human transferrin receptor 1 cDNA (pCDTR-1; ATCC, Manassas, VA), free of 3′ and 5′ regulatory elements (hereafter called hTfR) (Luttropp et al., 1998), was inserted into the Inv promoter cassette provided by Lorne Taichman (Carroll et al., 1993) or into a K14 promoter cassette provided by Elaine Fuchs (Hutton et al., 1998).
Genomic dot blots
Genomic DNA, spotted on Hybond-N+ (RPN203B; Amersham, Piscataway, NJ), was probed using a 32P-labeled, 800 bp, Xba/HindIII fragment from the human transferrin cDNA that does not hybridize to murine DNA.
PCR
The hTfR transgene was detected by 30 cycles of PCR on genomic DNA, using Taq polymerase (M1661, Promega) and primers for the hTfR coding sequence that do not amplify murine DNA. Reverse transcriptase-PCR for hTfR in cellular RNA (TRIzol, Invitrogen, Carlsbad, CA) used the same primers and primers for glyceraldehyde-3-phosphate dehydrogenase (details in Figures S1 and S2).
Human transferrin receptor synthesis
Keratinocytes from transgenic pups or BALB/c keratinocytes transfected with K14-hTfR and INV-hTfR transgenes using Lipofectamine (Invitrogen, Carlsbad, CA) were labeled with Tran35S-Label (370 MBq/ml; ICN, Aurora, OH). Six hours later, cells were extracted in TENP (50 mm Tris, pH 8.0, 5 mm EDTA, 1% NP-40, 0.15 m NaCl, and protease inhibitors). Protein (500 μg) was incubated with 1 μg of antibody B3/25 (Omary et al., 1980), specific for human TfR (Roche, Indianapolis, IN) and protein A beads (Amersham, Piscataway, NJ) overnight at 4°C. After washing three times with TENP containing 0.4 m NaCl, the eluted receptor was resolved by 10% SDS-PAGE and detected by autoradiography.
Histology and immunohistochemistry
Tissues were fixed in Bouin's solution, embedded in paraffin, sectioned at 5 μm, deparaffinized and stained with hematoxylin and eosin or an anti-human transferrin receptor antibody (MS-1096-S1; NeoMarkers, Freemont, CA) and a silver-enhanced secondary antibody (RPN471; Amersham).
Iron measurement
Plucked hair and dispase-separated epidermis from 1 × 2 cm pieces of skin were rinsed in PBS, blotted, and then dried overnight at 65°C. Non-heme iron was measured using a dye binding assay (Torrance and Bothwell, 1980), adapted to a microtiter plate format (Adams BD et al. J Invest Dermatol (in press)). Data for the epidermis and hair were pooled from males and females, as there was no sex difference when analyzed separately. Liver data were separated by sex.
Ferritin measurement
Epidermal sheets were extracted by sonication on ice in 10 mm Tris, pH 7.4. containing 1% SDS and 1 mm dithiothreitol and protease inhibitors. Protein was measured by a modification of the Bramhall assay (Milstone et al., 1982), and 20 μg separated by 15% SDS-PAGE. After protein transfer for 3.5 hours at 100 mA, the 21 kDa ferritin band was identified using a rabbit, anti-horse ferritin antibody at 1:1,750 (F-5762; Sigma, St Louis, MO), a goat anti-rabbit horseradish peroxidase-antibody at 1:5,000 (NA934; Amersham), and an ECL kit (RPN2106; Amersham) and X-ray film (870−1302; Kodak, Rochester, NY). Protein loading was monitored by stripping and reprobing the blot using a rabbit anti-mouse actin antibody at 1:2,000 (A-2066; Sigma). For the estimation of H- and L-chain of ferritin, epidermis was extracted with 20 mm Tris, pH 7.4 plus protease inhibitors and then 20 μg separated on non-denaturing, 7% polyacrylamide gels run for 3 hours at 100 V (Santambrogio et al., 2000). Protein was transferred to polyvinylidene difluoride membrane (162−0176; Biorad, Hercules, CA) for 4 hours at 215 mA. The H- and L-chain-specific rabbit, anti-ferritin antibodies, a kind gift from Paolo Santambrogio (Scientific Institute of Recovery and Care, Milan, Italy) were used at 1:2,000.
