Abstract
Little is known about plant circadian oscillators, in spite of how important they are to sessile plants, which require accurate timekeepers that enable the plants to respond to their environment. Previously, we identified a circadian clock-associated (CCA1) gene that encodes an Myb-related protein that is associated with phytochrome control and circadian regulation in plants. To understand the role CCA1 plays in phytochrome and circadian regulation, we have isolated an Arabidopsis line with a T DNA insertion that results in the loss of CCA1 RNA, of CCA1 protein, and of an Lhcb-promoter binding activity. This mutation affects the circadian expression of all four clock-controlled genes that we examined. The results show that, despite their similarity, CCA1 and LHY are only partially redundant. The lack of CCA1 also affects the phytochrome regulation of gene expression, suggesting that CCA1 has an additional role in a signal transduction pathway from light, possibly acting at the point of integration between phytochrome and the clock. Our results indicate that CCA1 is an important clock-associated protein involved in circadian regulation of gene expression.
Organisms have internal clocks to regulate physiological and cellular processes. The timekeepers, or oscillators, controlling these clocks are proposed to be negative autoregulatory feedback loops in which one or more gene products feed back and repress their own expression (1). The oscillator can be reset by input pathways from environmental cues, such as light and temperature, and, in turn, can regulate overt rhythmicity. Although much has been learned about circadian clocks in other organisms, little is known about the molecular basis of plant circadian oscillators. Plant homologs have not been identified yet for any of the known circadian genes from other organisms, including frq from Neurospora, per and tim from Drosophila, mper from mice, and the kai genes from the cyanobacterium Synechococcus (1, 2). An Arabidopsis mutant, toc1, that affects the period of clock-controlled processes has been isolated; however, the corresponding gene has not been cloned, and its role in the oscillator is not known (3).
Recently, two Arabidopsis Myb-related genes, CCA1 (circadian clock-associated) and LHY, have been shown to have an important function in the circadian control of a number of plant processes (4, 5). CCA1 and LHY are 60% similar at the amino acid level, with regions of identity extending throughout the protein. Constitutive expression of either gene affects multiple processes controlled by circadian rhythms, including oscillations of circadian clock-controlled genes (4, 5). Furthermore, both CCA1 and LHY RNAs showed circadian oscillations after a period of entrainment and showed feedback inhibition of their own synthesis. Constitutive expression of CCA1 represses the expression of the LHY gene (4). CCA1 also is regulated by the plant photoreceptor phytochrome (4).
To understand how CCA1 might function in the circadian control of plant processes and in the signal transduction pathway from phytochrome, we have identified a null mutation that results in the loss of CCA1. We show here that loss of CCA1 affects the expression of all the clock-controlled genes examined, as well as the phytochrome regulation of gene expression. These data show that CCA1 plays a central role in both phytochrome and circadian regulation of gene expression.
MATERIALS AND METHODS
Plant Materials and Growth.
Light-grown plants were grown on MS2S medium [Murashige and Skoog salts (GIBCO/BRL), 0.05% Mes (pH 5.7), 0.8% Phytagar (GIBCO/BRL), and 2% sucrose] or on soil (both methods gave the same results) under light:dark cycles [12 h of light (125 μE⋅m−2⋅sec−1):12 h of darkness; E = 1 mol of photons] at 23°C for 17 days before harvesting at dawn. For circadian experiments, plants were transferred to constant light (100 μE⋅m−2⋅sec−1) at 23°C. Etiolated plants were grown for 6 days in the dark before being given 1 min of red light and then being harvested at indicated intervals.
T DNA Insertion-Line Identification.
DNA prepared from T DNA insertion lines in a Wassilewskija background (Arabidopsis Biological Resource Center stock CS6502; seeds were donated by K. Feldmann, University of Arizona, Tucson, AZ; pools of DNA were prepared by C. Lin, University of California, Los Angeles) was screened by PCR by using T DNA left and right border-specific primers (6) and primers specific for CCA1 5′-TGAGATTTCTCCATTTCCGTAGCTTCTGG and 3′-ATCCGTTTGGGATCTTTCTGTTCCACATG. One pool was found to give a PCR product with the left T DNA border primer in conjunction with either the 3′ or the 5′ CCA1 primers. Southern blot analysis confirmed that this product included regions of the CCA1 gene. Progressively smaller pools of DNA containing the T DNA insertion in CCA1 were identified by PCR until a single line with a T DNA insert in the CCA1 gene was isolated.
