Abstract
We have developed an efficient strategy that combines immunoglobulin (Ig) gene repertoire analysis and Ig reactivity profiling at the single cell level. Based on surface marker expression individual cells at different stages of human B cell development are isolated by fluorescence-activated cell sorting. For each cell Ig heavy and corresponding Ig light chain gene transcripts are amplified by nested RT-PCR and cloned into eukaryotic expression vectors to produce monoclonal human antibodies of the same specificity in vitro. All reactions are performed in 96-well plates and allow cloning of large numbers of Ig genes. The recombinant antibodies are tested for reactivity with diverse self-and non-self antigens and the reactivity profile can be directly linked to the complete Ig heavy and IgL chain gene sequence information that is obtained as part of the cloning strategy. In summary, our method to clone and express human monoclonal antibodies is unbiased, highly efficient, requires only small cell numbers and the recombinant antibodies allow direct conclusions on the frequency of specific human B cells in a diverse repertoire.
Keywords: antibody, single cell PCR, self-reactivity, polyreactivity
Introduction
The diversity of the antibody repertoire is based on somatic recombination of variable (V), diversity (D) and joining (J) gene segments (Tonegawa, 1983). In humans, immunoglobulin (Ig) genes are randomly assembled from about 50 V, 25 D and 6 J gene segments for heavy (H) chains and over 30 potentially functional Vκ and Vλ light (L) chain genes and 5 Jκ and 4 Jλ genes, respectively (Ravetch et al., 1981; Hieter et al., 1982; Schable and Zachau, 1993; Corbett et al., 1997; Kawasaki et al., 1997; Matsuda et al., 1998). Basic understanding of the expressed human antibody repertoire at different stages during B cell development came from Ig gene analyses from cDNA libraries of bulk isolated human B cell subpopulations and single B cells of defined origin (Huang and Stollar, 1991; Huang et al., 1992; Shiokawa et al., 1999; Wang and Stollar, 2000). Other studies compared the Ig gene usage and Ig gene characteristics among distinct B cells subpopulations from healthy individuals and patients with autoimmune diseases and demonstrated that differences in the overall representation of individual Ig genes are part of the normal B cell repertoire (Stewart et al., 1993; Demaison et al., 1995; Pascual and Capra, 1995; Demaison et al., 1996; Dorner et al., 1998; Dorner et al., 1999; Hansen et al., 2000).
Analyses of Ig genes from single B cells on a genomic level gave further insight in the molecular basics of Ig gene recombination, allelic exclusion and selection events in the antibody repertoire (Ehlich et al., 1993; Kuppers et al., 1993; Brezinschek et al., 1995; Ghia et al., 1996; Brezinschek et al., 1997; Casellas et al., 2001). However, sequence analysis alone does not allow predictions on antibody reactivities. To date human antibodies of defined specificities were obtained in large by cloning Ig genes from hybridomas or Epstein-Barr virus (EBV) transformed human B cells. Obtaining these stable human B cell lines involves low efficiency and therefore highly selective procedures (Stahli et al., 1980; Aman et al., 1984; Redmond et al., 1986; Borrebaeck et al., 1988; Crain et al., 1989; Laffly and Sodoyer, 2005). Thus, they cannot be used to document the frequency of B cells with defined specificities or Ig gene usage in individual B cell populations. To overcome these limitations we developed a strategy to clone and express antibodies from single human B cells of defined origin (Wardemann et al., 2003). The recombinant antibodies can be generated in large amounts to provide sufficient material for testing in various assays (Wardemann et al., 2003; Meffre et al., 2004; Ng et al., 2004; Herve et al., 2005; Samuels et al., 2005; Yurasov et al., 2005; Tsuiji et al., 2006; Yurasov et al., 2006; Herve et al., 2007; Tiller et al., 2007).
Materials and methods
Isolation of single human B cells by Fluorescence Activated Cell Sorting
Human samples were collected after signed informed consent in accordance with Institutional Review Board (IRB)-reviewed protocols. Mononuclear cells were isolated from peripheral venous blood or bone marrow after enrichment with RosetteSep® human B cell enrichment cocktail (Stemcell Technologies Inc.) and purified by Ficoll-Paque (GE Healthcare) density gradient centrifugation according to the manufacturer’s instructions. The RosetteSep® human B cell enrichment antibody cocktail (Stemcell Technologies Inc.) crosslinks non-B cells to multiple red blood cells (RBCs), forming immunorosettes which pellet along with free RBCs when centrifuged over a buoyant density medium. If necessary, for example due to low B cell counts in samples from patients with autoimmune disease, B cells were enriched using anti-CD19 magnetic beads (Miltenyi Biotech). Purified mononuclear cells were stained on ice with anti-human antibodies (Becton Dickinson) directly coupled to fluorescein-isothiocyanate (FITC), phycoerythrin (PE), allophycocyanin (APC), or biotin to distinguish among individual B cell subpopulations (Table 1). Biotinylated antibodies were detected using Streptavidin-PECy7 (CALTAG). Single cells were sorted on a FACSVantage (Becton Dickinson) excluding cell duplets into 96-well PCR plates (Eppendorf) containing 4 μl/well of ice-cold 0.5× phosphate-buffered saline (PBS) containing 10 mM DTT, 8 U RNAsin (Promega), 0.4 U 5′-3′ Prime RNAse Inhibitor (Eppendorf). Plates were sealed with Microseal® ‘F’ Film (BioRad) and immediately frozen on dry ice before storage at −80°C.
Table 1.
Phenotype of single isolated human B cells.
| Source | B cell population | Phenotype |
|---|---|---|
| Bone marrow | Early immature | CD19+CD34−CD10+IgM− |
| Immature | CD19+CD34−CD10+IgM+ | |
| Peripheral blood | New emigrant | CD19+CD10+CD27−IgM+ |
| Mature naive | CD19+CD10−CD27−IgM+ | |
| IgM+ memory | CD19+CD10−CD27+IgM+ | |
| IgG+ memory | CD19+CD10−CD27+IgG+ |
Single cell RT-PCR and Ig gene amplification
The RT-PCR protocol was carried out manually. cDNA was synthesized in a total volume of 14 μl/well in the original 96-well sorting plate. Total RNA from single cells was reverse transcribed in nuclease-free water (Eppendorf) using 150 ng random hexamer primer (pd(N)6, GE Healthcare), 0.5 μl of 10 mM each nucleotide dNTP-Mix (Invitrogen), 1 μl 0.1 M DTT (Invitrogen), 0.5% v/v Igepal CA-630 (Sigma), 4 U RNAsin (Promega), 6 U Prime RNAse Inhibitor (Eppendorf) and 50 U Superscript® III reverse transcriptase (Invitrogen). Reverse transcription (RT) reaction was performed at 42°C for 10 min, 25°C for 10 min, 50°C for 60 min and 94°C for 5 min. cDNA was stored at −20°C. IgH, Igλ and Igκ V gene transcripts were amplified independently by nested PCR starting from 3.5 μl of cDNA as template. All PCR reactions were performed in 96-well plates in a total volume of 40 μl per well containing 20 nM each primer or primer mix (Table 2), 300 nM each dNTP (Invitrogen) and 1.2 U HotStar® Taq DNA polymerase (Qiagen). HotStar®Taq DNA polymerase with an error rate of ~2 ×10−5/nucleotide and cycle was preferred over high fidelity enzymes with a 10× lower error rate because of its high amplification efficiency for low copy templates from single cells. All primers were stored in small aliquots to avoid repeated freezing and thawing and all PCRs were performed with nuclease-free water. All nested PCR reactions with gene-specific primers or primer mixes were performed with 3.5 μl of unpurified first PCR product (Fig. 1 and Table 2). Each round of PCR was performed for 50 cycles at 94°C for 30 sec, 58°C (IgH/Igκ) or 60°C (Igλ) for 30 sec, 72°C for 55 sec (1st PCR) or 45 sec (2nd PCR).
