Abstract
The proton-translocating NADH-quinone oxidoreductase (EC 1.6.99.3) is the largest and least understood enzyme complex of the respiratory chain. The mammalian mitochondrial enzyme (also called complex I) contains more than 40 subunits, whereas its structurally simpler bacterial counterpart (NDH-1) in Paracoccus denitrificans and Thermus thermophilus HB-8 consists of 14 subunits. A major unsolved question is the location and mechanism of the terminal electron transfer step from iron–sulfur cluster N2 to quinone. Potent inhibitors acting at this key region are candidate photoaffinity probes to dissect NADH-quinone oxidoreductases. Complex I and NDH-1 are very sensitive to inhibition by a variety of structurally diverse toxicants, including rotenone, piericidin A, bullatacin, and pyridaben. We designed (trifluoromethyl)diazirinyl[3H]pyridaben ([3H]TDP) as our photoaffinity ligand because it combines outstanding inhibitor potency, a suitable photoreactive group, and tritium at high specific activity. Photoaffinity labeling of mitochondrial electron transport particles was specific and saturable. Isolation, protein sequencing, and immunoprecipitation identified the high-affinity specifically labeled 23-kDa subunit as PSST of complex I. Immunoprecipitation of labeled membranes of P. denitrificans and T. thermophilus established photoaffinity labeling of the equivalent bacterial NQO6. Competitive binding and enzyme inhibition studies showed that photoaffinity labeling of the specific high-affinity binding site of PSST is exceptionally sensitive to each of the high-potency inhibitors mentioned above. These findings establish that the homologous PSST of mitochondria and NQO6 of bacteria have a conserved inhibitor-binding site and that this subunit plays a key role in electron transfer by functionally coupling iron–sulfur cluster N2 to quinone.
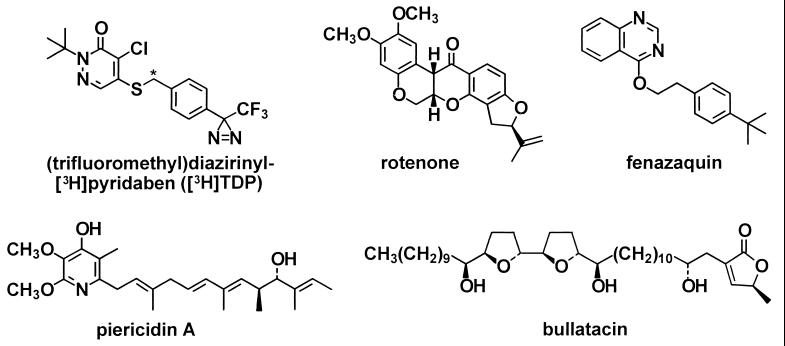
NADH-ubiquinone oxidoreductase (complex I: EC 1.6.99.3) is the first of three multisubunit enzyme complexes in the inner membranes of mitochondria forming the electron transport chain from NADH to oxygen. It is one of the most complicated enzyme complexes known, containing one noncovalently bound flavin mononucleotide and at least five iron–sulfur clusters recognized by their electron paramagnetic resonance signals. Complex I consists of more than 40 protein subunits, 7 of which (ND1 to ND6 plus ND4L) are encoded in the mitochondrial genome and the remainder (including PSST) of which originate from the nuclear DNA (1). Structural and functional defects of complex I are involved in many mitochondria-derived diseases (1, 2). Leber’s hereditary optical neuropathy is related to point mutations in the three mitochondrially encoded subunits ND1, ND4, and ND6 (3, 4). Chemically induced Parkinson’s disease from 1-methyl-4-phenylpyridinium ion (MPP+) is associated with the inhibition of complex I (5, 6). NADH-ubiquinone oxidoreductase inhibitors block induced ornithine decarboxylase activity and are thereby candidate cancer chemopreventive agents (7, 8). Complex I inhibitors are also important botanical and synthetic pesticides, including insecticides, miticides, and piscicides. Among the natural products, rotenone has been used for more than 300 years, and piericidin A and various annonaceous acetogenins (including bullatacin and rolliniastatin I) were candidate pesticides (9, 10). Pyridaben is one of four important synthetic heterocyclic insecticides and miticides with NADH-ubiquinone oxidoreductase as the target (9, 10).
