Abstract
We have applied a newly developed SYBR green based real-time RT-PCR assay for quantification of full-length and truncated neurokinin-1 receptor (NK1R) mRNA expression in 9 regions of human brain tissues obtained from 23 subjects who died with no evidence of neurological or neurodegenerative disease. The following brain regions were examined: cingulate cortex, cerebellum, nucleus accumbens, caudate nucleus, putamen, pons, hippocampus, locus coeruleus, and basal ganglia. The SYBR green based-real-time PCR was more sensitive than TaqMan probe based real-time PCR in amplifying both full-length and truncated NK1R mRNA. The real-time RT-PCR assay had excellent specificity and sensitivity, with a dynamic range of detection between 100 and 1000,000 copies of the NK1R cDNA per reaction. The truncated NK1R mRNA levels were more abundant than those of the full-length NK1R in most of the regions examined and there was no significant difference in the truncated NK1R mRNA levels among the nine regions studied. There was, however, a significant difference in the expression of full-length NK1R mRNA levels among the nine regions (P=0.0024), and the putamen region expressed the highest full-length NK1R mRNA. Further studies are needed in order to examine the differences between full-length and truncated NK1R in signal transduction and functional consequences in order to delineate the significance of the copresence of the two forms of NK1R in the human brain.
1. Introduction
Substance P (SP), a member of the tachykinin family, is widely distributed in the mammalian CNS and peripheral tissue. The biologic responses to SP are mediated by its preferring neurokinin-1 receptor (NK1R). The human NK-1R gene (Fong et al., 1992; Gerard et al., 1991; Hopkins et al., 1991; Takeda et al., 1991), localized to chromosome 2, has been cloned. NK1R has been identified on CNS and immune cells (Bost, 2004; Ho and Douglas, 2004; Ho et al., 1997; Lai et al., 1998a; Lai et al., 1998b; Lai et al., 1999; Lai et al., 2000; Lucey et al., 1994; Payan et al., 1984; Shanahan et al., 1985; Stanisz et al., 1987; Wozniak et al., 1989). Mammalian NK1R has been studied by several methods and has been found in the dorsal striatum, nucleus accumbens, hippocampus, raphe nuclei, and medulla oblongata (Otsuka and Yoshioka, 1993). NK1R localization in specific regions of the human brain was observed in the striatum (Aubry et al., 1994) and cerebral cortex (Kus et al., 1998; Tooney et al., 2000).
NK1R, a member of the G-protein coupled receptor (GPCR) superfamily (Gq/11), has a typical structure of seven transmembrane domains, an extracellular N-terminus and an intracellular C-terminus (Fong et al., 1992). A splice variant of the human NK1R mRNA with a truncated C-terminus (truncated NK1R) has been cloned and identified (Fong et al., 1992). The full-length NK1R is the predominant form reported to be expressed at sites in the human brain, whereas the truncated NK-1R is widespread in peripheral tissues (Caberlotto et al., 2003). Both the full-length and the truncated NK1R protein have been detected in rat submaxillary glands (Kage et al., 1993) and parotid (Mantyh et al., 1996). A distinct distribution of these two NK1Rs has been observed in rat striatum, submaxillary glands and parotid (Mantyh et al., 1996). The only sequence difference between the two forms NK1R is in the length of the C-terminal tail (Fong et al., 1992). The C-terminal region has been postulated to be of importance to the coupling to different second messenger systems and targeting of receptors on neurons. Our group has demonstrated that the C-terminal tail of NK1R is required for SP-induced intracellular calcium increase in a monocyte-macrophage cell line (THP-1) (Lai et al., 2006). Although activation of the truncated NK1R did not trigger a calcium increase in these cells, SP primes CCL5-mediated calcium increases via activation of the truncated NK1R (Lai et al., 2006). The binding and signaling properties of the truncated receptor and NK1R mutants have been compared with those of the full-length receptor that is expressed in CHO cells, xenopus oocytes and rat Kirsten virus-transformed kidney epithelial cells (KNRK) (Bohm et al., 1997; Fong et al., 1992; Li et al., 1997; Sasakawa et al., 1994). The truncated NK1R undergoes a less rapid desensitization (Li et al., 1997), suggests a longer duration of the response evoked by SP agonists in those brain regions where the truncated form predominates. Thus, it is very likely that there are differences in signal transduction pathways and functional consequences for the expression of differences between the full-length and truncated NK1R in CNS. In order to delineate the role of these two forms of NK1R in the brain, we investigated the expression of the full-length and truncated NK1R in different brain regions.