Table 2.
Primer Sequences
| Forward Primer | 5′ - 3′ sequence |
|---|---|
| 5′ L-VH 1 | ACAGGTGCCCACTCCCAGGTGCAG |
| 5′ L-VH 3 | AAGGTGTCCAGTGTGARGTGCAG |
| 5′ L-VH 4/6 | CCCAGATGGGTCCTGTCCCAGGTGCAG |
| 5′ L-VH 5 | CAAGGAGTCTGTTCCGAGGTGCAG |
|
| |
| 5′ AgeI VH1 | CTGCAACCGGTGTACATTCCCAGGTGCAGCTGGTGCAG |
| 5′ AgeI VH1/5 | CTGCAACCGGTGTACATTCCGAGGTGCAGCTGGTGCAG |
| 5′ AgeI VH3 | CTGCAACCGGTGTACATTCTGAGGTGCAGCTGGTGGAG |
| 5′ AgeI VH3-23 | CTGCAACCGGTGTACATTCTGAGGTGCAGCTGTTGGAG |
| 5′ AgeI VH4 | CTGCAACCGGTGTACATTCCCAGGTGCAGCTGCAGGAG |
| 5′ AgeI VH 4-34 | CTGCAACCGGTGTACATTCCCAGGTGCAGCTACAGCAGTG |
|
| |
| 5′ AgeI VH 1-18 | CTGCAACCGGTGTACATTCCCAGGTTCAGCTGGTGCAG |
| 5′ AgeI VH 1-24 | CTGCAACCGGTGTACATTCCCAGGTCCAGCTGGTACAG |
| 5′ AgeI VH3-33 | CTGCAACCGGTGTACATTCTCAGGTGCAGCTGGTGGAG |
| 5′ AgeI VH 3-9 | CTGCAACCGGTGTACATTCTGAAGTGCAGCTGGTGGAG |
| 5′ AgeI VH4-39 | CTGCAACCGGTGTACATTCCCAGCTGCAGCTGCAGGAG |
| 5′ AgeI VH 6-1 | CTGCAACCGGTGTACATTCCCAGGTACAGCTGCAGCAG |
|
| |
| 5′ L Vκ 1/2 | ATGAGGSTCCCYGCTCAGCTGCTGG |
| 5′ L Vκ 3 | CTCTTCCTCCTGCTACTCTGGCTCCCAG |
| 5′ L Vκ 4 | ATTTCTCTGTTGCTCTGGATCTCTG |
|
| |
| 5′ Pan Vκ | ATGACCCAGWCTCCABYCWCCCTG |
|
| |
| 5′ AgeI Vκ 1-5 | CTGCAACCGGTGTACATTCTGACATCCAGATGACCCAGTC |
| 5′ AgeI Vκ 1-9 | TTGTGCTGCAACCGGTGTACATTCAGACATCCAGTTGACCCAGTCT |
| 5′ AgeI Vκ 1D-43 | CTGCAACCGGTGTACATTGTGCCATCCGGATGACCCAGTC |
| 5′ AgeI Vκ 2-24 | CTGCAACCGGTGTACATGGGGATATTGTGATGACCCAGAC |
| 5′ AgeI Vκ 2-28 | CTGCAACCGGTGTACATGGGGATATTGTGATGACTCAGTC |
| 5′ AgeI Vκ 2-30 | CTGCAACCGGTGTACATGGGGATGTTGTGATGACTCAGTC |
| 5′ Age Vκ 3-11 | TTGTGCTGCAACCGGTGTACATTCAGAAATTGTGTTGACACAGTC |
| 5′ Age Vκ 3-15 | CTGCAACCGGTGTACATTCAGAAATAGTGATGACGCAGTC |
| 5′ Age Vκ 3-20 | TTGTGCTGCAACCGGTGTACATTCAGAAATTGTGTTGACGCAGTCT |
| 5′ Age Vκ 4-1 | CTGCAACCGGTGTACATTCGGACATCGTGATGACCCAGTC |
|
| |
| 5′ L Vλ 1 | GGTCCTGGGCCCAGTCTGTGCTG |
| 5′ L Vλ 2 | GGTCCTGGGCCCAGTCTGCCCTG |
| 5′ L Vλ 3 | GCTCTGTGACCTCCTATGAGCTG |
| 5′ L Vλ 4/5 | GGTCTCTCTCSCAGCYTGTGCTG |
| 5′ L Vλ 6 | GTTCTTGGGCCAATTTTATGCTG |
| 5′ L Vλ 7 | GGTCCAATTCYCAGGCTGTGGTG |
| 5′ L Vλ 8 | GAGTGGATTCTCAGACTGTGGTG |
|
| |
| 5′ AgeI Vλ 1 | CTGCTACCGGTTCCTGGGCCCAGTCTGTGCTGACKCAG |
| 5′ AgeI Vλ 2 | CTGCTACCGGTTCCTGGGCCCAGTCTGCCCTGACTCAG |
| 5′ AgeI Vλ 3 | CTGCTACCGGTTCTGTGACCTCCTATGAGCTGACWCAG |
| 5′ AgeI Vλ 4/5 | CTGCTACCGGTTCTCTCTCSCAGCYTGTGCTGACTCA |
| 5′ AgeI Vλ 6 | CTGCTACCGGTTCTTGGGCCAATTTTATGCTGACTCAG |
| 5′ AgeI Vλ 7/8 | CTGCTACCGGTTCCAATTCYCAGRCTGTGGTGACYCAG |
|
| |
| 5′ Ab sense | GCTTCGTTAGAACGCGGCTAC |
|
| |
| Reverse Primer | 5′ - 3′ sequence |
|
| |
| 3′ Cγ CH1 | GGAAGGTGTGCACGCCGCTGGTC |
| 3′ Cμ CH1 | GGGAATTCTCACAGGAGACGA |
| 3′ IgG (internal) | GTTCGGGGAAGTAGTCCTTGAC |
|
| |
| 3′ SalI JH 1/2/4/5 | TGCGAAGTCGACGCTGAGGAGACGGTGACCAG |
| 3′ SalI JH 3 | TGCGAAGTCGACGCTGAAGAGACGGTGACCATTG |
| 3′ SalI JH 6 | TGCGAAGTCGACGCTGAGGAGACGGTGACCGTG |
|
| |
| 3′ Cκ 543 | GTTTCTCGTAGTCTGCTTTGCTCA |
|
| |
| 3′ Cκ 494 | GTGCTGTCCTTGCTGTCCTGCT |
|
| |
| 3′ BsiWI Jκ 1/4 | GCCACCGTACGTTTGATYTCCACCTTGGTC |
| 3′ BsiWI Jκ 2 | GCCACCGTACGTTTGATCTCCAGCTTGGTC |
| 3′ BsiWI Jκ 3 | GCCACCGTACGTTTGATATCCACTTTGGTC |
| 3′ BsiWI Jκ 5 | GCCACCGTACGTTTAATCTCCAGTCGTGTC |
|
| |
| 3′ Cλ | CACCAGTGTGGCCTTGTTGGCTTG |
|
| |
| 3′ XhoI Cλ | CTCCTCACTCGAGGGYGGGAACAGAGTG |
Restriction sites are underlined
Figure 1.
