Abstract
Central core disease is a rare, nonprogressive myopathy that is characterized by hypotonia and proximal muscle weakness. In a large Mexican kindred with an unusually severe and highly penetrant form of the disorder, DNA sequencing identified an I4898T mutation in the C-terminal transmembrane/luminal region of the RyR1 protein that constitutes the skeletal muscle ryanodine receptor. All previously reported RYR1 mutations are located either in the cytoplasmic N terminus or in a central cytoplasmic region of the 5,038-aa protein. The I4898T mutation was introduced into a rabbit RYR1 cDNA and expressed in HEK-293 cells. The response of the mutant RyR1 Ca2+ channel to the agonists halothane and caffeine in a Ca2+ photometry assay was completely abolished. Coexpression of normal and mutant RYR1 cDNAs in a 1:1 ratio, however, produced RyR1 channels with normal halothane and caffeine sensitivities, but maximal levels of Ca2+ release were reduced by 67%. [3H]Ryanodine binding indicated that the heterozygous channel is activated by Ca2+ concentrations 4-fold lower than normal. Single-cell analysis of cotransfected cells showed a significantly increased resting cytoplasmic Ca2+ level and a significantly reduced luminal Ca2+ level. These data are indicative of a leaky channel, possibly caused by a reduction in the Ca2+ concentration required for channel activation. Comparison with two other coexpressed mutant/normal channels suggests that the I4898T mutation produces one of the most abnormal RyR1 channels yet investigated, and this level of abnormality is reflected in the severe and penetrant phenotype of affected central core disease individuals.
Central core disease (CCD) is an autosomal dominant congenital myopathy (1). Both clinical and histological variability is observed, but affected individuals typically display hypotonia and proximal muscle weakness in infancy, leading to the delay of motor milestones (2). The clinical course of the disorder is usually slow or nonprogressive in adulthood, and the severity of the symptoms may vary from normal to significant muscle weakness. A range of skeletal defects is associated with the disorder, including congenital hip dislocation, thoracic deformities, pes cavus, clubfoot, and kyphoscoliosis (2). Microscopic examination of CCD-affected skeletal muscle reveals a predominance of type I fibers containing amorphous-looking areas (cores) that do not stain with oxidative and phosphorylase histochemical techniques, and this assay forms the basis of a diagnostic test (3). Electron microscopy of the core regions has demonstrated a relative lack or even absence of mitochondria, proliferation of the sarcotubular system, less numerous glycogen granules, and changes in the contractile apparatus, including less well-defined myofibrils and Z disk streaming (4). The exact biochemical nature of the core region remains unknown. However, analysis of CCD muscle fibers by immunocytochemistry revealed that desmin intermediate filaments were reduced in the core regions and overexpressed in the extra core regions (5).
CCD is closely associated with malignant hyperthermia (MH) (6). MH is a pharmacogenetic disorder of skeletal muscle that is triggered in susceptible individuals by commonly used inhalational anesthetics and depolarizing muscle relaxants (7). MH susceptibility (MHS) is determined by an in vitro contracture test (IVCT) that measures the sensitivity of biopsied skeletal muscle fibers to caffeine and halothane (8). Both MH (9, 10) and CCD (11, 12) have been linked to the human RYR1 gene located on chromosome 19q13.1, which encodes the skeletal muscle Ca2+ release channel and is commonly known as the ryanodine receptor (RyR1). To date, some 17 missense mutations in RYR1 have been linked to MH, of which five also have been linked to CCD (13). Central cores also have been detected in the skeletal muscle of a subset of individuals affected with hypertrophic cardiomyopathy, which is linked to mutations in the β-myosin heavy chain gene on chromosome 14q1 (14). The MH status of these individuals has never been investigated.
Here we report a large Mexican kindred in which all affected members suffer from a clinically severe and highly penetrant form of CCD. Sequencing of the entire RYR1 cDNA in an affected member identified a single mutation in the C-terminal transmembrane/luminal domain of the protein. The introduction of this mutation into a recombinant RyR1 protein expressed in HEK-293 cells resulted in loss of channel activation by caffeine and halothane and a significant reduction in ryanodine binding. When mutant and normal channels were coexpressed in a 1:1 ratio to mimic the heterozygous state in CCD patients, transfected cells exhibited abnormal properties, including a reduction in maximal release of Ca2+ through the channel, reduced luminal Ca2+ storage, and increased cytoplasmic Ca2+ levels. These findings, which point to high basal activity of the mutant Ca2+ channel, could explain the muscle weakness and muscle atrophy observed in CCD patients in this family.
METHODS
Patients.
Thirty-five members of a Mexican CCD family participated in this study, and blood and tissue samples were obtained with informed consent. Muscle biopsies were taken from two members for MHS and CCD diagnosis. The IVCT was performed on biopsied vastus lateralis muscle in the Cork MH Testing Center as outlined by the European MH Group protocol (8). A patient is diagnosed as MHS if the muscle biopsy strips produce a sustained increase of 0.2 g in muscle tension at a caffeine concentration of 2.0 mM or less and a halothane concentration of 2% vol/vol or less. MH normal is diagnosed if the 0.2 g threshold is not attained at these concentrations, and MH equivocal (MHE) is diagnosed if the threshold value is attained with caffeine [MHE(c)] or with halothane [MHE(h)] but not with both. No anesthetic complications were recorded in the family despite the fact that 19 members, including six affected individuals not available for this study, were exposed to the MH triggering agent halothane.