Recently, Caberlotto et al., using a quantitative TaqMan PCR analysis, observed that the full-length NK1R was the most prevalent throughout the human brain, while in the peripheral tissue, the truncated form was most represented (Caberlotto et al., 2003). This study (Caberlotto et al., 2003) reported the distribution of NK1R mRNA in four post-mortem human brains. However, when the sensitivity of this TaqMan primer/probe assay (Caberlotto et al., 2003) was compared to our established real-time PCR assay which detects both full-length and truncated NK1R (Lai et al., 2005) using full-length NK1R plasmid cDNA as template, our assay was 4–16 times more sensitive than that of the TaqMan assay (Caberlotto et al., 2003) (data not shown). The sensitivity difference between these two assays is most likely due to the location of the primers and/or the Taqman probe used. We therefore modified this real-time PCR assay and examined the expression of the full-length and truncated NK1R in nine brain regions obtained from 23 post-mortem human brains. The regions studied included: (cingulate cortex, cerebellum, nucleus accumbens, caudate nucleus, putamen, pons, hippocampus, locus ceruleus, and basal ganglia.
2. Material and Methods
2.1 Subjects and Specimens
Twenty-three brain specimens were obtained from individuals who died with no evidence of neurological or neurodegenerative disease through the National NeuroAIDS Tissue Consortium (NNTC). Among the 23 subjects, 19 were HIV-positive and 4 were HIV-negative, 7 were female and 16 were male. These subjects died at ages 27–66 years old (average 46 years of age). Among these 23 subjects, 8 were Caucasian (35%) and 15 were non-Caucasian (65%). The specimens obtained from the 23 subjects, included: 23 cingulate cortex, 17 cerebellum, 9 nucleus accumbens, 8 caudate nucleus, 8 putamen, 8 pons with locus ceruleus, 7 hippocampus, 7 locus coeruleus, and 6 basal ganglia specimens.
2.2 RNA Extraction
Total RNA was extracted from these brain tissues using Tri-Reagent (Molecular Research Center, Cincinnati, OH), as instructed by the manufacturer. After centrifugation at 13,000x g for 15 min., RNA-containing aqueous phase was precipitated in isopropanol. RNA precipitates then were washed once in 75% ethanol and solubilized in 30 ul of RNase-free water (Lai et al., 2006). RNA concentration of pooled brain tissues was determined by UV spectroscopy at A260 nm and the RNA was used to generate GAPDH standard curve in order to quantitatively determine total RNA amount in each sample by real-time PCR.
2.3 Reverse Transcription
Total RNA (4 ul) was subjected to reverse transcription. The final reaction mixture (20 ul) contained the following elements: 5 mM MgCl2, 1X RT buffer, 500 uM of each dNTPs, 1 unit/ul recombinant RNasin, 10–15 units of AMV reverse transcriptase (Promega), and 50 ng random primers. Reverse transcriptase negative controls were used in order to control for genomic DNA contamination. RT was performed at 42°C for 1 h. The reaction was terminated by holding the reaction mixture at 99°C for 5 min. One tenth (2 ul) of the resulting cDNA was used as a template for real-time PCR amplification.
2.4 Real Time PCR Primers
The PCR primers used for quantitative measurement of the full-length and the truncated NK-1R mRNA were modified from Caberlotto et al. (Caberlotto et al., 2003). The sequences of the primer pair used to amplify the full-length of NK-1R were: (sense-L-F) 5′-TCTTCTTCCTCCTGCCCTACATC-3′; (antisense-L-R-967) 5′-AGCACCGGAAGGCATGCTTGAAGCCCA -3′, which is specific for the full-length NK-1R sequence. The sequences of the primer pair for the truncated NK-1R were: (sense-L-F) 5′-TCTTCTTCCTCCTGCCCTACATC-3′; (antisense-S-R-1083) 5′-TGGAGAGCTCATGGGGTTGGGATCCT-3′. The forward primer is identical to the one used by Caberlotto et al(Caberlotto et al., 2003). However, the reverse primers for both full-length and the truncated NK1R are located further downstream from the forward primer in order to avoid potential non-specific amplification due to the 50% sequence homology of reverse primers of the full-length and truncated NK1R (Diagram 1). The sequences of the primer pair for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were: 5′-GGTGGTCTCCTCTGACTTCAACA-3′ (sense); 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ (antisense) (Lai et al., 2003). The specificity of the modified primer pair for the truncated NK1R were confirmed by SYBR green dye dissociation curve and agarose gel electrophoresis in our preliminary experiments. The primers were re-suspended in TE buffer and stored at −30°C.
Diagram 1.
LR-967 and SR-1083 are redesigned reverse primers for the full-length and truncated NK1R, respectively. L-F/L-R and L-F/S-R were the primer pairs used for amplification of the full-length and truncated NK1R with the probe, respectively [Caberlotto et al., 2003].