Strategy to clone and express human monoclonal antibodies. IgH and IgL chain genes were amplified by nested RT-PCR from single cell cDNA generated by amplification with random hexamers before ligation into eukaryotic expression vectors. 1st PCRs were performed with forward primer mixes specific for the leader region and reverse primers specific for the respective IgH, Igκ or Igλ constant region. 2nd PCRs were performed with forward primer mixes specific for FWR1 and respective nested reverse primers specific for the IgH, Igκ and Igλ J genes or constant regions. If 2nd PCR primers contained restriction sites, the products were sequenced and directly cloned. Alternatively, to avoid the introduction of aa exchanges by the use of primer mixes and for the cloning of full-length Igκ genes, 2nd PCR products were sequenced to determine the respective V and J gene. After that V gene and J gene specific primers containing restriction sites were used in another round of amplification using the 1st PCR product as template. All PCR products were sequenced before and after expression vector cloning. For recombinant antibody production, plasmids containing inserts with 100% identity to the original PCR product were co-transfected into HEK 293 cells and antibodies were purified from supernatants after culture.
Ig gene sequence analysis
Aliquots of the VH, Vκ and Vλ chain second PCR products were purified with ExoSAP-IT (USB) according to the manufacturer’s instructions and sequenced with the respective reverse primer (Table 2). Sequences were analyzed by IgBLAST comparison with GenBank (http://www.ncbi.nlm.nih.gov/igblast/) to identify germline V(D)J gene segments with highest identity. IgH complementarity determining region (CDR)3 length was determined as indicated in IgBlast by counting the aa residues following framework region (FWR)3 up to the conserved tryptophan-glycine motif in all JH segments or up to the conserved phenylalanin-glycine motif in JL segments (Kabat, 1991). The number of positively (Histidine (H), Arginine (R), Lysine (K)) and negatively charged (Aspartate (D), Glutamate (E)) amino acids (aa) were determined for each IgH and IgL CDR3 region. IgH chain D genes and D gene reading frames (RF) were identified following the criteria of Corbett et al. (Corbett et al., 1997). Polymorphisms were identified by comparison with published germline sequences (http://imgt.cines.fr) or based on our own observations. In contrast to sequences from cloned Ig genes, 2nd PCR product sequences are unlikely to show the mutations that were introduced by the Taq polymerase and would do so only if the mutations were introduced early during the PCR. Analysis of Ig gene sequences from naive B cells lacking somatic mutations allows the detection of Taq-mediated misincorporated nucleotides by comparison to published germline sequences. Our own observations show that the single misincorporated nucleotides can be seen in less than 10% of all 2nd PCR product sequences analysed (Wardemann et al., 2003 and TT, unpublished observations).
Expression vector cloning
Before cloning all PCR products were purified using Qia-Quick 96 PCR Purification Kit (Qiagen) and QIAvac96. Samples were eluted with 50 μl nuclease-free water (Eppendorf) into 96-well plates. Typical volumes obtained after elution were 25–30 μl. Digests were carried out with the respective restriction enzymes AgeI, SalI and XhoI (all from NEB) in the same plate in a total volume of 35–40 μl and digested PCR products were purified as described before ligation into human Igγ1, Igκ and Igλ expression vectors containing a murine Ig gene signal peptide sequence (GenBank accession no. DQ407610) and a multiple cloning site upstream of the human Igγ1, Igκ or Igλ constant regions. Transcription is under the influence of the human cytomegalovirus (HCMV) promotor and clones can be selected based on resistance to ampicillin. Ligation was performed in a total volume of 10 μl with 1 U T4-Ligase (Invitrogen), 7.5 μl of digested and purified PCR product and 25 ng linearized vector. Competent E. coli DH10B bacteria (Clontech) were transformed at 42°C with 3 μl of the ligation product in 96-well plates. Colonies were screened by PCR using 5′Absense as forward primer and 3′IgGinternal, 3′Cκ494 or 3′Cλ as reverse primer, respectively (Table 2). PCR products of the expected size (650 bp for Igγ1, 700 bp for Igκ and 590 bp for Igλ) were sequenced to confirm identity with the original PCR products (Fig. 1). Due to the use of error-prone Taq-Polymerase approximately 10% of all cloned inserts contained mutations leading to aa exchanges if compared to the nucleotide sequences obtained from 2nd PCR products of the same Ig gene before cloning and were therefore excluded from further analyses (data not shown). Plasmid DNA was isolated from 3 ml bacteria cultures grown for 16 h at 37°C in Terrific Broth (Difco Laboratories) containing 75 μg/ml ampicillin (Sigma) using QIAprep Spin columns (Qiagen). From 1.5 ml baceria cultures, on average 35 μg plasmid DNA was recovered after elution with 75 μl of EB elution buffer (Qiagen).