RNA and Protein Analyses.
RNA and protein analyses were carried out as described (4, 7, 8). A DNA template for synthesis of an RNA probe for CAT2 was synthesized by PCR amplification of a genomic DNA fragment containing nucleotides 1,553–1,773 (9) with primers CAT2S (5′-AACGCGTGAAAGAATTCTTGATTGGCC-3′) and T7CAT2A (5′-TGTAATACGACTCACTATAGGGGACCTTTCATCAAGTAACACC-3′, with the T7 promoter region in bold).
Electrophoretic Mobility-Shift Assays.
Protein extracts were prepared from 3-week-old soil-grown plants as described (7), except that phosphatase inhibitors (50 mM NaF, 0.2 mM (NH4)6Mo7O24, 5 mM NH4VO3, and 1 mM EGTA) were added to all the solutions, and the whole-cell extracts were desalted on a Sephadex G-25 column (Amersham Pharmacia). The proteins were then preincubated at 30°C for 30 min with 50 mM Tris⋅HCl (pH 7.5), 0.1 mM Na2 EDTA, 5 mM DTT, and 2 mM MnCl2, and electrophoretic mobility-shift assays were carried out as described (7). The wild-type promoter fragment designated A2 (7) was used unless otherwise stated.
RESULTS
Isolation of a CCA1-Null Line.
We used a PCR-based procedure (6) to isolate a CCA1-null line (cca1-1) of Arabidopsis, in which the CCA1 gene was interrupted by a T DNA insertion. Fig. 1A shows a diagram of the large T DNA insertion that disrupts intron 4 of the CCA1 gene. Partial sequencing of the insertion site showed that the T DNA insert was in the fourth intron of the CCA1 gene. There is a small (24-bp) deletion in the CCA1 sequence and another small (26-bp) fragment of non-CCA1 or T DNA at the insertion site. Both ends of the T DNA insert have left borders, suggesting that two of the 17-kb T DNAs have been inserted in a head-to-head configuration, resulting in a 34-kb insertion in the CCA1 gene.
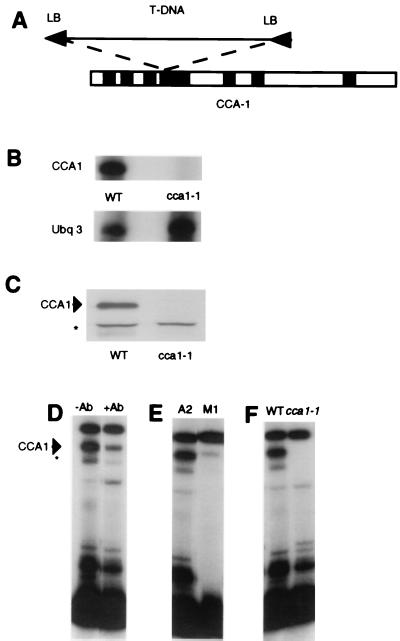
Figure 1.
Identification of the CCA1-null line. (A) T DNA insertion site in CCA1. Black boxes, introns; white boxes, coding regions; LB, left border. (B) The cca1-1 line lacks CCA1 RNA. RNA from wild-type (WT) Wassilewskija ecotype and cca1-1 plants was used as a template for RNase protection analysis with probes for CCA1 and a control, UBQ3. (C) CCA1 protein is absent in the cca1-1 plants. Western blots of wild-type (WT) and proteins from cca1-1 plants were assayed with CCA1 antibodies. The upper band (arrowhead) is CCA1; the lower band (asterisk) is a nonspecific band routinely seen in Western blot analyses carried out with CCA1 antibodies. (D–F) Electrophoretic mobility-shift assays showing a major (arrowhead) and a minor (asterisk) CCA1-binding activity; these activities are reduced after preincubation of protein extracts with CCA1 Abs and when a mutated promoter fragment (M1) is used in the binding assay, and the activities are missing completely in protein extracts from cca1-1 plants.