Nucleic Acid Isolation.
Genomic DNA was extracted from EDTA-preserved blood by nuclear isolation and lysis, protease digestion, and ethanol precipitation (15). Total RNA was extracted from a 100-mg muscle sample biopsied from individual III:4 by using RNA Isolator total RNA extraction reagent (Genosys Biotechnologies, Cambridge, U.K.). First-strand cDNA synthesis was performed by using 200 ng of random hexamer primer, 1 μg total RNA, and 15 units of avian myeloblastosis virus reverse-transcriptase in a 20-μl reaction volume (16).
Genotyping and Linkage Analysis.
The polymorphic markers D19S220 and D19S47 were amplified by PCR in the presence of [α-32P]dCTP, and the allele sizes were determined on denaturing 8% polyacrylamide 7 M urea sequencing gels. Logarithm of odds scores were calculated with the liped program by using a gene frequency of 0.001 for CCD (17).
DNA Sequencing.
The entire 15-kb RYR1 cDNA was amplified in 23 overlapping PCR fragments by using primers designed from the published human RYR1 cDNA sequence (18). After thermal cycling, primers and unincorporated deoxynucleotides were removed from amplification reactions using a Sephacryl S-300 spin column (Amersham Pharmacia). Purified DNA fragments were sequenced directly by using a ThermoSequenase radiolabeled terminator cycle sequencing protocol (Amersham Pharmacia).
Glycosylase-Mediated Polymorphism Detection (GMPD) Analysis.
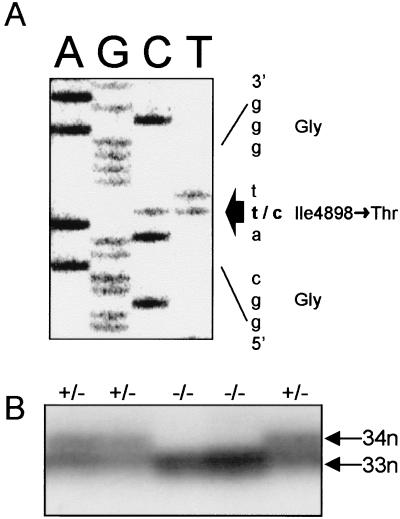
Genomic DNA samples were genotyped for the T14693C mutation by using the recently described GMPD method (19). A 122-bp DNA fragment, which contained the mutation site, was amplified from 100 ng of genomic DNA in the following 10-μl amplification reaction: 50 mM KCl, 10 mM Tris⋅HCl (pH 8.8), 0.1% Triton X-100, 0.2 mM dGTP, dATP, dUTP, and dCTP, 1.5 mM MgCl2, 20 ng 32P-labeled forward primer (5′-ACATGTACGTGGGTGTCCGGGCT-3′), 20 ng reverse primer (5′-GACGATGACGAAGAAGAAGAAGG-3′), and 0.8 units of Taq polymerase. The thermal profile of the PCR consisted of 95°C for 5 min, followed by 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min. The 3′ end of the forward primer was designed so that the first uracil residue incorporated downstream of the forward primer was at, or distal to, the 14693 position. After amplification, glycoslyase-mediated cleavage of the amplified product was performed (19). Uracil DNA-glycosylase (0.1 units, New England Biolabs) was added to 3 μl of PCR, and the solution was incubated for 30 min at 37°C. To cleave the resulting apyrimidinic sites, NaOH was added to a final concentration of 0.05 M followed by an incubation at 95°C for 15 min. The reaction was neutralized by the addition of Tris⋅HCl (pH 8.0) to a final concentration of 18 mM. Cleavage products were resolved by denaturing gel electrophoresis (20%, 19:1 polyacrylamide) and visualized by autoradiography. The normal T allele was detected as a 33-nt fragment and the mutant C allele as a 34-nt fragment.
A nested primer approach was necessary when screening cDNA samples because of the high levels of dTTP present in the cDNA synthesis reactions. Initially, a 702-bp fragment spanning the mutation site was amplified by using primers 14456 (5′-GCACCATCCTGTCCTCTGTCA-3′) and 15137 (5′-GGGCTTGCTGTGAGAATAAGG-3′). The reactions were carried out in a total volume of 25 μl containing 50 mM KCl, 10 mM Tris⋅HCl (pH 8.8), 0.1% Triton X-100, 1.5 mM MgCl2, 0.2 mM of dGTP, dATP, dTTP, and dCTP, 50 ng of each primer, 0.5 μl of cDNA synthesis reaction, and 1 unit of Taq polymerase. A thermal profile of 95°C for 5 min followed by 34 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, and a final soak of 72°C for 10 min was performed. After amplification, the PCR was diluted 1:500 with sterile water, and 1 μl served as template in the GMPD amplification reaction.
Construction of the I4897T Mutation in Rabbit RYR1 cDNA.