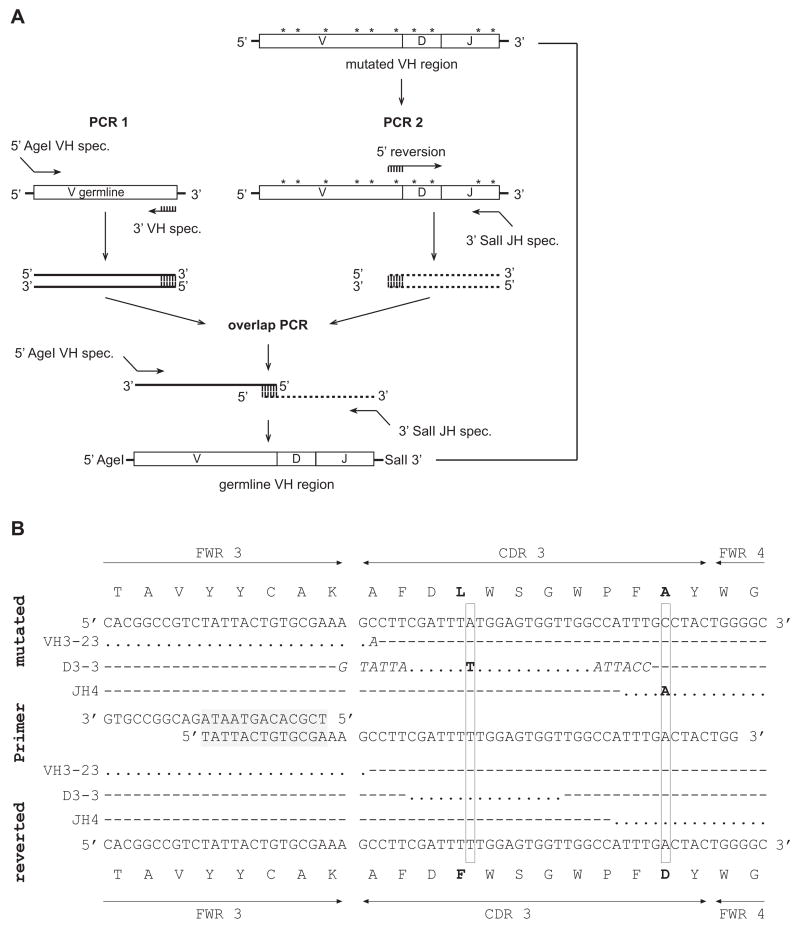
Strategy to revert mutated IgH and IgL chain genes
Somatically mutated Ig genes were reverted into their unmutated germline counterparts using an overlap PCR strategy (Fig. 2A; Herve et al., 2005; Tsuiji et al., 2006; Tiller et al., 2007). Unmutated germline V genes were amplified with gene-specific forward primers containing the restriction site AgeI and individual gene-specific reverse primers that anneal at the 3′ end of FWR3 (Table 2). Germline V genes were amplified from previously cloned plasmids encoding unmutated Ig genes with identical V genes or if not available from genomic DNA. Mutated CDR3-J sequences were reverted independently by PCR using forward primers with minimal complementarity of 10 nucleotides to the germline V gene PCR product. Reverse J gene specific primers included the respective restriction sites as indicated (Fig. 2B, Table 2). PCRs were performed at 94°C for 30 sec, 58°C for 30 sec and 72°C for 45 sec for 30 cycles. Equal ratios of the reverted V and CDR3-J gene PCR products were fused under the same conditions in a third 20-cycles overlap PCR as depicted (Fig. 2A). Full-length reverted V(D)J gene PCR products were gel extracted before digestion and purified before final expression vector cloning. The successful reversion of somatic mutations was confirmed by sequence analysis of the cloned products.
Figure 2.
Reversion of somatic mutations. (A) PCR strategy to revert mutated Ig genes into their germline counterparts. Asterics indicate somatic mutations. PCR 1 amplifies a germline VH gene corresponding to the VH in the mutated clone with gene specific primers. Primers used in PCR 2 revert somatic mutations in the mutated clone. Homology of the PCR 2 forward primer to the reverse primer used in PCR 1 is indicated. The PCR 2 reverse primer is JH-specific and contains the SalI restriction site. PCR products 1 and 2 are fused via the homologous region (indicated) in a subsequent overlap PCR using the same 5′AgeI VH specific forward primer as in PCR 1 and the 3′SalI JH specific reverse primer used in PCR 2 to generate the complete germline VDJ sequence. IgL chain somatic mutations are reverted following the same principle. Overlap PCR products are cloned into the respective expression vectors.
(B) Partial sequence of a representative mutated IgH gene (top) starting in FWR3 and including the CDR3 region and the conserved JH tryptophan (W) – glycine (G) motif (FWR4). The corresponding unmutated germline configuration after successful reversion of somatic mutations as indicated on the bottom. Dots represent nucleotide identity. Dashes are non-identical nucleotides. Nucleotide exchanges and aa exchanges are indicated in bold. Germline nucleotide sequences of full-length VH3-23, D3-3 and JH4 genes are indicated in italics.
Recombinant antibody production
Human embryonic kidney (HEK) 293 (ATCC, No.CRL-1573) or 293T (ATCC, No. CRL-11268) cells were cultured in 150 mm plates (Falcon, Becton Dickinson) under standard conditions in Dulbecco’s Modified Eagle’s Medium (DMEM; GibcoBRL) supplemented with 10% heat-inactivated ultralow IgG fetal calf serum (FCS) (Invitrogen), 1 mM sodium pyruvate (GibcoBRL), 100 μg/ml streptomycin, 100 U/ml penicillin G and 0.25 μg amphotericin (all from GibcoBRL).
Transient transfections of exponentially growing 293 cells were performed by calcium-phosphate precipitation at 80% cell confluency. Equal amounts (12.5–20 μg each) of IgH and corresponding IgL chain expression vector DNA and 0.7 mM chloroquine (Sigma) were mixed in 1 ml sterile water and 2.5 M CaCl2 was added drop-wise to a concentration of 250 mM. An equal volume of 2× HEPES-buffered saline (50 mM HEPES, 10 mM KCl, 12 mM Dextrose, 280 mM NaCl, 1.5 mM Na2HPO4-7H2O, pH 7.05) was mixed with the calcium-DNA solution under slow vortexing and incubated at room temperature for 10 min to allow formation of precipitates. The precipitation mixture was distributed evenly to the culture dish. The cells were washed with 10 ml serum-free DMEM after 8–12 h and cultured for 6 d in 25 ml DMEM supplemented with 1% Nutridoma-SP (Roche) before supernatants were harvested and analyzed by enzyme-linked immunosorbent assay (ELISA) for recombinant antibody production (see below).
Recombinant antibody purification
Cell debris was removed by centrifugation at 800 × g for 10 min and culture supernatants were stored at 4°C with 0.05% sodium azide. Recombinant antibodies were purified with Protein G beads (GE Healthcare) according to the manufacturer’s instructions. In brief, 25 ml cell culture supernatant were incubated with 25 μl Protein G beads for at least 14 h at 4°C under rotation. Supernatants were removed after centrifugation at 800 × g for 10 min and the beads were transferred to a chromatography spin column (BioRad) equilibrated with PBS. After two rounds of washing with 1 ml PBS, antibodies were eluted in 3–4 fractions (200 μl each) with 0.1 M glycine (pH 3.0). Eluates were collected in tubes containing 20 μl 1 M Tris (pH 8.0) with 0.5% sodium azide.