Many prokaryotes possess a structurally simpler but highly homologous counterpart of NADH-ubiquinone oxidoreductase designated NDH-1. NDH-1 from Paracoccus denitrificans and Thermus thermophilus HB-8 has the same number of prosthetic groups as the mammalian enzyme and 14 homologous subunits (11). The bacterial enzymes are also inhibited by rotenone and piericidin A (12). The multiple components of NADH-quinone oxidoreductase from both prokaryotes and eukaryotes catalyze the transfer of electrons from NADH to quinone through the protein-bound prosthetic groups. A major unsolved question is the location and mechanism of the terminal step in this energy conservation process involving iron–sulfur cluster N2 and one or more subunits in electron transfer to quinone (1, 13, 14). This study uses a very potent inhibitor as a specific photoaffinity ligand to identify this key region or subunit, which was then found to be the common target for many potent inhibitors and toxicants.
The probe to dissect complex I was selected on the basis of introducing a suitable photoreactive group and tritium at high specific activity while retaining outstanding inhibitor potency. Each of the pesticides mentioned above inhibits NADH-ubiquinone oxidoreductase activity at nanomolar levels (9, 10) and was therefore a candidate prototype for a photoaffinity probe. Earlier studies with two rotenone-derived photoaffinity probes and isolated complex I recognized a single inhibitor-binding site localized in a 33-kDa protein (15, 16). We selected (trifluoromethyl)diazirinyl[3H]pyridaben ([3H]TDP) (Fig. 1) as our probe because it is more potent than rotenone as an NADH oxidase inhibitor, and the observed photoreactivity and high specific activity (56 Ci/mmol; 1 Ci = 37 GBq) were suitable to proceed (17). Electron transport particles (ETP) and bacterial membranes were used with the target enzyme in situ rather than as the isolated complex to ensure the intactness of mitochondrial complex I and bacterial NDH-1 (13).
Figure 1.
Structures of the photoaffinity probe (trifluoromethyl)diazirinyl[S-C3H2]pyridaben ([3H]TDP) and other NADH-quinone oxidoreductase inhibitors. Pyridaben is an analog of the photoaffinity probe with a t-butyl group instead of the (trifluoromethyl)diazirinyl substituent. Rolliniastatin I (structure not shown) is a stereoisomer of bullatacin.
MATERIALS AND METHODS
Materials.
Standard procedures were used to prepare ETP (18), cholate-treated membranes of P. denitrificans (19) and membranes of T. thermophilus HB-8 (20). The synthesis of [3H]TDP has been described (17). Sources for the inhibitors were rolliniastatin I from E. Estornell (University of Valencia, Spain); bullatacin from J. McLaughlin (Purdue University, West Lafayette, IN); others from the authors’ laboratories.
Enzyme Activity.
NADH oxidase activity and the effect of inhibitors were determined according to Singer (21) at 30°C for ETP and membranes of P. denitrificans and at 65°C for membranes of T. thermophilus.
Photoaffinity Labeling.
ETP (1 mg of protein) in 10 ml of buffer (250 mM sucrose in 50 mM sodium phosphate at pH 7.5) was treated with [3H]TDP (final concentration 0.6 to 240 nM, actual value determined by radioassay, introduced in 10–100 μl of ethanol) for 10 min at room temperature, and then with NADH (400 μM) and sodium pyruvate (1 mM) for 10 min. l-Lactate dehydrogenase (3 units) was added to destroy NADH, and after 2 min the suspension was cooled to 10°C and irradiated in a Rayonet photochemical reactor equipped with a merry-go-round apparatus and four RPR-3500A lamps at a distance of 3 cm for 20 min at 10°C; this irradiation condition completely photodecomposes the radioligand. The proteins were pelleted (12 min, 144,000 × g, 4°C) for analysis by electrophoresis and immunoprecipitation. Competitive inhibitors (in ethanol or dimethyl sulfoxide, <1% final concentration) were added 10 min before [3H]TDP.