The construction and expression of single CCD/MH mutant forms of the full-length rabbit skeletal muscle ryanodine receptor have been described (20). Because of differences in the protein sequences, the human mutation site occurs at Ile-4897 in the rabbit RyR1 sequence. The I4897T mutation was introduced into RyR1 cassette 11 (pCS11) by using the QuickChange site-directed mutagenesis protocol (Strategene). The ClaI/HindIII fragment of pCS11-I4897T then was exchanged for the same region in full-length pcDNA-RyR1.
Cell Culture, DNA Transfection, and Immunostaining.
DNA transfection into HEK-293 cells was achieved by using the calcium phosphate precipitation method (20). In a routine transfection, 20 μg of RYR1 cDNA in the pcDNA3 vector were added to each 10-cm dish. In the coexpression experiments, cDNAs were cotransfected in equimolar ratios while maintaining the total amount of cDNA at 20 μg per dish. Immunostaining with mAb 34C was carried out as described (20). Samples of whole-cell extracts containing 100 μg of protein were separated by 7.5% SDS/PAGE and transferred to nitrocellulose. The samples were probed with mAb 34C and secondary horseradish peroxidase-conjugated anti-mouse IgG.
[3H]Ryanodine Binding Assay.
[3H]Ryanodine binding to supernatants from transfected HEK-293 cells was measured as described (21). Supernatants from 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)-solubilized cells were centrifuged at 14,000 rpm for 30 min (0.1–0.5 mg protein/ml) and incubated with various concentrations (1 nM–100 μM) of Ca2+ for 2 hr at 37°C in 0.25 ml of a solution containing 0.2 M KCl, 20 mM Pipes, pH 7.2, 0.1% CHAPS, 1 mM EGTA, and 10 nM [3H]ryanodine. The amount of [3H]ryanodine bound was determined by filtration through Whatman GF/B membranes presoaked with 1% polyethylenimine. The filters were washed three times with 5 ml of an ice-cold solution containing 20 mM Hepes, pH 7.2, and 0.2 M KCl. Radioactivity associated with the filters was determined by liquid scintillation counting. Nonspecific binding was determined by measuring [3H]ryanodine binding in the presence of 10 μM unlabeled ryanodine.
Fluorescence Measurements.
Ca2+ photometry was carried out with a Photon Technology International (Princeton) microfluorimetry system as described (20). In the Ca2+ imaging assay (22), cells were loaded with 4 μM fura-2 acetoxymethyl ester/0.02% pluronic F-127 for 45 min, and emitted fluorescence was fed into a charge-coupled device camera instead of a photomultiplier tube. Digital images acquired at two frames per 3 sec were analyzed with image master 2.0 (Photon Technology). The ratio of fluorescence emitted at 340 and 380 nm (340:380 ratio) of single cells was converted to Ca2+ concentration by the method of Grynkiewicz et al. (23): [Ca2+] = Kd⋅[(R − Rmin)/(Rmax − R)]⋅(Sf2/Sb2) (23). The value of 16 for Rmax was obtained by using 10 μM ionomycin; Rmin and the Sf2/Sb2 constant were 0.4 and 15.2, respectively. The value of 224 nM for the apparent Kd of Ca2+ binding to fura-2 was used. All other procedures were the same as for the Ca2+ photometric assay. In the thapsigargin-induced Ca2+ release studies, transfected cells initially were treated with 10 mM caffeine, washed, and then treated with 1.5 μM thapsigargin.
Statistical Methods.
All data are expressed as mean ± SE. Linear regression analysis was performed by using origin software (Microcal Software, Northampton, MA). An unpaired Student’s t test was used for statistical comparisons of mean values between samples. A value of P < 0.05 was taken to indicate statistical significance.
RESULTS
CCD Diagnosis and Linkage to the RYR1 Gene Locus.
Clinical examination led to the diagnosis of CCD in 21 individuals in the large Mexican kindred. Affected individuals presented with one or more of the following symptoms consistent with CCD: muscular weakness at birth, fetal hypotonia, delayed motor milestones, foot deformities, proximal muscle weakness, congenital hip dislocation, and kyphoscoliosis. The severity ranged from mild to severe. Histological analysis consisted of NADH-dehydrogenase enzyme histostaining of vastus lateralis muscle samples biopsied from individuals III:4 and IV:10. Eighty-five percent of fibers in the III:4 sample displayed cores while 95% of the fibers were classified as type I. Fifty-three percent of the muscle fibers in IV:10 displayed cores, and a type I fiber prevalence and mild fiber size variance was noted. Muscle fibers containing two or more cores were observed.
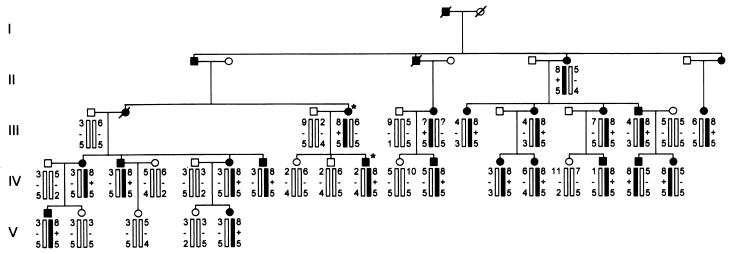
Linkage analysis generated a logarithm of odds score of 6.1 at θ = 0.0 favoring linkage between the marker D19S220 on chromosome 19q13.1 and CCD, and no recombinants were observed (Fig. 1). Two affected individuals, III:4 and IV:10, were diagnosed as MHS in the IVCT performed by using the European MH Group protocol (Table 1). No anesthetic complications were recorded in the medical history of the family, despite the exposure of 19 affected family members to MH-triggering agents.