Recombinant antibody concentrations were determined by ELISA using 3 μg/ml human serum IgG1 (Sigma) diluted in PBS as a standard. ELISAs were performed with high-binding capacity ELISA plates (Costar) coated with 50 μl goat anti-human IgG (Fcγ specific; Jackson) at a concentration of 2 μg/ml in PBS. Plates were washed 3× with 200 μl of water per well before incubation for 1h with 200 μl 2 mM EDTA and 0.05% Tween-20 in PBS and washed again. 50 μl of the standards and samples were transferred into the ELISA plate and incubated for 2 h. Unbound antibodies were removed by washing under the same conditions as above before incubation with 50 μl per well of horseradish peroxidase (HRP) coupled goat anti-human IgG (Jackson) at a concentration of 0.8 μg/ml in PBS with 2 mM EDTA and 0.05% Tween-20. Incubation with the secondary antibody was for 2 h and unbound antibodies were removed by washing followed by incubation with 200 μl PBS, 2 mM EDTA, 0.05% Tween-20 and a last washing step. Assays were developed using HRP chromogenic substrate (ABTS; Pierce) according to the manufacturer’s instructions. Optical densities (OD) were measured at 405nm and antibody concentrations were determined using SoftMax software (Molecular Devices). All steps were performed at ambient temperature.
Polyreactivity and HEp-2 ELISA
Antibody concentrations in supernatants or after purification were adjusted to 1 μg/ml and three consecutive 1:4 dilutions in PBS were prepared. ELISAs were performed as described above except high-binding-capacity ELISA plates were coated with 50 μl/well of individual antigens at concentrations of 5 μg/ml (insulin) or 10 μg/ml (double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), lipopolysaccharide (LPS)) in PBS. ssDNA was prepared from salmon sperm dsDNA after boiling at 95°C for 30 min and aliquots were immediately frozen at −20°C. Human recombinant insulin solution (Sigma) and LPS from E. coli Serotype 055:B5 (Sigma) were stored at +4°C. Controls for polyreactivity were the recombinant human monoclonal antibodies mGO53 (negative; Wardemann et al., 2003), eiJB40 (low polyreactive; Wardemann et al., 2003) and ED38 (highly polyreactive; Meffre et al., 2004) and were included on each plate. Bound antibodies were detected using 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid (ABTS) as substrate (BioRad). The cut-off OD at which antibodies were considered reactive was determined for each experiment based on the OD405 minus 2x the standard deviation for the low positive control antibody eiJB40 at a concentration of 1μg/ml. A minimum of 3 controls was included in each experiment to allow calculation of standard deviations. Antibodies were only considered reactive if positive reactivity was confirmed in at least two independent experiments. Antibodies were considered polyreactive if they bound to at least two different types of antigens (DNA, insulin, LPS).
Self-reactivity screens with HEp-2 cell lysates were performed using commercially available QUANTA Lite™ anti-nuclear antibody (ANA) ELISA plates enriched for nuclear antigens (INOVA Diagnostics). Low positive and negative controls were used as provided by the manufacturer and included sera from patients and healthy individuals, respectively. Controls were used according to the manufacturer’s instructions and at three consecutive 1:4 dilutions in PBS. Protein G-purified recombinant monoclonal human antibodies including eiJB40 (Wardemann et al., 2003) and ED38 (Meffre et al., 2004) as additional controls were used at 10–25 μg/ml and three consecutive 1:4 dilutions in PBS. Bound antibodies were detected using 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid (ABTS) as substrate (BioRad). Antibodies were considered self-reactive if positivity in the HEp-2 ELISA could be confirmed in the HEp-2 immunofluorescence assay or if they showed strong and specific HEp-2 immunofluorescence patterns (typically with cytoplasmic antigens such as cytosceleton components presumably not abundant in the HEp-2 cell ELISA with enriched nuclear antigens).
HEp-2 Immunofluorescence Assays
For immunofluorescence assays, HEp-2 cell coated slides (Bion Enterprises, LTD) were incubated in a moist chamber at ambient temperature with 20 μl purified antibodies at 50 –150 μg/ml for 30 min, washed in PBS and incubated for 30 min with FITC-labeled goat anti-human Ig according to the manufacturer’s instructions. Control stainings with PBS, ANA-negative and ANA-positive control sera were performed as suggested by the manufacturer and were included in all experiments. Samples were examined on a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss MicroImaging, Inc.). Positive staining was determined by comparison to the controls at equal exposure times. If immunofluorescence assays are performed, special care needs to be taken to discriminate between weak unspecific background staining patterns to subnuclear and cytoplasmic structures frequently observed for non-reactive antibodies if used at high concentrations and for polyreactive antibodies with similar but stronger binding patterns. It is important to note that subnuclear and cytoplasmic staining patterns are not sufficient to determine if an antibody is polyreactive. Hence, the HEp-2 immunofluorescence assay can be used to confirm polyreactivity as detected by ELISA with individual antigens and to identify specific antibody binding to diverse HEp-2 cell structures including the nucleus.
Statistical analysis
P-values for Ig gene repertoire analysis were calculated by 2 × 5 Fisher’s Exact Test and Chi-square test. P-values for analysis of charges in IgH CDR3, and antibody reactivity were calculated by 2 × 2 Fisher’s Exact Test. Statistical analyses for IgH CDR3 length and mutation numbers was performed using non-paired two-tailed Student’s t test.
Results
To be able to determine the frequency of B cells expressing autoreactive antibodies at different stages of their development we established an efficient method to produce recombinant monoclonal antibodies from single human B cells (Fig. 1). Isolation of individual cells is performed by single cell FACS sorting and allows the cloning of antibodies from defined B cell populations from various sources even if the cells are represented at low frequency or if the absolute samples size is small.
The strategy is based on the amplification and cloning of full-length IgH and Igκor Igλ L chain V regions into eukaryotic expression vectors containing the human Igγ1, Igκ1 or Igλ2 constant regions, respectively. All PCR, purification and cloning reactions are performed in 96-well plates, which allows the fast and efficient handling of large numbers of clones. Full-length Ig gene transcripts were amplified in two nested PCRs. The first PCR used forward primer mixes specific for the respective VH, Vκ or Vλ leader regions and a single reverse primer specific for the respective constant region. If desired reverse primer mixes can be used for example for the amplification of IgH chains with different isotypes such as μ, γ and α. Except for the amplification of Igκ genes that are amplified with a single forward primer (panVκ), nested IgH and Igλ PCRs are performed with mixes of forward primers which include the AgeI restriction site and anneal to the first 18 nucleotides of the respective V genes (Table 2). If used in combination with the reverse 3′Sal JH gene primer mix or the 3′Xho Cλ primer, respectively, these PCR products can be directly used for cloning. However, the use of primer mixes frequently leads to the introduction of aa exchanges in the annealing region due to cross-priming of non-identical primers in the mix. To avoid such alterations or if restriction sites were not introduced by the 2nd PCR primers as for the amplification of Igκ genes, nested PCR products were sequenced to identify the specific V and J gene combination for each gene. Furthermore, amplification of IgH chains using the nested reverse primers specific for the constant regions of all 4 human isotype subclasses (3′CγCH1, 3′IgG internal; Table 2) allowed the discrimination of each subclass antibody after sequencing.