Photoaffinity labeling of the bacterial membranes (1 mg of protein in 10 ml of buffer) was carried out in the same way except with 80 nM [3H]TDP.
Electrophoresis.
Sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) was performed according to Laemmli (22) on 13.5% slab gels (82 × 55 × 1.5 mm for analysis or 160 × 130 × 1.5 mm for preparative isolations). Blue native PAGE on 5–8% gels (160 × 130 × 1.5 mm) used n-dodecyl β-d-maltoside as the detergent (3% final concentration) according to Schägger and von Jagow (23). Their procedure was also used for electroelution of complex I from blue native PAGE gels (23). For localization and analysis of labeled protein bands the gels were cut into 1- to 2-mm slices, which were individually incubated with 0.5 ml of 30% hydrogen peroxide overnight at 55°C and acidified with 0.1 ml of 2 M hydrochloric acid, and the radioactivity was determined by liquid scintillation counting in 5 ml of Hionic Fluor (Packard Instrument, Meriden, CT) scintillation cocktail.
Immunoprecipitation.
The procedure of Anderson and Blobel (24) was applied to complex I purified by blue native PAGE or bacterial membranes, using antibodies against P. denitrificans (Pd) NQO6 (19), T. thermophilus (Tth) NQO6 (25), and ND1 (26). The PSST subunit shares 67% sequence identity with NQO6 of the P. denitrificans NDH-1, and antibodies raised against Pd NQO6 crossreact with its bovine counterpart (19). The labeled protein preparation in 240 μl of dilution buffer (1.25% Triton X-100/190 mM sodium chloride/6 mM EDTA/0.1 mM phenylmethanesulfonyl fluoride in 60 mM Tris⋅hydrochloride at pH 7.4) was treated with subunit-specific antiserum (5 μl) and incubated overnight at 4°C. Protein A-Sepharose CL-4B beads as a suspension (30 μl, equilibrated with 50 mM Tris⋅hydrochloride at pH 7.4) were added, and the mixture was incubated at room temperature for 2 hr. The beads were pelleted and washed four times with 1.0 ml of buffer A (0.1% Triton X-100/0.02% SDS/150 mM sodium chloride/5 mM EDTA/0.1 mM phenylmethanesulfonyl fluoride in 50 mM Tris⋅hydrochloride at pH 7.4) then once with buffer B (same as buffer A but without Triton X-100 and SDS). The samples were incubated in Laemmli’s 1× sample buffer [6.0% SDS/20% (vol/vol) glycerol/0.005% bromophenol blue in 80 mM Tris⋅hydrochloride at pH 6.8] for 2 hr at room temperature and pelleted, and the supernatants were applied to SDS/PAGE for analysis of the labeled immunoprecipitated proteins.
RESULTS
Photoaffinity Labeling of ETP.