Figure 1.
Pedigree of the Mexican family showing: CCD status, MHS status, haplotypes constructed with markers for loci D19S220 and D19S47, and the presence of the I4898T mutation. CCD-affected individuals are indicated by closed symbols and normal individuals are indicated by open symbols. The two individuals diagnosed as MHS are indicated by an asterisk next to the symbol. Diagonal bars through symbols denote deceased individuals. The haplotype segregating with the disease is indicated by the filled bar. The presence or absence of the I4898T mutation is indicated by + or −, respectively.
Table 1.
IVCT results
| Caffeine, mM | III:4 Tension, g | IV:10 Tension, g | Halothane, % v/v | III:4 Tension, g | IV:10 Tension, g |
|---|---|---|---|---|---|
| 0.5 | 0.105 | 0.00 | 0.5 | 0.05 | 0.00 |
| 1.0 | 0.1 | 0.00 | 1.0 | 0.1 | 0.14 |
| 1.5 | 0.16 | 0.1 | 2.0 | 0.3 | 1.14 |
| 2.0 | 0.20 | 0.40 |
The threshold muscle tension for diagnosis of MHS is greater than or equal to 0.2 g tension at, or below 2 mM caffeine and 2% halothane.
Identification of a RYR1 Mutation.
To establish whether a RYR1 mutation causes CCD in this family, the entire 15,117-bp RYR1 cDNA of individual III:4 was amplified by PCR. DNA sequence analysis revealed the presence of a single T to C transition at nucleotide 14693, resulting in the substitution of an Ile for Thr at codon 4898 (Fig. 2A). The sequence change neither created nor disrupted any known restriction site. An assay for the mutation was established by checking for the presence or absence of T14693 nucleotide by using the GMPD method (19). The mutation segregated precisely with the CCD phenotype in the family (Figs. 1 and 2B). The mutation was absent in 200 normal chromosomes, including 86 normal Mexican chromosomes, indicating that the mutation was not simply an innocuous polymorphism. Analysis of 80 unrelated MHS individuals, including six CCD individuals, did not reveal the presence of the mutation, suggesting that the T14693C substitution is a private mutation restricted to this Mexican family.
Figure 2.
Sequence and detection of the I4898T mutation. (A) Nucleotide and deduced amino acid sequence of the RYR1 cDNA from individual III:4. The T14693C mutation changes the Ile-4898 codon, ATT, to a threonine codon, ACT. (B) Detection of the T14693C mutation by the GMPD method. The 33-nt digestion product represents the normal T14693 allele, while the 34-nt digestion product represents the mutant C14693 allele. Samples yielding only the 33-nt product are normal, while samples yielding both 33- and 34-nt products are heterozygous for the T14693C mutation.
Expression and Functional Analysis of I4898T Mutant.
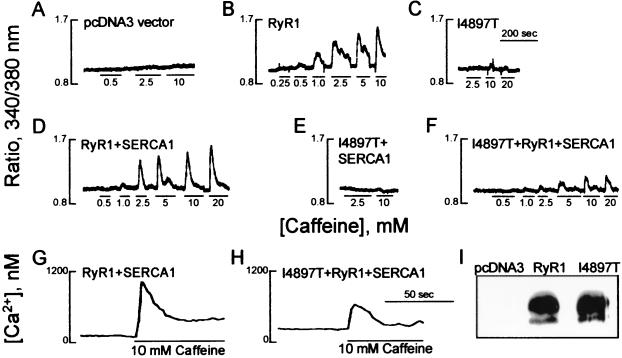
To investigate the effect of the mutation on channel function, the C14693 mutation was introduced into the corresponding position of a rabbit RYR1 cDNA, Ile-4897, and expressed in HEK-293 cells. The caffeine and halothane sensitivities of the mutant channel then were determined in the cellular Ca2+ photometry assay used previously to demonstrate that 15 MHS mutant channels were functional and were significantly more sensitive to caffeine- and halothane-induced Ca2+ release by comparison with cells expressing the normal channel (20). By contrast, the I4897T mutant channel exhibited little or no caffeine-induced Ca2+ release in the assay (Fig. 3 A–C). Western blot analysis confirmed that there was no difference in expression levels of the mutant channel by comparison with normal channel expression (Fig. 3I) and sucrose gradient centrifugation confirmed tetramer formation for both the normal and mutant channels (J.T. and D.H.M., unpublished data).
Figure 3.