Based on the sequence information, all nested PCRs were repeated using the respective V gene-specific forward and J gene-specific reverse primers with restriction sites and the first PCR product as template (specific PCR). Although this strategy reverts all somatic mutations present in the primer annealing regions it prevents the introduction of largely random aa exchanges at the beginning of FWR1 or the end of FWR4 as it is the case if primer mixes were used. All PCR products were sequenced after cloning to confirm identity with the original PCR product and to ensure that clones with mutations introduced by the error-prone Taq polymerase were excluded from the analyses (Fig. 1). V regions were cloned in frame with the respective human Igγ1, Igκ1 or Igλ2 constant region genes encoded by the eukaryotic expression vectors.
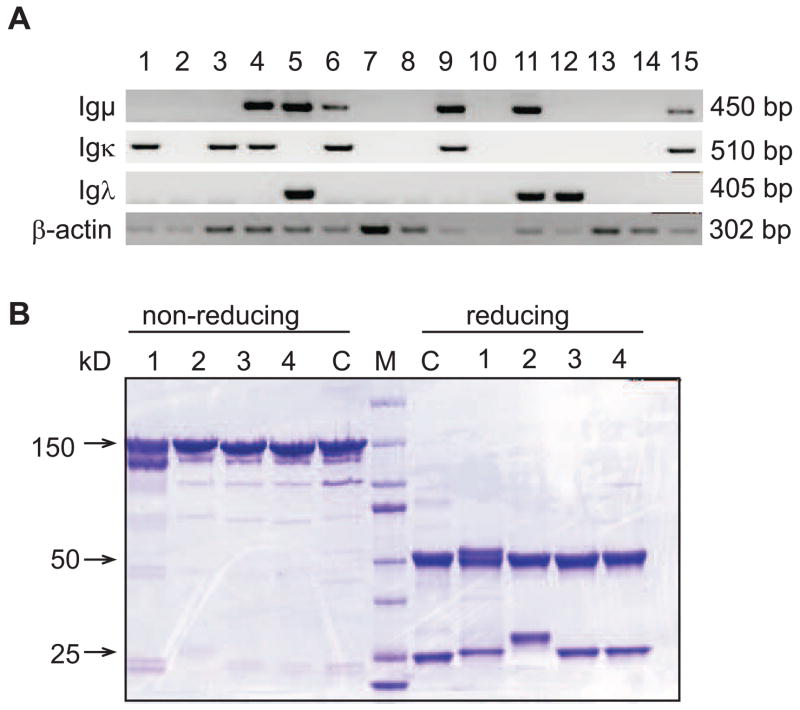
Clonally related sequences with identical IgH and IgL chain rearrangements were not detected in naïve and memory B cells from healthy individuals and patients and V, D and J genes from almost all Ig gene families, and nearly all Ig gene family members were amplified (Wardemann et al., 2003; Meffre et al., 2004; Ng et al., 2004; Herve et al., 2005; Samuels et al., 2005; Yurasov et al., 2005; Tsuiji et al., 2006; Yurasov et al., 2006; Herve et al., 2007; Tiller et al., 2007). The generation of single cell cDNA libraries with random hexamers allows RT-PCR mediated amplification of all expressed genes. We used the housekeeping gene beta-actin as positive control for the sorting and RT reaction, which typically can be amplified from >95% of all wells (data not shown and Fig. 3A). Throughout our analyses of different B cell compartments the overall efficiency for amplification of corresponding IgH and IgL chain gene pairs from single cells typically ranged between 30–60% and the amplification of Igκ and Igλ light chain genes typically resembled the approximate ratio of 60% Igκ and 40% Igλ expressing B cells in humans (Fig. 3A and Wardemann et al., 2003). In about 5% of the cases IgH chains were amplified with both Igκ and Igλ light chain. Surprisingly, in half of these cases both IgL chain alleles were functionally rearranged (data not shown and Wardemann et al., 2003; Yurasov et al., 2005; Tsuiji et al., 2006).
Figure 3.
(A) Representative gel picture showing RT-PCR products of Igμ (450 bp), Igκ (510 bp) and Igλ (405 bp) V genes and beta-actin (302 bp) amplified from single mature naive human B cells. (B) Representative SDS gel picture of purified recombinant monoclonal human antibodies (lane #1–3) under non-reducing (left) and reducing conditions (right). For reduction, 5% β-mercaptoethanol was added to the denaturating sample buffer and all samples were incubated for 10 min at 70°C. 15 μg of proteins were subjected to a 4–12% BisTris polyacrylamide gel (BioRad). Protein bands were visualized by Coomassie blue staining. C: Control human IgG1κ antibody (Sigma). M: Molecular weight marker (BioRad).
For recombinant antibody production IgH chain and corresponding IgL chain encoding plasmids were co-transfected by calcium-phosphate-precipitation but transfections based on liposome-DNA complex formation were also highly efficient (data not shown). Typically, 85% of individual transfections produced monoclonal recombinant antibodies at concentrations from 1 μg/ml to 20 μg/ml with an average concentration of 5 μg/ml. Low amounts of antibody production (<1 μg/ml) are frequently indicative of production of non-functional Igs (data not shown). Functional Ig production is tested by sodium-dodecylsulfate (SDS) gel electrophoresis (Fig 3B).
To determine if antibodies showed self-reactivity, commercially available ELISA and immunofluorescence assays were used. Both assays measure reactivity with the human epithelial cell line HEp-2 and are used in clinical diagnostics for autoimmunity (Egner, 2000) (Fig 4A). To test for specificity of self-reactive antibodies reactivity with structurally diverse individual antigens such as ssDNA, dsDNA, insulin and LPS were performed. Control antibodies with strong (ED38; Meffre et al., 2004), weak (eiJB40; Wardemann et al., 2003) or no detectable levels of self- and polyreactivity (mGO53; Wardemann et al., 2003) were included in each experiment and were originally cloned from B cells of healthy human donors. Thresholds at which sample antibodies were determined reactive or non-reactive were set based on relative levels of reactivity compared to the control antibodies. Polyreactive antibodies typically show nuclear and cytoplasmic staining patterns in HEp-2 cell immunoflourescence assays and react with HEp-2 cells in ELISA (Fig. 4A and Wardemann et al., 2003).
Figure 4.
Detection of self-reactive and polyreactive antibodies. (A) Typical HEp-2 cell reactivity profiles of IgG memory B cell antibodies (VB13, VB56, VB72, VB108, VB148) of healthy donor VB as determined by ELISA and immunofluorescence assay (Tiller et al., 2007). High-positive control antibody ED38 (dotted line; Meffre et al., 2004) and low ANA control serum (red line; INOVA) are shown for comparison. (B) Reactivity profiles of the high-positive control antibody ED38 (dotted line; Meffre et al., 2004), low-positive control antibody eiJB40 (red line; Wardemann et al., 2003) and negative control antibody mGO53 (green line; Wardemann et al., 2003) as tested by ELISA with dsDNA, ssDNA, LPS and insulin.