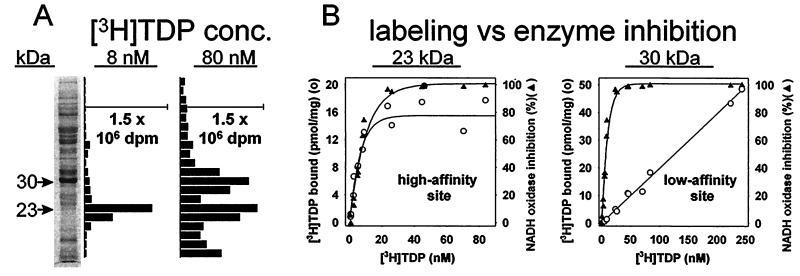
Photoaffinity labeling of ETP with a low concentration (8 nM) of [3H]TDP results in only one labeled protein region, with an apparent molecular mass of 23 kDa on SDS-PAGE. At a [3H]TDP concentration of 80 nM an additional labeled region of 30 kDa molecular mass is also evident (Fig. 2A). Labeling at the 23-kDa site reaches half of the maximum value at a [3H]TDP concentration of ≈6 nM, approximating the level required for 50% inhibition of NADH oxidase activity; in fact, binding of [3H]TDP to the 23-kDa protein correlates with inhibition of enzyme activity (Fig. 2B). The second labeled region, the 30-kDa protein, does not saturate up to 240 nM, far beyond the level completely inhibiting the enzyme (Fig. 2B).
Figure 2.
Photoaffinity labeling with [3H]TDP of specific binding sites at apparent molecular masses of 23 and 30 kDa in ETP. (A) SDS/PAGE analysis with a 13.5% gel establishes labeling of only the 23-kDa region at 8 nM [3H]TDP and of 23- and 30-kDa regions at 80 nM. (B) Labeling of the 23-kDa region (high-affinity site) correlates with inhibition of NADH oxidase activity. Labeling of the 30-kDa region (low-affinity site) does not undergo saturation up to 240 nM [3H]TDP. Conditions as in A but varying the [3H]TDP level. Values for [3H]TDP bound at above saturation levels are less reliable than at lower concentrations because of increasing background.
Identification of the High-Affinity Specific Binding Site Protein and the Second Labeled Protein in ETP.
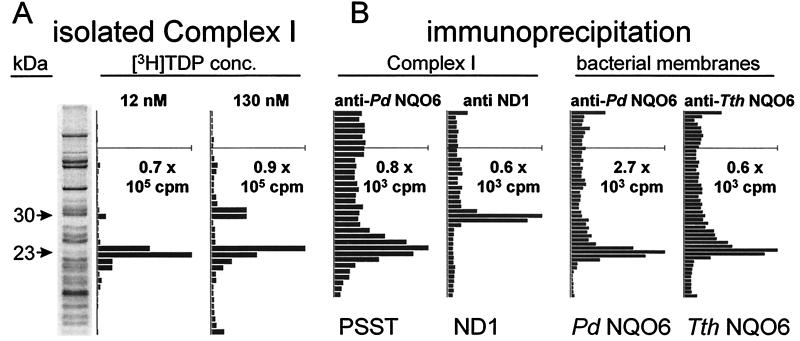
Identification of the labeled 23- and 30-kDa proteins required purification of complex I from ETP followed by protein sequencing or immunoassay of the proposed subunits. Separation by blue native PAGE of ETP photoaffinity-labeled with [3H]TDP at 8 nM gave only the complex I region with high radioactivity relative to protein (data not shown). Fractionation by SDS/PAGE of purified complex I labeled at 12 and 130 nM [3H]TDP gave the same 23- and 30-kDa-labeled subunits as determined with ETP directly (Fig. 3A), demonstrating that both radiolabeled sites reside in complex I. Tryptic digestion of the 23-kDa subunit thus obtained yielded peptides with sequences of PSST (27), namely (Leu)33-(Asp)34-Asp35-Leu36-Ile37-Asn38-(Trp)39-Ala40 and Phe72-Gly73-Val74-(Val)75-Phe76 (parentheses indicate amino acids predicted but not conclusively identified).
Figure 3.
Identification of the photoaffinity-labeled complex I high-affinity binding site protein (molecular mass 23 kDa) as PSST and the 30-kDa protein as ND1 and of the photoaffinity-labeled bacterial NDH-1 site protein (molecular mass 23 kDa) as NQO6. (A) SDS/PAGE analysis as in Fig. 2A but complex I isolated from ETP by blue native PAGE after labeling. Two of the darkest protein bands at 52 and 56 kDa are from complex V. (B) Immunoprecipitation of labeled complex I involved blue native PAGE followed by treatment with anti-Pd NQO6 antibody for PSST and anti-bovine ND1 antibody for ND1. Immunoprecipitation of labeled bacterial membranes involved direct treatment of P. denitrificans and T. thermophilus preparations with anti-Pd NQO6 antibody and anti-Tth NQO6 antibody, respectively.