2.5 Real-Time PCR Assay
The MyiQ iCycler system (Bio-Rad Laboratories, Inc., Hercules, CA) was used for real-time PCR analysis. Thermal cycling conditions were designed as follows: initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Fluorescent measurements were recorded during each annealing step. At the end of each PCR run, data were automatically analyzed by the system and amplification plots were obtained. For each PCR, 2 ul of cDNA template was added to 23 ul of iQ™ SYBR Supermix or iQ Supermix (for TaqMan probe) (Bio-Rad) containing the primer pair for either the full-length or the truncated NK-1R, or for GAPDH. An NK-1R full-length plasmid (a gift from Dr. Norma Gerard, Harvard University, Boston, MA) was used to prepare standard curves and used as a specificity control for real-time PCR. The full-length plasmid was amplified by the primer pair for the full-length NK-1R, but not by the primer pair for the truncated NK-1R. In our preliminary experiments, using serial diluted cDNA derived from pooled brain tissues, the amplification efficiency of the full-length and truncated NK1R primer pairs were very close to each other (103% for truncated and 102% for the full-length NK1R, respectively), which allow us to use the full-length NK1R standards to measure the truncated NK1R expression levels. All amplification reactions were performed in duplicate and the average threshold cycle (Ct) numbers of the duplicates were used to calculate the expression levels (copy numbers) of full-length and truncated NK-1R using the standard curve generated with the full-length NK1R plasmid. In order to control the integrity of the RNA and normalize NK-1R mRNA levels in these samples, a GAPDH mRNA fragment in the RNA also was amplified using our established real-time RT-PCR with iQ™ SYBR green Supermix (Bio-Rad), as previously reported (Lai et al., 2003). In brief, in order to generate a total RNA standard curve with GAPDH primers, a known amount of total RNA standard extracted from pooled brain tissues was reverse transcribed and serial diluted (ranging from 32 to 2000 ng), and the GAPDH fragment was amplified for 40 cycles. The message in the brain samples was amplified in the same plate with the standard under the identical conditions. The quantity (ng) of total RNA in the samples was automatically calculated by The MyiQ iCycler system based on the data obtained from the total RNA standard curve. All amplification reactions were performed in duplicate and an average RNA quantity (ng) of the duplicates was used to normalize the full-length and truncated NK1R mRNA levels in the brain tissue samples examined.
The expression levels of the full-length and truncated NK-1R cDNA were normalized to an endogenous cDNA, GAPDH. In order to normalize the full-length and truncated NK1R mRNA levels, the full-length and truncated NK1R copy numbers in brains tissues samples were divided by the total RNA quantity (ng) determined by the GAPDH real-time RT-PCR in the same sample and then multiplied by 1000 to convert the unit to the copy number of NK-1R full-length or truncated mRNA per microgram (ug) of total RNA. The full-length and truncated NK1R mRNA levels in these tissue samples are expressed as the mean copy number of NK1R mRNA per ug of total RNA.
2.6 Statistical Methods
NK-1R expression levels from nine regions of human brain tissues were used for data analysis. The distributions of the NK-1R expression values were examined. The distribution of the data was significantly skewed from the normal distribution; therefore, a Wilcoxon signed-rank matched pairs test was used in order to test whether the expression levels were different between full length and truncated isoforms within the same region of the human brain. Stata 9 (StataCorp, College Station, TX) was used. P<0.05 was used to define significance.
3. Results and discussion
3.1 Verification of the modified primer pairs for full length and truncated NK1R amplification
When 50% of the sequence in the reverse primers (full-length and truncated) is identical, the potential for non-specific amplification increases. In order to prevent this non-specific amplification and to be certain that the primers will bind only to the full-length or truncated NK1R cDNA, we redesigned these two reverse primers further downstream of the mRNA for the full-length and truncated NK1R mRNA, respectively (Diagram 1). As validation of the redesigned reverse primers, we used a full-length NK1R plasmid as templates with or without the TaqMan probe. A cDNA derived from IM9 cells (human lymphoblastoid cell line (Gerard et al., 1991)) which expresses both full-length and truncated NK1R mRNA was also used for this comparison. The redesigned primers had comparable sensitivity with original primers (Caberlotto et al., 2003) in both SYBR green based- and TaqMan probe-based real-time PCR assay for the full-length NK1R (Table 1). SYBR green based real-time PCR was more sensitive in amplification both full-length (NK-1R plasmid and IM9 cDNA) and truncated NK1R (IM9 cells derived cDNA) than TaqMan probe based real-time PCR, regardless of the primer set used. There was non-specific amplification with Caberlotto’s (Caberlotto et al., 2003) reverse primer for truncated NK1R mRNA when the NK1R full-length plasmid was used as template with SYBR green based real-time PCR, but not using the redesigned reverse primer (see Table 1). The sensitivity of this SYBR green based-real-time PCR assay is significantly improved in comparison to the TaqMan probe (Table 1). This may be due to the fact that the probe used was suboptimal for the amplification with these primer sets (such as the sequence composition and the location of the probe). These two sets of primers (L-F/L-R and L-F/L-R-967) had a very similar sensitivity in both TaqMan PCR and SYBR green PCR, although the sensitivity of SYBR green PCR is much higher than that of the TaqMan PCR amplification of full-length NK1R with both primer sets (Table 1).
Table I.