To determine the relative contribution of somatic mutations to self-reactivity we used an overlap PCR strategy to revert the IgH and IgL chain genes to the corresponding germline form. Unmutated V genes were amplified from germline genes by PCR and mutations in CDR3-J gene regions were removed in an independent PCR using primers that revert the mutations to the germline sequence. Both PCR products show homology at the respective 3′ (V gene) and 5′ (CDR3-J region) end that allows fusion of both products in a third overlap PCR using equal amounts of both products as template. Clones for reversion experiments were chosen randomly but clear identification of D genes in IgH chain CDR3 regions was a prerequisite to best allow discrimination between Ig gene encoded nucleotides and N/P nucleotides in CDR3 regions. Somatically mutated recombinant antibodies and their corresponding unmutated counterparts were produced and tested by ELISA and immunofluorescence assays for self-and polyreactiviy (Herve et al., 2005; Tsuiji et al., 2006; Tiller et al., 2007).
Discussion
Hybridomas and EBV immortalized B cell lines are the most commonly used technologies to generate human monoclonal antibodies. However, overall transformation efficiencies are extremely low (about 1–3%), largely depend on the maturation status of the cell and are frequently performed after stimulation (Stahli et al., 1980; Aman et al., 1984; Redmond et al., 1986; James and Bell, 1987; Borrebaeck et al., 1988; Crain et al., 1989). Furthermore, detailed Ig gene information depends on additional cloning of the IgH and IgL chain genes (Larrick et al., 1989). Recently, the Lanzavecchia laboratory reported a protocol for the efficient EBV-mediated transformation of human B cells but this is limited to CpG-activated memory B cells (Traggiai et al., 2004). Our strategy to clone Ig genes from single human B cells shows comparable efficiency. However, the production of recombinant monoclonal antibodies as described here is not limited to distinct B cell populations and does not depend on prior stimulation. Additionally, detailed sequence information is obtained as part of the cloning process and includes not only the V region but also part of the constant regions to allow discrimination of isotype subclasses.
To date we have amplified IgH chain and corresponding IgL chain genes from more than 2000 unique antibodies expressed by B cells from healthy human donors and patients with autoimmunity and immunodeficiencies that allow us to directly compare Ig gene sequence information with antibody reactivities (Wardemann et al., 2003; Meffre et al., 2004; Ng et al., 2004; Herve et al., 2005; Samuels et al., 2005; Yurasov et al., 2005; Tsuiji et al., 2006; Yurasov et al., 2006; Herve et al., 2007; Tiller et al., 2007). However, our analyses showed no direct correlations between individual Ig genes or Ig gene characteristics and antibody reactivities suggesting that the human antibody repertoire may be too diverse to allow predictions on antibody reactivities based on sequence analysis alone.
Irrespective of their original isotype, all recombinant antibodies are generated as IgG1 molecules to facilitate the detection and comparability in reactivity assays. The in vitro production efficacy after transient transfection of HEK 293 cells is high and depending on the transfection system (1–20 μg antibody per ml supernatant for calcium-phosphate and 10–80 μg/ml for liposome based transfection protocols, TT and HW unpublished observations) easily compares to the amounts produced by stable B cell lines or hybridomas. Furthermore, expression of IgH and IgL chain genes is driven by independent vectors and thus allows replacement of IgH or IgL chains to determine their relative contribution to antibody reactivity levels (Wardemann et al., 2004).
In conclusion, we have described an unbiased and highly efficient method for the production of human recombinant monoclonal antibodies derived from single B cells at any maturation stage. This method is well suited to study the expressed antibody repertoire in humans.
Acknowledgments
We thank all members of the Nussenzweig and Wardemann laboratory for discussion and S. Boscardin for helpful comments on the manuscript. We are indebted to Patrick Wilson and Anne Schaefer for their help and Aaron B. Kantor for advice on cDNA synthesis from single cells. This work was supported by grants from the National Institutes of Health (to M.C. Nussenzweig, E. Meffre) and the Dana Foundation Human Immunology Program (to E. Meffre, H. Wardemann). S. Yurasov is a Charles H. Revson Fellow in Biomedical Research and is supported by the Charles A. Dana Foundation. M.C. Nussenzweig is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- Ig
immunoglobulin
- EBV
Epstein-Barr virus
- FWR
framework region
- CDR
complementarity determining region
- ANA
anti-nuclear antibody
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aman P, Ehlin-Henriksson B, Klein G. Epstein-Barr virus susceptibility of normal human B lymphocyte populations. J Exp Med. 1984;159:208–20. doi: 10.1084/jem.159.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrebaeck CA, Danielsson L, Moller SA. Human monoclonal antibodies produced by primary in vitro immunization of peripheral blood lymphocytes. Proc Natl Acad Sci U S A. 1988;85:3995–9. doi: 10.1073/pnas.85.11.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- Brezinschek HP, Foster SJ, Brezinschek RI, Dorner T, Domiati-Saad R, Lipsky PE. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(−)/IgM+ B cells. J Clin Invest. 1997;99:2488–501. doi: 10.1172/JCI119433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R, Shih TA, Kleinewietfeld M, Rakonjac J, Nemazee D, Rajewsky K, Nussenzweig MC. Contribution of receptor editing to the antibody repertoire. Science. 2001;291:1541–4. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- Corbett SJ, Tomlinson IM, Sonnhammer EL, Buck D, Winter G. Sequence of the human immunoglobulin diversity (D) segment locus: a systematic analysis provides no evidence for the use of DIR segments, inverted D segments, “minor” D segments or D-D recombination. J Mol Biol. 1997;270:587–97. doi: 10.1006/jmbi.1997.1141. [DOI] [PubMed] [Google Scholar]
- Crain MJ, Sanders SK, Butler JL, Cooper MD. Epstein-Barr virus preferentially induces proliferation of primed B cells. J Immunol. 1989;143:1543–8. [PubMed] [Google Scholar]
- Demaison C, David D, Fautrel B, Theze J. V(H) gene-family representation in peripheral activated B cells from systemic lupus erythematosus (SLE) patients. Clin Exp Immunol. 1996;104:439–45. doi: 10.1046/j.1365-2249.1996.56763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaison C, David D, Letourneur F, Theze J, Saragosti S, Zouali M. Analysis of human VH gene repertoire expression in peripheral CD19+ B cells. Immunogenetics. 1995;42:342–52. doi: 10.1007/BF00179395. [DOI] [PubMed] [Google Scholar]
- Dorner T, Farner NL, Lipsky PE. Ig lambda and heavy chain gene usage in early untreated systemic lupus erythematosus suggests intensive B cell stimulation. J Immunol. 1999;163:1027–36. [PubMed] [Google Scholar]