Immunoprecipitation of the complex I high-affinity binding site protein with anti-Pd NQO6 antibody established its identity as PSST, and precipitation of the 30-kDa labeled protein with anti-ND1 antibody defined that it is ND1 (Fig. 3B).
Identification of the Specific Binding Site Protein in Bacterial Membranes.
The enzyme complexes of P. denitrificans and T. thermophilus are sensitive to the same inhibitors as complex I with strong inhibition by pyridaben, rotenone, piericidin A, bullatacin, and rolliniastatin I at 100 nM and 4′-decyl-MPP+ at 100 μM. The inhibition site of bacterial NADH-quinone oxidoreductase is therefore a suitable model for that of the mammalian enzyme. The photoaffinity probe with both bacterial membranes labeled the NQO6 subunit (molecular mass 23 kDa) of NDH-1—i.e., the counterpart of PSST of mitochondria, based on immunoprecipitation assays (Fig. 3B).
Common High-Affinity Binding Site for Structurally Diverse Inhibitors.
We compared the binding site(s) of various unlabeled inhibitors with that of [3H]TDP by allowing them to compete before irradiation and analysis of the potentially reduced labeling for the 23-kDa subunit. High-affinity binding was completely prevented by pyridaben, rotenone, piericidin A, bullatacin, and rolliniastatin I (individually tested at 2 μM) and 4′-decyl-MPP+ (100 μM) (data not shown), levels at which each of these compounds fully blocked NADH oxidase activity. No competition with [3H]TDP was observed for antimycin A at 2 μM or MPP+ at 1 mM, concentrations that did not inhibit activity at complex I. Photoaffinity labeling of the ND1 subunit was not reduced by pyridaben, rotenone, piericidin A, bullatacin, or rolliniastatin I at 2 μM.
DISCUSSION
There is a common high-affinity binding domain for structurally diverse inhibitors associated with the inhibition of NADH-ubiquinone oxidoreductase activity. This has been shown with beef heart ETP using [14C]rotenone (28), [14C]piericidin A (29), and [3H]fenazaquin (30) and with insect submitochondrial particles and partially purified complex I by using [3H]dihydrorotenone and a [3H]aminopyrimidine (31, 32). In all cases inhibition of radioligand binding by a variety of compounds was proportional to inhibition of enzyme activity but the lack of a suitable photoaffinity probe and high nonspecific binding ruled out subunit localization in complex I.
NADH-ubiquinone oxidoreductase in ETP is photoaffinity-labeled by [3H]TDP at low concentration (e.g., 8 nM) at a specific site on a protein of 23 kDa. The labeling is high affinity and saturable and correlates with inhibition of enzyme activity. The 23-kDa protein that contains the high-affinity binding site is therefore the primary target of [3H]TDP action in mitochondrial membranes. Isolation and sequencing of this labeled protein identified it as the PSST subunit of complex I, a finding confirmed by immunoprecipitation with anti-Pd NQO6, a crossreacting antibody developed against the bacterial equivalent of PSST. Bacterial membranes of P. denitrificans and T. thermophilus are also photoaffinity labeled by [3H]TDP, and this occurs specifically at the NQO6 subunit as determined by immunoprecipitation. All of these findings support the labeling of an equivalent specific high-affinity site in ETP and bacterial membranes—i.e., this is a conserved binding site in mitochondrial complex I and its bacterial counterpart NDH-1. The labeling of PSST with [3H]TDP is displaced by pyridaben, rotenone, piericidin A, bullatacin, rolliniastatin-I, and 4-decyl-MPP+. Hence, the photoaffinity labeling and identification of the high-affinity binding site in PSST establishes that diverse inhibitors bind in a mutually exclusive way—i.e., at the same or closely coupled sites or overlapping subsites similar to the observation by x-ray crystallography for the Qo site inhibitors of complex III (33).