Results of Ca2+ photometry and imaging assays and expression of the normal and I4897T mutant RYR1 cDNAs in HEK-293 cells. Traces of the responses in the Ca2+ photometry assay to incremental doses of caffeine in HEK-293 cells expressing: (A) the pcDNA3 vector, (B) normal RyR1, (C) I4897T mutant, (D) RyR1 plus SERCA1, (E) I4897T plus SERCA1, and (F) I4897T plus RyR1 plus SERCA1. Changes in cytosolic Ca2+ concentration are recorded as the ratio of fluorescence at 340/380 nm. The Ca2+ imaging assay demonstrates caffeine-induced Ca2+ release in a single HEK-293 cell expressing: (G) RyR1 plus SERCA1 or (H) I4897T plus RyR1 plus SERCA1. (I) Western blot of HEK-293 cell extracts 48 hr after transfection with pcDNA3 vector, normal RyR1, and I4897T mutant constructs.
Lack of caffeine-induced Ca2+ release could be explained by loss of channel function or, conversely, by enhanced Ca2+ channel activity, which would dissipate luminal endoplasmic reticulum Ca2+ stores. Overexpression of the sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA1) might be expected to ameliorate a leaky effect. When SERCA1 was coexpressed with normal RYR1 cDNA, the expressed channel exhibited a 2-fold increase in the ED50 for caffeine-induced Ca2+ release (Fig. 3D, Table 2). Coexpression of SERCA1 with the I4897T mutant, however, did not re-establish caffeine-induced Ca2+ release (Fig. 3E, Table 2).
Table 2.
Comparison of caffeine- and halothane-induced Ca2+ photometry responses for normal and I4897T mutant ryR1 receptors
| Construct | Caffeine-induced Ca2+ release
|
Halothane-induced Ca2+ release
|
||
|---|---|---|---|---|
| ED50, mM | Δ ratio (max) (n) | ED50, % v/v | Δ ratio (max) (n) | |
| RyR1 | 1.40 ± 0.11 | 0.49 ± 0.07 (6) | ||
| I4897T | N.D. | N.D. (8) | ||
| RyR1 + | 3.13 ± 0.44 | 0.64 ± 0.15 (6) | 0.51 ± 0.13 | 0.8 ± 0.09 (5) |
| SERCA1 | ||||
| I4897T + | N.D. | N.D. (3) | ||
| SERCA1 | ||||
| I4897T + | 3.29 ± 0.45 | 0.21 ± 0.05* (6) | 0.49 ± 0.06 | 0.32 ± 0.04* (4) |
| RyR1 + | ||||
| SERCA1 | ||||
ED50, the caffeine or halothane concentration required to achieve half maximal caffeine-induced Ca2+ release; Δ ratio (max), the maximal change in the ratio of fluorescence at 340 nm and 380 nm. N.D., not detected. ∗, P < 0.05.
Analysis of high-affinity [3H]ryanodine binding showed that maximal ligand binding was reduced by 96% in lysates produced from the mutant cell lines by comparison with lysates from cells expressing the normal channel (Table 3). As ryanodine binds preferentially to the open channel and as there was no difference in expression levels of mutant vs. normal channels, this result suggests that the mutant channel is in a closed conformation (24). However, the result also could be explained by perturbation, by the Thr-4897 substitution, of the ryanodine binding site located between residue 4475 and the C terminus of the protein (25).
Table 3.
Comparison of [3H]ryanodine binding for normal and I4897T mutant RyR1 receptors
| Construct | Bmax, fmol/mg protein (n) | Kd[Ca2+], nM (n) |
|---|---|---|
| RyR1 | 73.1 ± 3.0 (3) | 270 ± 30 (3) |
| I4897T | 3.0 ± 1.7* (4) | |
| I4897T + RyR1 | 11.3 ± 1.1* (3) | 69 ± 24* (3) |
Kd[Ca2+], Ca2+ concentration required for half-maximal ryanodine binding. ∗, P < 0.001.
Affected individuals in the CCD pedigree are heterozygous for the I4898T mutation. To mimic the heterozygous state, mutant and normal channels were coexpressed in a 1:1 ratio in HEK-293 cells. Caffeine and halothane ED50 values for the heterozygote channels were similar to that for the normal channel (Fig. 3F, Table 2), however, the maximal levels of Ca2+ release were reduced by 67% in the heterozygote. Maximal [3H]ryanodine binding was reduced by 84% in lysates from the cells coexpressing normal and mutant channels by comparison with cells expressing normal channels (Table 3). The Ca2+ dependence of high-affinity [3H]ryanodine binding demonstrated a 4-fold decrease in the Kd for Ca2+ stimulated ryanodine binding in co-transfected cell lysate compared with wild-type RyR1 cell lysate (Table 3).
A Ca2+ imaging assay was used to investigate the effect of the mutation on intracellular Ca2+ concentrations at the single-cell level (22). The resting intracellular Ca2+ concentration was increased significantly (P < 0.001) in cells coexpressing the normal and mutant RyR1 channels and SERCA1 pump (349 ± 25 nM, n = 17) than in cells coexpressing normal RyR1 and SERCA1 (127 ± 4 nM, n = 14). In the heterozygous mutant cells, a reduction in the maximal response to caffeine also was confirmed (Fig. 3 G and H, Table 4). This result suggests an increased rate of Ca2+ leakage through the mutant channels from the lumen of the endoplasmic reticulum into the cytoplasm.