ACKNOWLEDGMENTS
We are grateful to Nancy Andrews and her lab for ongoing advice and encouragement. We thank Elaine Fuchs for the K14 expression cassette, Lorne Taichman for the involucrin expression cassette, Kathryn Morton for the SV40-hTfR expression plasmid, Paolo Santambrogio and Sonia Levy for the anti-ferritin antibodies, and Julie Miao for contributions to standardizing hair analysis. This work was supported by a grant from NIAMS R01-AR47303.
Abbreviations
- hTfR
human transferrin receptor
- K14
keratin 14
- Inv
involucrin
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
Supplementary Material
SUPPLEMENTARY MATERIAL
Figure S1. (a) Transgene expression in transfected mouse keratinocytes. Cultured cells were labeled with 35S. Extracted protein was immunoprecipitated with antibody B3/25 and human transferrin receptor identified on SDS-PAGE by autoradiography. HK–positive control of human keratinocytes. MK – negative control of BALB/c keratinocytes. MK+K14hTFR – BALB/c keratinocytes transfected with the hTfR construct and labeled 72 hours later. (b) Synthesis of human transferrin receptor in keratinocytes isolated from K14-hTfR transgenic mice. Cell lines were established from six littermates, labeled with 35S, and analyzed for human transferrin receptor synthesis as in (a). PCR of genomic DNA using primers specific for the human transferrin receptor distinguished transgenic (pcr+) from wild-type (pcr–) littermates. The labeled band at approximately 92 kDa, the expected size of the human transferrin receptor (hTfR), was seen in only the pcr+ cells. A non-specifically precipitating protein was seen in all lanes, and serves as an internal loading control. (c and d) Expression of hTfR is limited to lower layers of epidermis in transgenic mice. Immunohistochemistry, using Neomarker antibody to human transferrin receptor, was done on tail skin from a K14-hTfR transgenic mouse. Expression is most intense in the basal layer of the epidermis and sebaceous glands and the outer root sheath of hair follicles of transgenic animals (c), but is absent from the upper layers of epidermis and totally absent from skin of non-transgenic littermates (d). (e) mRNA for the hTfR transgene is found in the epidermis only in K14-hTfR transgenic mice. Reverse transcriptase-PCR was performed on RNA extracted from epidermis and liver of littermates. The 180bp pcr-amplified cDNA product was identified in epidermis of two animals that were positive for the transgene in genomic DNA (+) and absent from two animals that were negative for the transgene in genomic DNA (−). RNA from liver (L), which should not express protein from a K14 promoter, did not yield amplified product. In separate reactions, the 525 bp band, using primers for glyceraldehyde-3-phosphate dehydrogenase, verified the presence of amplifiable cDNA. Positive pcr control=genomic DNA(D); negative pcr control (C)=water.
hTfR1: 5′-CTGCTATGGGACTATTGCTGTG-3′.
hTfR2: 5′-CCGACAACTTTCTCTTCAGGTC-3′.
GAPDH1: 5′CTCCTTGGAGGCCATGTAGGCCATG.
GAPDH2: 5′GCAACTGCTTAGGGGGGGTGGCCAG.