A second site in ETP is labeled at higher [3H]TDP levels (e.g., 80 nM) and is not saturated up to 240 nM radioligand, a concentration well above that required for complete inhibition of NADH-ubiquinone oxidoreductase activity. Labeling at the secondary site involves high specificity in binding and reaction with a single 30-kDa protein within a mixture of >100 proteins in ETP. Immunoprecipitation identifies this secondary binding site for [3H]TDP as residing in ND1. Interestingly, this is the same protein photoaffinity-labeled in isolated complex I by two rotenone analogues (15, 16). Thus, the site labeled with rotenone derivatives as photoaffinity probes was not the PSST high-affinity site identified here with [3H]TDP but instead the ND1 secondary site. There are three possible explanations for labeling ND1 instead of PSST with the rotenone derivatives: the specific activity of [3H]arylazidoamorphigenin (0.02 Ci/mmol; ref. 15) was too low to detect the high-affinity binding site; photoaffinity labeling with [3H]dihydrorotenone (58 Ci/mmol; ref. 16) was inefficient despite the arylketone moiety; NADH-ubiquinone oxidoreductase activity of isolated complex I is less sensitive than that of ETP to rotenone and piericidin A (13), possibly due to changes in the high-affinity site. An important observation is that photoaffinity labeling of the ND1 subunit with [3H]TDP is not displaced by the complex I inhibitors studied, including rotenone, at levels that totally block labeling of the high-affinity site. On this basis the ND1 subunit is not directly involved in the action of the complex I inhibitors studied here.
Two inhibitor-binding sites in ETP have been proposed for rotenone, piericidin A, and MPP+, in part on the basis of radioligand binding experiments in the presence of bovine serum albumin; these two sites have similar affinities but unequal contribution to the inhibition and both must be occupied for complete block of NADH oxidation (34–36). A single inhibitor-binding site is observed in other studies with partially purified insect complex I and bovine submitochondrial particles (32). The current photoaffinity studies without albumin show a single high-affinity binding region responsible for inhibition of enzyme activity. However, they do not define the stoichiometry of [3H]TDP binding sites relative to the PSST and ND1 subunits.
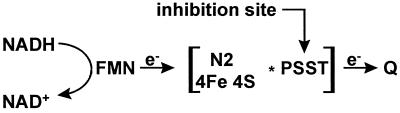
Several lines of evidence assign the PSST subunit as the final protein component involved in electron transfer from iron–sulfur cluster N2 to quinone (Fig. 4). Cluster N2 has the highest redox potential and is therefore considered to be the final iron–sulfur cluster. Electron paramagnetic resonance studies establish that rotenone and piericidin A interrupt electron transfer between cluster N2 and quinone (14). The PSST subunit and its bacterial counterpart are identified here as the target protein for [3H]TDP and in the former case also for rotenone, piericidin A, and other potent complex I inhibitors. PSST and NQO6 have highly conserved cysteine motifs in their primary structure and are therefore homologous candidate subunits for housing N2. These two subunits are located at the interface between the hydrophilic extramembrane portion and the hydrophobic intermembrane region (11, 37). We therefore propose that PSST or NQO6 is directly associated with iron–sulfur cluster N2 and serves as a conduit in the transfer of electrons to the quinone. Localization of the high-affinity inhibitor-binding site in the PSST and NQO6 subunits of mammalian and bacterial NADH-quinone oxidoreductases provides a crucial step in developing an architectural and mechanistic model for the electron and proton transport by complex I.
Figure 4.
Scheme showing proposed molecular architecture of complex I components involved in the terminal steps of electron transfer to quinone. See text for details.