Sensitivity and specificity of different primer/probe sets
| Primer set | Input NK1R Copy Numbers | SYBR Green Copy Numbers | With TaqMan Probe Copy numbers |
|---|---|---|---|
| L-F/L-R | 100,000 | 98,636 | 1,294 |
| 10,000 | 9,143 | 18 | |
| ** IM9 cDNA | 1,550 | 11 | |
| L-F/L-R-967 | 100,000 | 101,530 | 1,331 |
| 10,000 | 6,121 | 62 | |
| IM9 cDNA | 1,463 | 25 | |
| L-F/S-R | 100,000 | 5* | n.d |
| 10,000 | 1* | n.d | |
| IM9 cDNA | 601 | 4 | |
| L-F/S-R-1083 | 100,000 | n.d | n.d |
| 10,000 | n.d. | n.d | |
| IM9 cDNA | 608 | 13 |
A full-length NK1R plasmid DNA and cDNA derived from IM9 cells were used as templates in order to evaluate the sensitivity and specificity of the primer pairs (L-F/LR vs L-F/L-R-967, L-F/S-R vs L-F/S-R-1083) with or without using the TaqMan probe.
Non-specific amplification.
: cDNA derived from IM9 cells.
n.d.: not detectable
The full-length NK1R plasmid was used as a specificity control in order to examine the specificity of these two primer sets with or without the probe. This plasmid is amplified only by the primer pair for the full-length NK-1R and not by the primer pair for the truncated NK-1R. PCR amplification using the primer set L-F/S-R (for the truncated NK1R) (Caberlotto et al., 2003) demonstrated some non-specific amplification when only the full-length NK1R was used as the template (Table 1). Using the redesigned the reverse primers (S-R-1083) for the truncated NK1R, the specificity of the real-time PCR for the truncated NK1R is significantly improved and the primer pair does not non-specifically amplify full-length NK1R sequence (Table 1). These data demonstrated that the SYBR green based-real-time PCR assay with redesigned reverse primer sets is specific and more sensitive than that using TaqMan probe (Caberlotto et al., 2003).
3.2 Sensitivity of the SYBR green based- real-time RT-PCR
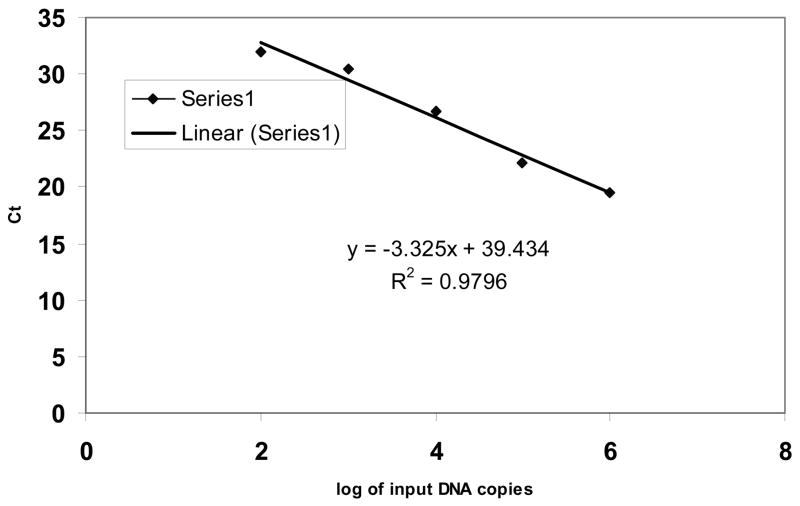
The analytical sensitivity of the SYBR green based-real-time PCR was determined using a series of dilutions of full-length NK1R plasmid DNA containing 0, 10, 100, 1,000, 10,000, 100,000 and 1000,000 molecules and tested three times, each in duplicate. The real-time PCR detected NK1R cDNA at abundance as low as 100 molecules, although the detection rate was 33% (2 out of 6 replicates) (data not shown). The detection rate, however, was 100% at an NK-1R copy number of 100 or higher (6 out of 6 replicates). The detection limit was 100 cDNA molecules per reaction mixture with linearity over 5 orders of magnitude (the correlation coefficient R2 = 0.979; a representative result is shown in Figure 1).
Figure 1.
The real-time RT-PCR sensitivity and linearity analysis with full-length NK1R plasmid DNA. Change in fluorescence (Rn) as a function of cycle numbers is demonstrated for a range of known input copy numbers of the NK1R plasmid. 10-fold serial dilutions of the DNA starting from 100 to 10 × 105 molecules per reaction were amplified by the SYBB green based real-time RT-PCR using the redesigned reverse primer. The standard curve of the serial dilutions of NK1R full-length plasmid DNA was generated with a correlation coefficient (R2) of 0.979.