Figure S2. (a) Transgene expression in transfected mouse keratinocytes. Cultured cells were labeled with 35S. Extracted protein was immunoprecipitated with antibody B3/25 and human transferrin receptor identified on SDS-PAGE by autoradiography. HK–positive control of human keratinocytes. MK – negative control of BALB/c keratinocytes. MK+Inv-hTFR – BALB/c keratinocytes transfected with the hTfR construct andþlabeled 72 hours later. (b) Synthesis of human transferrin receptor in keratinocytes isolated from Inv-hTfR transgenic mice is regulated by calcium. Cell lines were established from two transgenic animals, cultured for 2 days in either low calcium (lo) or high calcium (hi), labeled with 35S and analyzed for human transferrin receptor synthesis (hTfR) as in (a). The labeled band at approximately 92 kDa, the expected size of the human transferrin receptor, was faint in cells grown in low calcium but strong in cells grown in high calcium. (c and d) Expression of hTfR is limited to the upper layers of the epidermis in transgenic mice. Immunohistochemistry, using Neomarker antibody to human transferrin receptor, was done on sections of tail skin from an Inv-hTfR transgenic mouse. In transgenic animals (c), expression is most intense above the basal layer of the epidermis and the inner root sheath of hairs follicles. Staining was absent from sebaceous glands and the outer root sheath of hair follicles and totally absent from skin of non-transgenic littermates (d). (e) mRNA for the hTfR transgene is found in the epidermis only in Inv-hTfR transgenic mice. Reverse transcriptase-PCR was performed on RNA extracted from the epidermis and liver of littermates. The 180 bp pcr-amplified cDNA product was identified in the epidermis of two animals that were positive for the transgene in genomic DNA (+) and absent from two animals that were negative for the transgene in genomic DNA (−). RNA from liver (L), which should not express protein from an Inv promoter, did not yield amplified product. In separate reactions, the 525 bp band, using primers for glyceraldehyde-3-phosphate dehydrogenase, verified the presence of amplifiable cDNA. Positive pcr control=genomic DNA(D); (see Figure Supplementary 1e for primers used).
REFERENCES
- Applegate LA, Scaletta C, Panizzon R, Frenk E. Evidence that ferritin is UV inducible in human skin: part of a putative defense mechanism. J Invest Dermatol. 1998;111:159–63. doi: 10.1046/j.1523-1747.1998.00254.x. [DOI] [PubMed] [Google Scholar]
- Bhasin G, Kausar H, Sarwar Alam M, Athar M. Progressive iron overload enhances chemically mediated tumor promotion in murine skin. Arch Biochem Biophys. 2003;409:262–73. doi: 10.1016/s0003-9861(02)00616-1. [DOI] [PubMed] [Google Scholar]
- Bruegel Sanchez VL, Zhou J, LaCivita D, Milstone LM. Long-term murine keratinocyte cultures become tetraploid, yet maintain the ability to stratify. J Invest Dermatol. 2004;123:403–4. doi: 10.1111/j.0022-202X.2004.23218.x. [comment] [DOI] [PubMed] [Google Scholar]
- Brune M, Magnusson B, Persson H, Hallberg L. Iron losses in sweat. Am J Clin Nutr. 1986;43:438–43. doi: 10.1093/ajcn/43.3.438. [DOI] [PubMed] [Google Scholar]
- Callus BA, Iacopetta BJ, Kuhn LC, Morgan EH. Effects of overexpression of the transferrin receptor on the rates of transferrin recycling and uptake of non-transferrin-bound iron. Eur J Biochem. 1996;238:463–9. doi: 10.1111/j.1432-1033.1996.0463z.x. [DOI] [PubMed] [Google Scholar]
- Carroll JM, Albers KM, Garlick JA, Harrington R, Taichman LB. Tissue- and stratum-specific expression of the human involucrin promoter in transgenic mice. Proc Natl Acad Sci USA. 1993;90:10270–4. doi: 10.1073/pnas.90.21.10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslind B, Werner-Linde Y, Lindberg M, Pallon J. Elemental analysis mirrors epidermal differentiation. Acta Dermatol Venereol. 1999;79:12–7. doi: 10.1080/000155599750011624. [DOI] [PubMed] [Google Scholar]