To examine whether the insertion was sufficient to disrupt correct transcription and translation, we determined RNA and protein levels in the cca1-1 plants. Fig. 1B shows the results of an RNase protection analysis and shows that this line has no detectable CCA1 RNA. Because the T DNA insertion is located in the fourth intron of the CCA1 gene, we considered the possibility that a truncated RNA might be produced. Therefore, Northern analysis of the CCA1 RNA also was performed by using an RNA probe synthesized from a template made to the −68 to +186 end of the gene (7). The results of this experiment confirmed that there were no detectable CCA1 transcripts from the 5′ end of the CCA1 gene (data not shown). Furthermore, the cca1-1 line had no detectable CCA1 protein (Fig. 1C).
CCA1 was isolated originally as a factor that can bind well characterized regions of circadian-and phytochrome-regulated Lhcb (also known as CAB) promoters (7, 10–12). Analysis of the binding activity of proteins isolated from wild-type plants to an Lhcb1*3 promoter probe showed that two DNA–protein complexes were much reduced when they were preincubated with an anti-CCA1 antibody (Fig. 1D). Fig. 1E shows that these complexes were also reduced when the wild-type DNA probe A2 (7) was replaced by M1, which has mutations in the sequences that have been shown to be important for recombinant CCA1 binding (7). These results indicate that CCA1 is a component of these complexes. We, therefore, examined the binding activity of plant extracts isolated from cca1-1 plants. Fig. 1F shows that the Lhcb1*3-promoter binding complexes were undetectable in protein extracts isolated from cca1-1 plants. Taken together, these results confirm that cca1-1 is a null line lacking CCA1 RNA, CCA1 protein, and an Lhcb1*3-promoter binding activity.
Circadian Control of Gene Expression Is Altered in cca1-1 Plants.
We next tested whether the circadian regulation of gene expression was altered in the cca1-1 line. cca1-1 plants were transferred to constant light after entrainment in light:dark conditions (12 h of light:12 h of darkness). Fig. 2A shows that the period of Lhcb1*1 circadian oscillations was ≈3 h shorter (the average of the distances between the peaks measured for three independent experiments, carried out over four cycles) than that in wild-type plants grown under the same conditions. On the first day of the experiment, the timing of the increase of Lhcb1*1 RNA in cca1-1 plants was almost identical to that in wild-type plants. However, by the fourth day of the experiment, the Lhcb1*1 RNA peaked much earlier in cca1-1 than in wild-type plants.
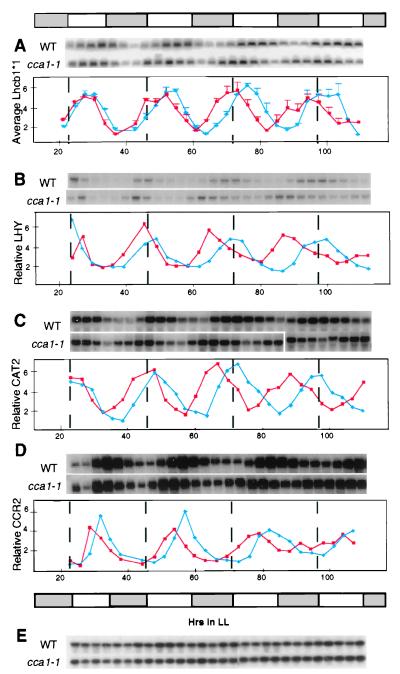
Figure 2.
Circadian oscillations of clock-controlled genes in the cca1-1 line. The expression of Lhcb1*1 (A), LHY (B), CAT2 (C), CCR2 (D), and UBQ10 (E) genes. Each experiment was repeated at least three times with identical results. One representative autoradiogram is shown for Lhcb1*1, LHY, CAT2, and CCR2. Quantitation based on expression of the UBQ10 control for the Northern blots for LHY, CAT2, and CCR2 is shown. Because quantitation of individual experiments with Lhcb1*1 showed some smaller secondary peaks, the results of the three experiments were averaged, and are shown ±SEM. Red lines, cca1-1; blue lines, wild-type (WT) Wassilewskija ecotype. The bar represents the subjective light conditions.