Table 4.
Caffeine-induced Ca2+ release and thapsigargin-induced Ca2+ release in single transfected HEK-293 cells
| Construct | Caffeine-induced Ca2+ release, nM | Thapsigargin-induced Ca2+ release, nM (n) |
|---|---|---|
| RyR1 | 874 ± 68 | 348 ± 28 (27) |
| R615L | 517 ± 50** | 273 ± 27 (19)* |
| Y523S | 212 ± 41** | 120 ± 34 (12)** |
| RyR1 + R615L | 609 ± 27* | 320 ± 31 (12) |
| RyR1 + Y523S | 578 ± 50* | 236 ± 41 (10)* |
| RyR1 + I4897T | 249 ± 39** | 110 ± 21 (17)** |
The intracellular responses of single cells to 10 mM caffeine followed by 1.5 μM thapsigargin were measured by Ca2+ imaging. ∗, P < 0.05; ∗∗, P < 0.001.
To further investigate this possibility, the luminal Ca2+ stores were measured by thapsigargin-induced Ca2+ release. Thapsigargin, a sesquiterpene lactone, inhibits SERCA enzymes irreversibly and arrests the repletion of luminal Ca2+ stores (26). The resulting level of Ca2+ leakage into the cytoplasm is a measure of the luminal Ca2+ concentration. Analysis of luminal Ca2+ concentration by using this approach showed a significant decrease in the size of luminal Ca2+ stores in cells cotransfected with normal RYR1 and I4897T cDNAs (Table 4). Analysis of several RyR1 mutants demonstrated a strong correlation (r = 0.94, P < 0.05) between the caffeine-induced Ca2+ release and the thapsigargin-induced Ca2+ release (Table 4). This result suggests that the decreased agonist-induced Ca2+ release, which was observed in the heterozygous I4897T cells, was caused by a reduced luminal Ca2+ concentration resulting from a leaky mutant channel. Furthermore, the cells expressing the I4897T mutant channels had the lowest thapsigargin-induced Ca2+ release (Table 4). This observation is consistent with the penetrance and severity of the clinical phenotype associated with the mutation.
DISCUSSION
All 17 of the previously described RYR1 MHS mutations lie between amino acids 35 and 614 (MH/CCD region 1) or between 2163 and 2458 (MH/CCD region 2) in the 5,038-aa receptor, and all of these are considered to reside in the myoplasmic region of the protein (13). By contrast, I4898T represents a novel type of MHS mutation as it resides in the C terminus of the RyR1 protein that forms the Ca2+ channel (27). According to the Takeshima et al. topology model (28), the mutation exists within a luminal loop flanked by proposed transmembrane sequences M3 and M4, or, according to the Zorzato et al. (18) topology model, is situated near the luminal end of proposed transmembrane sequence M9. Antibody binding studies suggest that the region contiguous to Ile-4898 is located within the lumen of the sarcoplasmic reticulum (29).
Ile-4898 is conserved across the 14 RyR molecules sequenced to date from species ranging from human to Drosophilia and is maintained in all known cardiac (RYR2) and brain (RYR3) isoforms. Furthermore, Ile-4898 and the surrounding amino acid region are conserved in a closely related Ca2+ release channel, the IP3 receptor. This region contains basic amino acid residues that may interact with acidic luminal proteins such as triadin. The luminal Ca2+ binding protein calsequestrin and RyR1 are linked functionally, possibly via triadin (30), as activation of RyR1 induces Ca2+ dissociation from calsequestrin (31). Therefore, perturbation of the putative triadin binding site by the I4898T mutation may disrupt molecular signaling between RyR1 and calsequestrin resulting in abnormal RyR1 channel gating.
The normal caffeine and halothane sensitivities observed in the I4897T/normal heterozygous mutant cells were unexpected, because the two patients investigated by the IVCT had increased sensitivity to caffeine- and halothane-induced muscle contractures. However, the threshold for the IVCT observed for both patients investigated was 2 mM caffeine and 2% halothane, indicating that the caffeine and halothane sensitivity of these human CCD channels was closer to normal than the majority of other MHS mutations investigated previously (13). In human skeletal muscle, the mutant channels are located in the sarcoplasmic reticulum where they are regulated by other muscle proteins, including FKBP12, calmodulin, and calsequestrin (32). Thus, the expression of other muscle-specific proteins in HEK-293 cells may be required to observe an increased agonist sensitivity.
Unlike previously characterized RyR1 mutations (33, 34), the I4897T mutation reduces [3H]ryanodine binding, possibly by perturbing the ligand binding site that is located in the C terminus of the protein (25). However analysis of the Ca2+ dependence of ryanodine binding in the I4897T/normal heterozygous cell lysates demonstrated that the mutant channel is half-activated by a 4-fold lower Ca2+ concentration than the normal channel. This result indicates that the mutation leads to a significant increase in the sensitivity of the channel to the activating effects of Ca2+. The observed lower luminal Ca2+ content and elevated cytoplasmic Ca2+ concentrations could result from this hypersensitivity to Ca2+ as the I4897T mutant channel is activated at the resting cellular Ca2+ concentration while the normal RyR1 channel remains closed.