3.3 Quantitation of full-length and truncated NK1R from different brain tissues
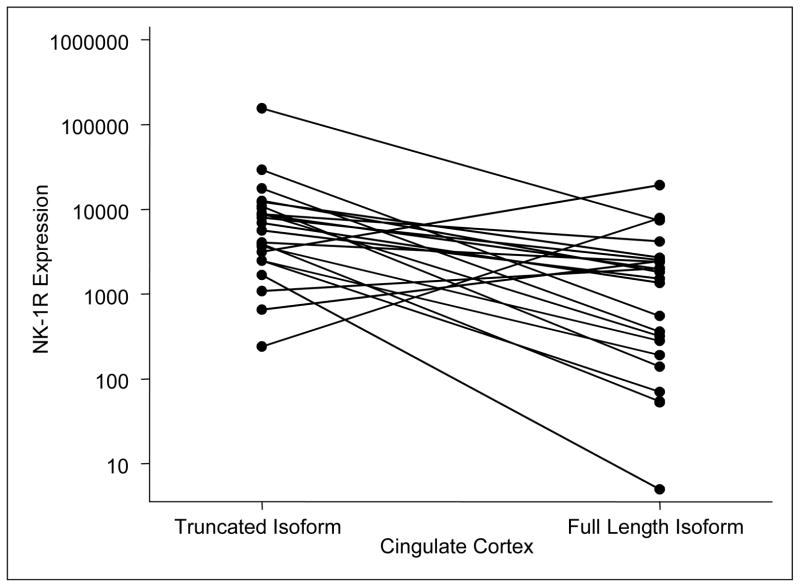
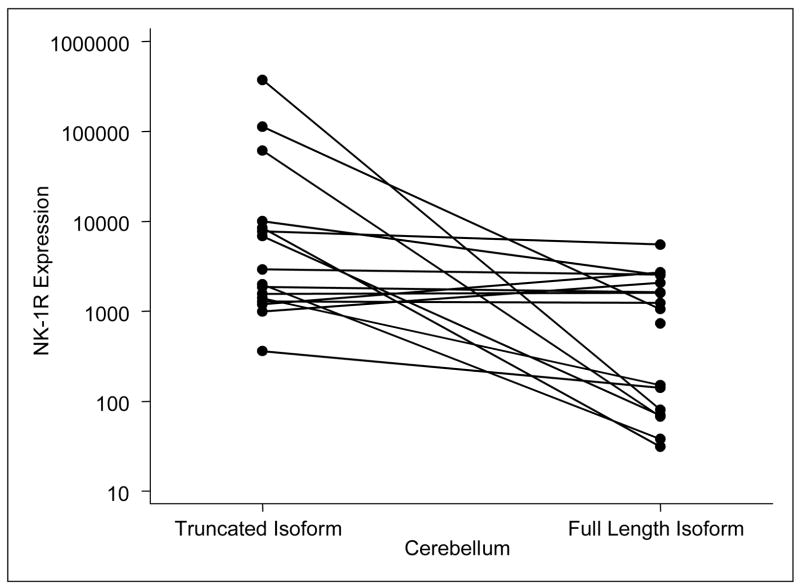
In order to measure the expression of full-length and truncated NK-1R mRNA levels in human brains tissues, total RNA isolated from these tissues was reverse transcribed. The cDNA were then amplified by the SYBR green based-real-time PCR using the L-F primer paired with redesigned reverse primers, respectively, for the full-length or the truncated NK1R mRNA. The expression levels of the full-length and truncated NK1R mRNA in different brain regions examined are summarized in Table 2. The data show the means and standard deviations of the expression levels (log10 of the copy numbers per ug total RNA) for full-length and truncated NK1R mRNA for each of nine brain regions. The truncated NK1R mRNA levels are significantly higher than those of full-length NK1R in cingulate cortex (P=0.0005, N = 23), cerebellum (P=0.0029, N = 17), nucleus accumbens (P = 0.09, N =9), and locus coeruleus (P=0.0093, N = 7). In the nucleus accumbens, there was a nearly significant trend towards superabundance of expression of the truncated NK1R in comparison with those in full-length NK1R (P= 0.09, N=9). The other brain regions examined, including hippocampus, caudate nucleus, and pons with locus ceruleus, revealed numeric differences, which, however, were not statistically significant. Sample size, however, precludes further interpretation of the findings in these regions. In the basal ganglia and the putamen regions, however, there were no observed differences in expression levels of these two forms of the receptor. Plots for NK1R expression levels in the cingulate cortex (N=23) and the cerebellum (N=17) are presented for both full-length and truncated NK1R (Figure 2 and 3). Four of the 23 samples had a higher expression level of the full-length NK1R in comparison to the truncated form, whereas 19 samples show a higher expression level of the truncated form in cingulate cortex (Figure 2). Two of the 17 samples had a higher expression level of the full-length NK1R in comparison to the truncated form in cerebellum. The expression levels of the two forms of NK1R among the nine regions were further compared. There was no significant difference in the truncated NK1R mRNA levels among the nine regions; however, there were significant differences in full-length NK1R mRNA levels among the nine regions studied (P=0.0024, Table 2). The highest full-length NK1R mRNA expression was observed in the putamen. In addition, the full-length NK1R mRNA expression levels in the putamen was significantly higher than that in the cerebellum (P= 0.025). Considering that the binding affinity of the full-length NK1R to SP, its preferred agonist, is much higher (at least 10 fold) than that of the truncated NK1R (Fong et al, 1992), our data is in agreement with a recent report (Nyman et al., 2007) which demonstrated that putamen had the highest binding potential in the living human brain using PET and [18F]SPA-RQ, a highly specific NK1R antagonist. Although the existence and expression of both the full-length and truncated NK1R have been documented, the differences in their expression in human brain tissue have been investigated only to a limited extent. Caberlotto et al (Caberlotto et al., 2003) reported that the full length NK1R was the most prevalent throughout the human brain using a quantitative PCR (Taqman) with four brain samples (Caberlotto et al., 2003). Our data, however, showed that the truncated NK1R mRNA levels were more abundant than that of the full-length NK1R in most of the regions examined (Table 2). There are several possible explanations for the differences between these observations. The improved sensitivity and specificity of the SYBR green based- real-time PCR assay may be the primary reason for the observed findings. The functional and signaling differences