Acknowledgments
Advice was provided by Bachir Latli, Gary Quistad, Manfred Schneider, and Dominik Hainzl (University of California, Berkeley) and assistance by Karina Lichtenstein (The Scripps Research Institute). Bruce Cochran and Brian A. C. Ackrell (Veterans Affairs Medical Center, San Francisco) prepared the mitochondria. Peptide sequencing was carried out by Young-Moo Lee and Tara Martinez (University of California, Davis). Robert Hollingworth of Michigan State University and Graham Palmer of Rice University provided useful comments. This work was supported by National Institutes of Health Grants P01 ES00049 and R01 ES04863 (to J.E.C.) and R01 GM33712 (to T.Y.) and by a Department of Veterans Affairs Merit Review Grant (to T.P.S.).
ABBREVIATIONS
- complex I
mitochondrial proton-translocating NADH-ubiquinone oxidoreductase
- ETP
electron transport particles
- MPP+
1-methyl-4-phenylpyridinium ion
- ND1
36-kDa subunit of complex I
- NDH-1
bacterial proton-translocating NADH-quinone oxidoreductase
- PSST
20-kDa subunit of complex I
- TDP and [3H]TDP
(trifluoromethyl)diazirinylpyridaben and tritiated form used as a photoaffinity ligand
- Pd
Paracoccus denitrificans
- Tth
Thermus thermophilus
References
- 1.Walker J E. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 2.Ernster, L., Luft, R. & Orrenius, S., eds. (1995) Mitochondrial Diseases, Nobel Symposium 90, Biochim. Biophys. Acta (Special Issue) 1271, 1–292. [PubMed]
- 3.Howell N. J Bioenerg Biomembr. 1997;29:165–173. doi: 10.1023/a:1022690030664. [DOI] [PubMed] [Google Scholar]
- 4.Robinson B H. Biochim Biophys Acta. 1998;1364:271–286. doi: 10.1016/s0005-2728(98)00033-4. [DOI] [PubMed] [Google Scholar]
- 5.Singer T P, Castagnoli N, Jr, Ramsay R R, Trevor A J. J Neurochem. 1987;49:1–8. doi: 10.1111/j.1471-4159.1987.tb03384.x. [DOI] [PubMed] [Google Scholar]
- 6.Ramsay R R, Youngster S K, Nicklas W J, McKeown K A, Jin Y-Z, Heikkila R E, Singer T P. Proc Natl Acad Sci USA. 1989;86:9168–9172. doi: 10.1073/pnas.86.23.9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerhäuser C, Mar W, Lee S K, Suh N, Luo Y, Kosmeder J, Luyengi L, Fong H H S, Kinghorn A D, Moriarty R M, et al. Nat Med. 1995;1:260–266. doi: 10.1038/nm0395-260. [DOI] [PubMed] [Google Scholar]
- 8.Fang N, Casida J E. Proc Natl Acad Sci USA. 1998;95:3380–3384. doi: 10.1073/pnas.95.7.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollingworth R M, Ahammadsahib K I. Rev Pestic Toxicol. 1995;3:277–302. [Google Scholar]
- 10.Lümmen P. Biochim Biophys Acta. 1998;1364:287–296. doi: 10.1016/s0005-2728(98)00034-6. [DOI] [PubMed] [Google Scholar]
- 11.Yagi T, Yano T, Di Bernardo S, Matsuno-Yagi A. Biochim Biophys Acta. 1998;1364:125–133. doi: 10.1016/s0005-2728(98)00023-1. [DOI] [PubMed] [Google Scholar]
- 12.Yagi T. Biochim Biophys Acta. 1993;1141:1–17. doi: 10.1016/0005-2728(93)90182-f. [DOI] [PubMed] [Google Scholar]
- 13.Singer T P, Ramsay R R. In: Molecular Mechanisms in Bioenergetics. Ernster L, editor. Amsterdam: Elsevier; 1992. pp. 145–162. [Google Scholar]