The I4898T mutation is the first RYR1 mutation to be associated with a highly penetrant and severe form of CCD in a large family. The location of the mutation identifies a region of the RYR1 gene (MH/CCD region 3) for mutation screening in CCD and MHS and also highlights the importance of this transmembrane/luminal region in normal RyR1 channel function. Cellular Ca2+ photometry and imaging assays indicate that the I4898T mutation produces the most abnormal disease-associated RyR1 channel yet investigated, mirroring the severe CCD phenotype associated with the mutation. The effect of the mutation is greatest on the Ca2+ sensitivity of the channel but does not affect the caffeine and halothane sensitivities to the same extent.
The results of the functional analysis provide support for a mechanism for the pathophysiology of CCD, proposed earlier (35), and supply insights into the aetiology of the disease. Depleted internal Ca2+ stores would be expected to manifest as muscle weakness, because contractile force in normal excitation-contraction coupling is proportional to activating Ca2+ concentrations and these would be predicted to be reduced significantly in the affected individuals. Depleted internal Ca2+ stores are also consistent with the lack of MH episodes in affected individuals in this family despite exposure to many MH-triggering agents. Elevated myoplasmic Ca2+ could lead to atrophy through the activation of known Ca2+-dependent proteases in skeletal muscle. Furthermore, it has been shown that prevention of myoplasmic Ca2+ transients by the specific blockage of Ca2+ release through the RyR1 channel disrupts skeletal sarcomere formation by interfering with myosin thick filament (A-band) assembly (36). Thus, if Ca2+ transients in affected individuals bearing this mutation are reduced, the consequence of this reduced signal also would be atrophy. It is of interest that exercise might be expected to counteract this effect and indeed, CCD is one of the few myopathies where exercise is of major benefit.
Acknowledgments
We thank Dr. Katie Keohane and Dr. Deirdre Ryan for muscle histological examination; Dr. John Mackrill and Sean O’Driscoll for reviewing of results; Dr. Grainne O’ Sullivan and Noreen Casey for performance of Ca2+ release experiments and IVCT; Dr. Susan Hamilton for family sample coordination, and the family members who participated in this study. This work was funded by an European Union BIOMED program grant to T.V.M. and by grants from the Medical Research Council of Canada, the Muscular Dystrophy Association of Canada, and the Canadian Genetic Diseases Network of Centers of Excellence (to D.H.M.). J.T. was supported by a studentship from the Medical Research Council of Canada.
ABBREVIATIONS
- CCD
central core disease
- MH
malignant hyperthermia
- RyR1
skeletal muscle ryanodine receptor
- IVCT
in vitro contracture test
- MHS
malignant hyperthermia susceptible
- GMPD
glycosylase-mediated polymorphism detection
- SERCA1
skeletal muscle Ca2+-ATPase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
A Commentary on this article begins on page 3345.
References
- 1.Shy G M, Magee K R. Brain. 1956;79:610–621. doi: 10.1093/brain/79.4.610. [DOI] [PubMed] [Google Scholar]
- 2.Shuaib A, Paasuke R T, Brownell K W. Medicine. 1987;66:389–396. [PubMed] [Google Scholar]
- 3.Dubowitz V, Brooke M H. In: Major Problems in Neurology. Walton J N, editor. London: Saunders; 1973. pp. 92–119. [Google Scholar]
- 4.Hayashi K, Miller R G, Brownell K W. Muscle Nerve. 1989;12:95–102. doi: 10.1002/mus.880120203. [DOI] [PubMed] [Google Scholar]
- 5.Vita G, Migliorato A, Baradello A, Mazzeo A, Rodolico C, Falsaperla R, Messina C. J Neurol Sci. 1994;124:71–76. doi: 10.1016/0022-510x(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 6.Denborough M A, Dennett X, Anderson R M. Br Med J. 1973;1:272–273. doi: 10.1136/bmj.1.5848.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mickelson J R, Louis C F. Physiol Rev. 1996;76:537–592. doi: 10.1152/physrev.1996.76.2.537. [DOI] [PubMed] [Google Scholar]
- 8.European Malignant Hyperthermia Group. Br J Anaesth. 1984;56:1267–1269. doi: 10.1093/bja/56.11.1267. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy T V, Healy J M S, Heffron J J A, Lehane M, Deufel T, Lehmann-Horn F, Farrall M, Johnson K. Nature (London) 1990;343:562–564. doi: 10.1038/343562a0. [DOI] [PubMed] [Google Scholar]