of the full-length and truncated NK1R have not been fully investigated. The sequence difference between the two forms of NK1R is in the length of the C-terminal tail (Fong et al., 1992). The presence of the C-terminal tail is the structural basis for SP-induced calcium increase (Lai et al., 2006; Lai et al., 2007). Activation of the truncated NK1R primes CCL5-mediated calcium increase, indicating that the truncated NK1R also has functional activity (Lai et al., 2006). Thus, the truncated NK1R most likely has different signal transduction pathway(s) in comparison to the full-length NK1R. We speculate that the expression and function of the truncated NK1R is universal in the regions studied, while the expression levels and function of the full-length NK1R is region-dependent and/or inducible. In addition, this modified real-time PCR assay could also be extended to in situ RT-PCR and applied to brain tissue microarrays in order to determine whether there is any anatomical distribution pattern of the full-length and truncated NK1R mRNA in human brain regions. Further studies are needed in order to elucidate the exact physiological significance of this differential expression of the full-length and truncated NK1R in the human brain.
Table II.
Comparisons of NK-1R expression levels (log10) within brain region
| Region of Brain Tissues | N | Truncated NK1R Mean (SD) | Full Length NK1R Mean (SD) | P-value |
|---|---|---|---|---|
| Cingulate Cortex | 23 | 8.61 (1.326) | 6.62 (1.969) | 0.0005 |
| Cerebellum | 17 | 8.66 (1.939) | 6.18 (1.746)* | 0.0029 |
| Nucleus Accumbens | 9 | 9.20 (1.133) | 7.23 (2.328) | 0.09 |
| Caudate Nucleus | 8 | 9.05 (1.760) | 8.34 (1.844) | 0.53 |
| Putamen | 8 | 8.86 (1.216) | 8.86 (2.079)* | 0.99 |
| Pons with Locus Ceruleus | 8 | 7.48 (1.538) | 7.17 (1.128) | 0.62 |
| Hippocampus | 7 | 9.26 (1.326) | 8.44 (1.035) | 0.33 |
| Locus Coeruleus | 7 | 8.67 (1.469) | 6.45 (0.930) | 0.0093 |
| Basal Ganglia | 6 | 8.64 (0.951) | 8.56 (1.533) | 0.92 |
| P-value | 0.0024 |
The means (log10 of copy number per ug of total RNA) and standard deviations for full-length and truncated NK-1R mRNA levels are showed within each of nine brain regions investigated.
P = 0.025.
Figure 2. NK-1R expression levels in the cingulate cortex by isoform.
Four of the 23 samples show a higher expression level of the full-length NK1R mRNA in comparison to the truncated form.
Figure 3. NK-1R expression levels in the cerebellum by isoform.
The plots of the full-length and truncated NK1R mRNA levels in the cerebellum. Two of the17 samples show a higher expression level of the full-length NK1R in comparison to the truncated form. The samples showing a higher level of expression of the full-length form than the truncated form were not the same samples in the two brain regions (i.e., six different samples showed a higher level of expression in the full-length NK1R in these two brain regions combined, while 34 samples showed a higher level of expression in the truncated form).
In summary, we developed a real-time PCR assay which detected both full-length and truncated NK1R mRNA in all brain regions studied. We demonstrated that the truncated NK1R has higher expression levels than that of the full-length NK1R. Among the regions studied, there was significant difference in the levels of the full-length NK1R mRNA, whereas significant differences in expression of the truncated NK1R were not observed. Further studies are needed in order to examine the differences of these two NK1Rs in G-protein coupling, signal transduction and functional consequences in order to understand the significance of the co-presence of these two forms of NK1R in human brain.
Acknowledgments
Supported by NIH P01-MH076388 and R01-MH049981 to S.D.D. Brain tissue specimens, data, and support were provided by the National NeuroAIDS Tissue Consortium (request R085). The NNTC is funded by the National Institutes of Health N01MH32002.