- 14.Ohnishi T. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 15.Earley F G P, Ragan C I. Biochem J. 1984;224:525–534. doi: 10.1042/bj2240525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Earley F G P, Patel S D, Ragan C I, Attardi G. FEBS Lett. 1987;219:108–112. doi: 10.1016/0014-5793(87)81200-0. [DOI] [PubMed] [Google Scholar]
- 17.Latli B, Morimoto H, Williams P G, Casida J E. J Labelled Compd Radiopharm. 1998;41:191–199. [Google Scholar]
- 18.Crane F L, Glenn J L, Green D E. Biochim Biophys Acta. 1956;22:475–487. doi: 10.1016/0006-3002(56)90058-0. [DOI] [PubMed] [Google Scholar]
- 19.Takano S, Yano T, Yagi T. Biochemistry. 1996;35:9120–9127. doi: 10.1021/bi9605853. [DOI] [PubMed] [Google Scholar]
- 20.Yagi T, Hon-nami K, Ohnishi T. Biochemistry. 1988;27:2008–2013. doi: 10.1021/bi00406a030. [DOI] [PubMed] [Google Scholar]
- 21.Singer T P. In: Methods of Biochemical Analysis. Glick D, editor. Vol. 22. New York: Wiley; 1974. pp. 123–175. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Schägger H, von Jagow G. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 24.Anderson D J, Blobel G. Methods Enzymol. 1983;96:111–120. doi: 10.1016/s0076-6879(83)96012-3. [DOI] [PubMed] [Google Scholar]
- 25.Yano T, Chu S S, Sled’ V D, Ohnishi T, Yagi T. J Biol Chem. 1997;272:4201–4211. doi: 10.1074/jbc.272.7.4201. [DOI] [PubMed] [Google Scholar]
- 26.Yagi T, Hatefi Y. J Biol Chem. 1988;263:16150–16155. [PubMed] [Google Scholar]
- 27.Arizmendi J M, Runswick M J, Skehel J M, Walker J E. FEBS Lett. 1992;301:237–242. doi: 10.1016/0014-5793(92)80248-f. [DOI] [PubMed] [Google Scholar]
- 28.Horgan D J, Singer T P, Casida J E. J Biol Chem. 1968;243:834–843. [PubMed] [Google Scholar]
- 29.Horgan D J, Casida J E. Biochem J. 1968;108:153–154. doi: 10.1042/bj1080153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood E, Latli B, Casida J E. Pestic Biochem Physiol. 1996;54:135–145. [Google Scholar]
- 31.Jewess P J. Biochem Soc Trans. 1994;22:247–251. doi: 10.1042/bst0220247. [DOI] [PubMed] [Google Scholar]
- 32.Okun J G, Lümmen P, Brandt U. J Biol Chem. 1999;274:2625–2630. doi: 10.1074/jbc.274.5.2625. [DOI] [PubMed] [Google Scholar]
- 33.Kim H, Xia D, Yu C-A, Xia J-Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Proc Natl Acad Sci USA. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutman M, Singer T P, Casida J E. J Biol Chem. 1970;245:1992–1997. [PubMed] [Google Scholar]
- 35.Ramsay R R, Krueger M J, Youngster S K, Gluck M R, Casida J E, Singer T P. J Neurochem. 1991;56:1184–1190. doi: 10.1111/j.1471-4159.1991.tb11409.x. [DOI] [PubMed] [Google Scholar]
- 36.Gluck M R, Krueger M J, Ramsay R R, Sablin S O, Singer T P, Nicklas W J. J Biol Chem. 1994;269:3167–3174. [PubMed] [Google Scholar]
- 37.Finel M, Skehel J M, Albracht S P J, Fearnley I M, Walker J E. Biochemistry. 1992;31:11425–11434. doi: 10.1021/bi00161a022. [DOI] [PubMed] [Google Scholar]