- Gatter KC, Brown G, Trowbridge IS, Woolston RE, Mason DY. Transferrin receptors in human tissues: their distribution and possible clinical relevance. J Clin Pathol. 1983;36:539–45. doi: 10.1136/jcp.36.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kuhn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci USA. 1996;93:8175–82. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–97. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- Ho PT, King I, Sartorelli AC. Transcriptional regulation of the transferrin receptor in differentiating HL-60 leukemic cells. Biochem Biophys Res Commun. 1986;138:995–1000. doi: 10.1016/s0006-291x(86)80594-0. [DOI] [PubMed] [Google Scholar]
- Hutton E, Paladini RD, Yu QC, Yen M, Coulombe PA, Fuchs E. Functional differences between keratins of stratified and simple epithelia. J Cell Biol. 1998;143:487–99. doi: 10.1083/jcb.143.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob RA, Sandstead HH, Munoz JM, Klevay LM, Milne DB. Whole body surface loss of trace metals in normal males. Am J Clin Nutr. 1981;34:1379–83. doi: 10.1093/ajcn/34.7.1379. [DOI] [PubMed] [Google Scholar]
- Kitano Y, Okada N. Separation of the epidermal sheet by dispase. Br J Dermatol. 1983;108:555–60. doi: 10.1111/j.1365-2133.1983.tb01056.x. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Rouault TA, Harford JB. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- Luttropp CA, Jackson JA, Jones BJ, Sohn MH, Lynch RE, Morton KA. Uptake of gallium-67 in transfected cells and tumors absent or enriched in the transferrin receptor. J Nucl Med. 1998;39:1405–11. [PubMed] [Google Scholar]
- Milstone LM, McGuire J, LaVigne JF. Retinoic acid causes premature desquamation of cells from confluent cultures of stratified squamous epithelia. J Invest Dermatol. 1982;79:253–60. doi: 10.1111/1523-1747.ep12500073. [DOI] [PubMed] [Google Scholar]
- Omary MB, Trowbridge IS, Minowada J. Human cell-surface glycoprotein with unusual properties. Nature. 1980;286:888–91. doi: 10.1038/286888a0. [DOI] [PubMed] [Google Scholar]
- Owen D, Kuhn LC. Noncoding 3′ sequences of the transferrin receptor gene are required for mRNA regulation by iron. EMBO J. 1987;6:1287–93. doi: 10.1002/j.1460-2075.1987.tb02366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reelfs O, Tyrrell RM, Pourzand C. Ultraviolet a radiation-induced immediate iron release is a key modulator of the activation of NF-kappaB in human skin fibroblasts. J Invest Dermatol. 2004;122:1440–7. doi: 10.1111/j.0022-202X.2004.22620.x. [see comment] [DOI] [PubMed] [Google Scholar]
- Santambrogio P, Cozzi A, Levi S, Rovida E, Magni F, Albertini A, et al. Functional and immunological analysis of recombinant mouse H- and L-ferritins from Escherichia coli. Protein Expr Purif. 2000;19:212–8. doi: 10.1006/prep.2000.1212. [DOI] [PubMed] [Google Scholar]
- Seite S, Popovic E, Verdier MP, Roguet R, Portes P, Cohen C, et al. Iron chelation can modulate UVA-induced lipid peroxidation and ferritin expression in human reconstructed epidermis. Photodermatol Photoimmunol Photomed. 2004;20:47–52. doi: 10.1111/j.1600-0781.2004.00064.x. [DOI] [PubMed] [Google Scholar]
- Tani H, Morris RJ, Kaur P. Enrichment for murine keratinocyte stem cells based on cell surface phenotype. Proc Natl Acad Sci USA. 2000;97:10960–5. doi: 10.1073/pnas.97.20.10960. [see comment] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrance J, Bothwell T, editors. Tissue iron stores (Methods in Hematology, vol 1) Churchill Livingstone; New York: 1980. p. 104. [Google Scholar]
- Weintraub L, Demis D, Conrad M, Crosby W. Iron excretion by the skin. Selective localization of iron in epithelia cells. Am J Pathol. 1965;46:121–7. [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Haggerty JG, Milstone LM. Growth and differentiation regulate CD44 expression on human keratinocytes. In Vitro Cell Dev Biol Anim. 1999;35:228–35. doi: 10.1007/s11626-999-0031-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIAL
Figure S1. (a) Transgene expression in transfected mouse keratinocytes. Cultured cells were labeled with 35S. Extracted protein was immunoprecipitated with antibody B3/25 and human transferrin receptor identified on SDS-PAGE by autoradiography. HK–positive control of human keratinocytes. MK – negative control of BALB/c keratinocytes. MK+K14hTFR – BALB/c keratinocytes transfected with the hTfR construct and labeled 72 hours later. (b) Synthesis of human transferrin receptor in keratinocytes isolated from K14-hTfR transgenic mice. Cell lines were established from six littermates, labeled with 35S, and analyzed for human transferrin receptor synthesis as in (a). PCR of genomic DNA using primers specific for the human transferrin receptor distinguished transgenic (pcr+) from wild-type (pcr–) littermates. The labeled band at approximately 92 kDa, the expected size of the human transferrin receptor (hTfR), was seen in only the pcr+ cells. A non-specifically precipitating protein was seen in all lanes, and serves as an internal loading control. (c and d) Expression of hTfR is limited to lower layers of epidermis in transgenic mice. Immunohistochemistry, using Neomarker antibody to human transferrin receptor, was done on tail skin from a K14-hTfR transgenic mouse. Expression is most intense in the basal layer of the epidermis and sebaceous glands and the outer root sheath of hair follicles of transgenic animals (c), but is absent from the upper layers of epidermis and totally absent from skin of non-transgenic littermates (d). (e) mRNA for the hTfR transgene is found in the epidermis only in K14-hTfR transgenic mice. Reverse transcriptase-PCR was performed on RNA extracted from epidermis and liver of littermates. The 180bp pcr-amplified cDNA product was identified in epidermis of two animals that were positive for the transgene in genomic DNA (+) and absent from two animals that were negative for the transgene in genomic DNA (−). RNA from liver (L), which should not express protein from a K14 promoter, did not yield amplified product. In separate reactions, the 525 bp band, using primers for glyceraldehyde-3-phosphate dehydrogenase, verified the presence of amplifiable cDNA. Positive pcr control=genomic DNA(D); negative pcr control (C)=water.
hTfR1: 5′-CTGCTATGGGACTATTGCTGTG-3′.
hTfR2: 5′-CCGACAACTTTCTCTTCAGGTC-3′.
GAPDH1: 5′CTCCTTGGAGGCCATGTAGGCCATG.
GAPDH2: 5′GCAACTGCTTAGGGGGGGTGGCCAG.
Figure S2. (a) Transgene expression in transfected mouse keratinocytes. Cultured cells were labeled with 35S. Extracted protein was immunoprecipitated with antibody B3/25 and human transferrin receptor identified on SDS-PAGE by autoradiography. HK–positive control of human keratinocytes. MK – negative control of BALB/c keratinocytes. MK+Inv-hTFR – BALB/c keratinocytes transfected with the hTfR construct andþlabeled 72 hours later. (b) Synthesis of human transferrin receptor in keratinocytes isolated from Inv-hTfR transgenic mice is regulated by calcium. Cell lines were established from two transgenic animals, cultured for 2 days in either low calcium (lo) or high calcium (hi), labeled with 35S and analyzed for human transferrin receptor synthesis (hTfR) as in (a). The labeled band at approximately 92 kDa, the expected size of the human transferrin receptor, was faint in cells grown in low calcium but strong in cells grown in high calcium. (c and d) Expression of hTfR is limited to the upper layers of the epidermis in transgenic mice. Immunohistochemistry, using Neomarker antibody to human transferrin receptor, was done on sections of tail skin from an Inv-hTfR transgenic mouse. In transgenic animals (c), expression is most intense above the basal layer of the epidermis and the inner root sheath of hairs follicles. Staining was absent from sebaceous glands and the outer root sheath of hair follicles and totally absent from skin of non-transgenic littermates (d). (e) mRNA for the hTfR transgene is found in the epidermis only in Inv-hTfR transgenic mice. Reverse transcriptase-PCR was performed on RNA extracted from the epidermis and liver of littermates. The 180 bp pcr-amplified cDNA product was identified in the epidermis of two animals that were positive for the transgene in genomic DNA (+) and absent from two animals that were negative for the transgene in genomic DNA (−). RNA from liver (L), which should not express protein from an Inv promoter, did not yield amplified product. In separate reactions, the 525 bp band, using primers for glyceraldehyde-3-phosphate dehydrogenase, verified the presence of amplifiable cDNA. Positive pcr control=genomic DNA(D); (see Figure Supplementary 1e for primers used).