The authors thank Dr. Norma Gerard, Harvard University, for her gift of an NK-1R full-length plasmid. The authors also thank Stephen Jasionowski for his editorial assistance and thank Yan-Jian Wang for technical assistance. Brain tissue specimens, data, and support were provided by the National NeuroAIDS Tissue Consortium. The NNTC is funded by the National Institutes of Health N01MH32002.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aubry JM, Lundstrom K, Kawashima E, Ayala G, Schulz P, Bartanusz V, Kiss JZ. NK1 receptor expression by cholinergic interneurones in human striatum. Neuroreport. 1994;5:1597–600. doi: 10.1097/00001756-199408150-00014. [DOI] [PubMed] [Google Scholar]
- Bohm SK, Khitin LM, Smeekens SP, Grady EF, Payan DG, Bunnett NW. Identification of potential tyrosine-containing endocytic motifs in the carboxyl-tail and seventh transmembrane domain of the neurokinin 1 receptor. J Biol Chem. 1997;272:2363–72. doi: 10.1074/jbc.272.4.2363. [DOI] [PubMed] [Google Scholar]
- Bost KL. Tachykinin-mediated modulation of the immune response. Front Biosci. 2004;9:3331–2. doi: 10.2741/1484. [DOI] [PubMed] [Google Scholar]
- Caberlotto L, Hurd YL, Murdock P, Wahlin JP, Melotto S, Corsi M, Carletti R. Neurokinin 1 receptor and relative abundance of the short and long isoforms in the human brain. Eur J Neurosci. 2003;17:1736–46. doi: 10.1046/j.1460-9568.2003.02600.x. [DOI] [PubMed] [Google Scholar]
- Fong TM, Anderson SA, Yu H, Huang RR, Strader CD. Differential activation of intracellular effector by two isoforms of human neurokinin-1 receptor. Mol Pharmacol. 1992;41:24–30. [PubMed] [Google Scholar]
- Gerard NP, Garraway LA, Eddy RL, Jr, Shows TB, Iijima H, Paquet JL, Gerard C. Human substance P receptor (NK-1): organization of the gene, chromosome localization, and functional expression of cDNA clones. Biochemistry (Mosc) 1991;30:10640–6. doi: 10.1021/bi00108a006. [DOI] [PubMed] [Google Scholar]
- Ho WZ, Douglas SD. Substance P and neurokinin-1 receptor modulation of HIV. J Neuroimmunol. 2004;157:48–55. doi: 10.1016/j.jneuroim.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Ho WZ, Lai JP, Zhu XH, Uvaydova M, Douglas SD. Human monocytes and macrophages express substance P and neurokinin-1 receptor. J Immunol. 1997;159:5654–60. [PubMed] [Google Scholar]
- Hopkins B, Powell SJ, Danks P, Briggs I, Graham A. Isolation and characterisation of the human lung NK-1 receptor cDNA. Biochem Biophys Res Commun. 1991;180:1110–7. doi: 10.1016/s0006-291x(05)81181-7. [DOI] [PubMed] [Google Scholar]
- Kage R, Leeman SE, Boyd ND. Biochemical characterization of two different forms of the substance P receptor in rat submaxillary gland. J Neurochem. 1993;60:347–51. doi: 10.1111/j.1471-4159.1993.tb05857.x. [DOI] [PubMed] [Google Scholar]
- Kus L, Mazzone SB, Paxinos G, Geraghty DP. Autoradiographic localisation of substance P (NK1) receptors in human primary visual cortex. Brain Res. 1998;794:309–12. doi: 10.1016/s0006-8993(98)00270-4. [DOI] [PubMed] [Google Scholar]
- Lai J-P, Douglas SD, Wang Y-J, Ho W-Z. Real-time reverse transcription-PCR quantitation of substance P receptor (NK-1R) mRNA. Clin Diagn Lab Immunol. 2005;12:537–41. doi: 10.1128/CDLI.12.4.537-541.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JP, Douglas SD, Ho WZ. Human lymphocytes express substance P and its receptor. J Neuroimmunol. 1998a;86:80–6. doi: 10.1016/s0165-5728(98)00025-3. [DOI] [PubMed] [Google Scholar]
- Lai JP, Douglas SD, Rappaport E, Wu JM, Ho WZ. Identification of a delta isoform of preprotachykinin mRNA in human mononuclear phagocytes and lymphocytes. J Neuroimmunol. 1998b;91:121–8. doi: 10.1016/s0165-5728(98)00170-2. [DOI] [PubMed] [Google Scholar]
- Lai JP, Douglas SD, Zhao M, Ho WZ. Quantification of substance P mRNA in human mononuclear phagocytes and lymphocytes using a mimic-based RT-PCR. J Immunol Methods. 1999;230:149–57. doi: 10.1016/s0022-1759(99)00120-9. [DOI] [PubMed] [Google Scholar]