- Dorner T, Foster SJ, Farner NL, Lipsky PE. Immunoglobulin kappa chain receptor editing in systemic lupus erythematosus. J Clin Invest. 1998;102:688–94. doi: 10.1172/JCI3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner W. The use of laboratory tests in the diagnosis of SLE. J Clin Pathol. 2000;53:424–32. doi: 10.1136/jcp.53.6.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlich A, Schaal S, Gu H, Kitamura D, Muller W, Rajewsky K. Immunoglobulin heavy and light chain genes rearrange independently at early stages of B cell development. Cell. 1993;72:695–704. doi: 10.1016/0092-8674(93)90398-a. [DOI] [PubMed] [Google Scholar]
- Ghia P, ten Boekel E, Sanz E, de la Hera A, Rolink A, Melchers F. Ordering of human bone marrow B lymphocyte precursors by single-cell polymerase chain reaction analyses of the rearrangement status of the immunoglobulin H and L chain gene loci. J Exp Med. 1996;184:2217–29. doi: 10.1084/jem.184.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Dorner T, Lipsky PE. Use of immunoglobulin variable-region genes by normal subjects and patients with systemic lupus erythematosus. Int Arch Allergy Immunol. 2000;123:36–45. doi: 10.1159/000024422. [DOI] [PubMed] [Google Scholar]
- Herve M, Isnardi I, Ng YS, Bussel JB, Ochs HD, Cunningham-Rundles C, Meffre E. CD40 ligand and MHC class II expression are essential for human peripheral B cell tolerance. J Exp Med. 2007;204:1583–1593. doi: 10.1084/jem.20062287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herve M, Xu K, Ng YS, Wardemann H, Albesiano E, Messmer BT, Chiorazzi N, Meffre E. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest. 2005;115:1636–43. doi: 10.1172/JCI24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter PA, Maizel JV, Jr, Leder P. Evolution of human immunoglobulin kappa J region genes. J Biol Chem. 1982;257:1516–22. [PubMed] [Google Scholar]
- Huang C, Stewart AK, Schwartz RS, Stollar BD. Immunoglobulin heavy chain gene expression in peripheral blood B lymphocytes. J Clin Invest. 1992;89:1331–43. doi: 10.1172/JCI115719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Stollar BD. Construction of representative immunoglobulin variable region cDNA libraries from human peripheral blood lymphocytes without in vitro stimulation. J Immunol Methods. 1991;141:227–36. doi: 10.1016/0022-1759(91)90149-a. [DOI] [PubMed] [Google Scholar]
- James K, Bell GT. Human monoclonal antibody production. Current status and future prospects. J Immunol Methods. 1987;100:5–40. doi: 10.1016/0022-1759(87)90170-0. [DOI] [PubMed] [Google Scholar]
- Kabat EA, Wu TT, Perry HM, Gotteman KS, Foeller C. Sequences of proteins of immunological interest. NIH Publication; 1991. [Google Scholar]
- Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits JL, Wang J, Shimizu N. One-megabase sequence analysis of the human immunoglobulin lambda gene locus. Genome Res. 1997;7:250–61. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Zhao M, Hansmann ML, Rajewsky K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. Embo J. 1993;12:4955–67. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffly E, Sodoyer R. Monoclonal and recombinant antibodies, 30 years after. Hum Antibodies. 2005;14:33–55. [PubMed] [Google Scholar]
- Larrick JW, Danielsson L, Brenner CA, Abrahamson M, Fry KE, Borrebaeck CA. Rapid cloning of rearranged immunoglobulin genes from human hybridoma cells using mixed primers and the polymerase chain reaction. Biochem Biophys Res Commun. 1989;160:1250–6. doi: 10.1016/s0006-291x(89)80138-x. [DOI] [PubMed] [Google Scholar]
- Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med. 1998;188:2151–62. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre E, Schaefer A, Wardemann H, Wilson P, Davis E, Nussenzweig MC. Surrogate light chain expressing human peripheral B cells produce self-reactive antibodies. J Exp Med. 2004;199:145–50. doi: 10.1084/jem.20031550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng YS, Wardemann H, Chelnis J, Cunningham-Rundles C, Meffre E. Bruton’s tyrosine kinase is essential for human B cell tolerance. J Exp Med. 2004;200:927–34. doi: 10.1084/jem.20040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual V, Capra JD. Immunoglobulin heavy chain variable region gene usage in human autoimmune diseases. Adv Exp Med Biol. 1995;386:133–9. doi: 10.1007/978-1-4613-0331-2_11. [DOI] [PubMed] [Google Scholar]
- Ravetch JV, Siebenlist U, Korsmeyer S, Waldmann T, Leder P. Structure of the human immunoglobulin mu locus: characterization of embryonic and rearranged J and D genes. Cell. 1981;27:583–91. doi: 10.1016/0092-8674(81)90400-1. [DOI] [PubMed] [Google Scholar]
- Redmond MJ, Leyritz-Wills M, Winger L, Scraba DG. The selection and characterization of human monoclonal antibodies to human cytomegalovirus. J Virol Methods. 1986;14:9–24. doi: 10.1016/0166-0934(86)90003-0. [DOI] [PubMed] [Google Scholar]
- Samuels J, Ng YS, Coupillaud C, Paget D, Meffre E. Impaired early B cell tolerance in patients with rheumatoid arthritis. J Exp Med. 2005;201:1659–67. doi: 10.1084/jem.20042321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schable KF, Zachau HG. The variable genes of the human immunoglobulin kappa locus. Biol Chem Hoppe Seyler. 1993;374:1001–22. [PubMed] [Google Scholar]
- Shiokawa S, Mortari F, Lima JO, Nunez C, Bertrand FE, 3rd, Kirkham PM, Zhu S, Dasanayake AP, Schroeder HW., Jr IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J Immunol. 1999;162:6060–70. [PubMed] [Google Scholar]
- Stahli C, Staehelin T, Miggiano V, Schmidt J, Haring P. High frequencies of antigen-specific hybridomas: dependence on immunization parameters and prediction by spleen cell analysis. J Immunol Methods. 1980;32:297–304. doi: 10.1016/0022-1759(80)90194-5. [DOI] [PubMed] [Google Scholar]
- Stewart AK, Huang C, Stollar BD, Schwartz RS. High-frequency representation of a single VH gene in the expressed human B cell repertoire. J Exp Med. 1993;177:409–18. doi: 10.1084/jem.177.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in Human IgG(+) Memory B Cells. Immunity. 2007;26:205–13. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, Murphy BR, Rappuoli R, Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–5. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuiji M, Yurasov S, Velinzon K, Thomas S, Nussenzweig MC, Wardemann H. A checkpoint for autoreactivity in human IgM+ memory B cell development. J Exp Med. 2006;203:393–400. doi: 10.1084/jem.20052033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Stollar BD. Human immunoglobulin variable region gene analysis by single cell RT-PCR. J Immunol Methods. 2000;244:217–25. doi: 10.1016/s0022-1759(00)00260-x. [DOI] [PubMed] [Google Scholar]
- Wardemann H, Hammersen J, Nussenzweig MC. Human autoantibody silencing by immunoglobulin light chains. J Exp Med. 2004;200:191–9. doi: 10.1084/jem.20040818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–7. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Yurasov S, Tiller T, Tsuiji M, Velinzon K, Pascual V, Wardemann H, Nussenzweig MC. Persistent expression of autoantibodies in SLE patients in remission. J Exp Med. 2006;203:2255–61. doi: 10.1084/jem.20061446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V, Nussenzweig MC. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med. 2005;201:703–11. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]