- 10.MacLennan D H, Duff C, Zorzato F, Fujii J, Phillips M, Korneluk R G, Frodis W, Britt B A, Worton R G. Nature (London) 1990;343:559–561. doi: 10.1038/343559a0. [DOI] [PubMed] [Google Scholar]
- 11.Mulley J C, Kozman H M, Phillips H A, Gedeon A K, McCure J A, Iles D E, Gregg R G, Hogan K, Couch F J, MacLennan D H, et al. Am J Hum Genet. 1993;52:398–405. [PMC free article] [PubMed] [Google Scholar]
- 12.Schwemmle S, Wolff K, Palmucci L M, Grimm T, Lehmann-Horn F, Hubner C H, Hauser E, Iles D E, MacLennan D H, Muller C R. Genomics. 1993;17:205–207. doi: 10.1006/geno.1993.1302. [DOI] [PubMed] [Google Scholar]
- 13.Loke J, MacLennan D H. Am J Med. 1998;104:470–486. doi: 10.1016/s0002-9343(98)00108-9. [DOI] [PubMed] [Google Scholar]
- 14.Fananapazir L, Dalakas M C, Cyran F, Cohn G, Epstein N D. Proc Natl Acad Sci USA. 1993;90:3993–3997. doi: 10.1073/pnas.90.9.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller S A, Dykes D D, Polesky H F. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quane K A, Healy J M S, Keating K A, Manning B M, Couch F J, Palmucci L M, Doriguzzi C, Fagerlund T H, Berg K, Ording H, et al. Nat Genet. 1993;5:51–55. doi: 10.1038/ng0993-51. [DOI] [PubMed] [Google Scholar]
- 17.Lathrop G M, Lalouel J M. Am J Hum Genet. 1984;36:460–465. [PMC free article] [PubMed] [Google Scholar]
- 18.Zorzato F, Fujii J, Otsu K, Phillips M, Green N M, Lai F A, Meissner G, MacLennan D H. J Biol Chem. 1990;265:2244–2256. [PubMed] [Google Scholar]
- 19.Vaughan P, McCarthy T V. Nucleic Acids Res. 1998;26:810–815. doi: 10.1093/nar/26.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong J, Oyamada H, Demaurex N, Grinstein S, McCarthy T V, MacLennan D H. J Biol Chem. 1997;272:26332–26339. doi: 10.1074/jbc.272.42.26332. [DOI] [PubMed] [Google Scholar]
- 21.Du G G, Imredy J P, MacLennan D H. J Biol Chem. 1998;273:33259–33266. doi: 10.1074/jbc.273.50.33259. [DOI] [PubMed] [Google Scholar]
- 22.Tong J, McCarthy T V, MacLennan D H. J Biol Chem. 1999;274:693–702. doi: 10.1074/jbc.274.2.693. [DOI] [PubMed] [Google Scholar]
- 23.Grynkiewicz G, Poenie M, Tsien R Y. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 24.Valdivia H H, Hogan K, Coronado R. Am J Physiol. 1991;261:C237–C245. doi: 10.1152/ajpcell.1991.261.2.C237. [DOI] [PubMed] [Google Scholar]
- 25.Callaway C, Seryshev A, Wang J P, Slavik K J, Needleman D H, Cantu III C, Wu Y, Jayaraman T, Marks A R, Hamilton S L. J Biol Chem. 1994;269:15876–15884. [PubMed] [Google Scholar]
- 26.Thastrup O, Cullen P J, Drobak B K, Hanley M R, Dawson A P. Proc Natl Acad Sci USA. 1990;87:2466–2470. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhat M B, Zhao J, Takeshima H, Ma J. Biophys J. 1997;73:1329–1336. doi: 10.1016/S0006-3495(97)78166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeshima H, Nishimura S, Matsumoto T, Ishida H, Kangawa K, Minamino N, Matsuo H, Ueda M, Hanaoka M, Hirose T, et al. Nature (London) 1989;339:439–445. doi: 10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- 29.Grunwald R, Meissner G. J Biol Chem. 1995;270:11338–11347. doi: 10.1074/jbc.270.19.11338. [DOI] [PubMed] [Google Scholar]
- 30.Guo W, Campbell K P. J Biol Chem. 1995;270:9027–9030. doi: 10.1074/jbc.270.16.9027. [DOI] [PubMed] [Google Scholar]
- 31.Ikemoto N, Antoniu B, Kang J J, Meszaros L G, Ronjat M. Biochemistry. 1991;30:5230–5237. doi: 10.1021/bi00235a017. [DOI] [PubMed] [Google Scholar]
- 32.Coronado R, Morrissette J, Sukhareva M, Vaughan D M. Am J Physiol. 1994;266:C1485–C1504. doi: 10.1152/ajpcell.1994.266.6.C1485. [DOI] [PubMed] [Google Scholar]
- 33.Mickelson J R, Gallant E M, Litterer L A, Johnson K M, Rempel W E, Louis C F. J Biol Chem. 1988;263:9310–9315. [PubMed] [Google Scholar]
- 34.Richter M, Schleithoff L, Deufel T, Lehmann-Horn F, Herrmann-Frank A. J Biol Chem. 1997;272:5256–5260. doi: 10.1074/jbc.272.8.5256. [DOI] [PubMed] [Google Scholar]
- 35.MacLennan D H, Phillips M S, Zhang Y. In: Molecular Biology of Membrane Transport Disorders. Schultz S G, Andreoli T E, Brown A M, Fambrough D, Hoffman J, Welsh M J, editors. New York: Plenum; 1996. pp. 181–200. [Google Scholar]
- 36.Ferrari M B, Ribbeck K, Hagler D J, Spitzer N C. J Cell Biol. 1998;141:1349–1356. doi: 10.1083/jcb.141.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]