- Lai JP, Ho WZ, Kilpatrick LE, Wang X, Tuluc F, Korchak HM, Douglas SD. Full-length and truncated neurokinin-1 receptor expression and function during monocyte/macrophage differentiation. Proc Natl Acad Sci U S A. 2006;103:7771–6. doi: 10.1073/pnas.0602563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JP, Lai S, Luo X, Tuluc F, Tansky MD, Leeman SE, Douglas SD. The C-terminus Tail of NK1R is Required for SP-induced Intracellular Calcium Increase, PKC delta Phosphorylation, and IL-8 Expression. Proceedings of Neuropeptides 2007: Function, Dysfunctions & Therapeutic Options; Santorini Island, Greece. 2007. [Google Scholar]
- Lai JP, Yang JH, Douglas SD, Wang X, Riedel E, Ho WZ. Quantification of CCR5 mRNA in human lymphocytes and macrophages by real-time reverse transcriptase PCR assay. Clin Diagn Lab Immunol. 2003;10:1123–8. doi: 10.1128/CDLI.10.6.1123-1128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JP, Zhan GX, Campbell DE, Douglas SD, Ho WZ. Detection of substance P and its receptor in human fetal microglia. Neuroscience. 2000;101:1137–44. doi: 10.1016/s0306-4522(00)00398-5. [DOI] [PubMed] [Google Scholar]
- Li H, Leeman SE, Slack BE, Hauser G, Saltsman WS, Krause JE, Blusztajn JK, Boyd ND. A substance P (neurokinin-1) receptor mutant carboxyl-terminally truncated to resemble a naturally occurring receptor isoform displays enhanced responsiveness and resistance to desensitization. Proc Natl Acad Sci U S A. 1997;94:9475–80. doi: 10.1073/pnas.94.17.9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucey DR, Novak JM, Polonis VR, Liu Y, Gartner S. Characterization of substance P binding to human monocytes/macrophages. Clin Diagn Lab Immunol. 1994;1:330–5. doi: 10.1128/cdli.1.3.330-335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantyh PW, Rogers SD, Ghilardi JR, Maggio JE, Mantyh CR, Vigna SR. Differential expression of two isoforms of the neurokinin-1 (substance P) receptor in vivo. Brain Res. 1996;719:8–13. doi: 10.1016/0006-8993(96)00050-9. [DOI] [PubMed] [Google Scholar]
- Nyman MJ, Eskola O, Kajander J, Vahlberg T, Sanabria S, Burns D, Hargreaves R, Solin O, Hietala J. Gender and age affect NK1 receptors in the human brain - a positron emission tomography study with [18F]SPA-RQ. Int J Neuropsychopharmacol. 2007;10:219–29. doi: 10.1017/S1461145706006572. [DOI] [PubMed] [Google Scholar]
- Otsuka M, Yoshioka K. Neurotransmitter functions of mammalian tachykinins. Physiol Rev. 1993;73:229–308. doi: 10.1152/physrev.1993.73.2.229. [DOI] [PubMed] [Google Scholar]
- Payan DG, Brewster DR, Missirian-Bastian A, Goetzl EJ. Substance P recognition by a subset of human T lymphocytes. J Clin Invest. 1984;74:1532–9. doi: 10.1172/JCI111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakawa N, Sharif M, Hanley MR. Attenuation of agonist-induced desensitization of the rat substance P receptor by progressive truncation of the C-terminus. FEBS Lett. 1994;347:181–4. doi: 10.1016/0014-5793(94)00532-x. [DOI] [PubMed] [Google Scholar]
- Shanahan F, Denburg JA, Fox J, Bienenstock J, Befus D. Mast cell heterogeneity: effects of neuroenteric peptides on histamine release. J Immunol. 1985;135:1331–7. [PubMed] [Google Scholar]
- Stanisz AM, Scicchitano R, Dazin P, Bienenstock J, Payan DG. Distribution of substance P receptors on murine spleen and Peyer’s patch T and B cells. J Immunol. 1987;139:749–54. [PubMed] [Google Scholar]
- Takeda Y, Chou KB, Takeda J, Sachais BS, Krause JE. Molecular cloning, structural characterization and functional expression of the human substance P receptor. Biochem Biophys Res Commun. 1991;179:1232–40. doi: 10.1016/0006-291x(91)91704-g. [DOI] [PubMed] [Google Scholar]
- Tooney PA, Au GG, Chahl LA. Localisation of tachykinin NK1 and NK3 receptors in the human prefrontal and visual cortex. Neurosci Lett. 2000;283:185–8. doi: 10.1016/s0304-3940(00)00979-4. [DOI] [PubMed] [Google Scholar]
- Wozniak A, McLennan G, Betts WH, Murphy GA, Scicchitano R. Activation of human neutrophils by substance P: effect on FMLP-stimulated oxidative and arachidonic acid metabolism and on antibody-dependent cell-mediated cytotoxicity. Immunology. 1989;68:359–64. [PMC free article] [PubMed] [Google Scholar]