Thus, loss of CCA1 affects the period of Lhcb1*1 gene expression. However, oscillations were not lost completely. We have shown that CCA1 binds directly to Lhcb promoters (ref. 7; Fig. 1D), and we have found that LHY, a protein that is highly homologous to CCA1 (4, 5), also binds the Lhcb promoter (C. Andronis, S. Sugano, and E.M.T., unpublished data). Therefore, LHY may be responsible for the control of Lhcb1*1 expression in the absence of CCA1. Fig. 2B shows that the period of LHY oscillations was indeed also ≈3 h shorter in the cca1-1 plants, resembling the period of Lhcb1*1 oscillations. By the fourth day in continuous light, the peak of LHY expression in cca1-1 plants occurred approximately 12 h earlier than it did in the wild-type plants. These results show not only that there is a strong correlation between the period of oscillations of LHY and that of Lhcb1*1 but also that the absence of CCA1 affects the period of the LHY RNA rhythm. In contrast, the elf3 mutation, which has been postulated to affect the input pathway to the circadian oscillator (13, 14), causes a loss of LHY RNA oscillations in constant-light conditions (5).
If both LHY and CCA1 are associated closely with a central oscillator in Arabidopsis cells, then a shortened period of oscillations also would be expected in the expression of other clock-controlled genes. Indeed, CAT2 RNA (catalase 2; ref. 15), which peaks around subjective dawn in wild-type plants, showed a shorter period of oscillation in cca1-1 plants than in wild-type plants (Fig. 2C). CCR2 (also known as AtGRP7), which encodes an RNA binding protein, is a clock-controlled gene whose RNA peaks considerably later in the cycle than the RNA of LHY, CAT2, and Lhcb1*1 (16, 17). Fig. 2D shows that circadian oscillations of CCR2 clearly were affected in the cca1-1 plants. However, it is difficult to determine conclusively whether this alteration is a phase or period shift.
The cca1-1 Line Shows a Reduced Phytochrome Induction of Lhcb1*3 Expression.
Environmental light perception in plants is mediated by several families of photoreceptors, including the phytochromes and cryptochromes. Photoreceptors, in turn, entrain one or more circadian oscillators (18) but are also able to induce gene expression directly. In addition to binding to a region of the Lhcb1*3 promoter required for phytochrome and circadian regulation, CCA1 has itself been shown to be induced transiently by the plant photoreceptor phytochrome (4). Thus, it also may play a role in the phototransduction pathway from phytochrome. Indeed, the phytochrome induction of Lhcb1*3 gene expression was reduced in cca1-1 plants (Fig. 3), reaching approximately 60% (average of three experiments) of the induction in the wild-type plants. Recently, it was reported (19) that imbibition of seeds is sufficient to start some circadian rhythms in Arabidopsis seedlings, and the timing of the light treatment might be expected to affect the magnitude of Lhcb1*3 induction observed. In our experiments, the times between imbibition and the red-light treatment given to the seedlings varied, but the amplitude of the induction of the Lhcb RNA did not. Our observations of the phytochrome induction of Lhcb1*3 RNA in the cca1-1 line are consistent with results obtained from plants transformed with a CCA1 antisense construct (7). They suggest that CCA1 may have a role not only in the circadian regulation of gene expression but also directly in a phototransduction pathway from phytochrome.
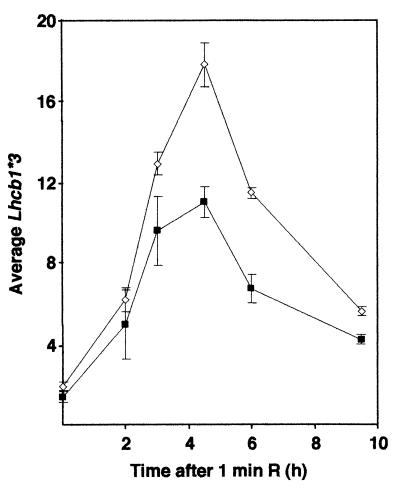
Figure 3.
Phytochrome induction in the cca1-1 line. Northern analysis of Lhcb1*3 RNA in 6-day-old etiolated plants after a 1-min exposure to red light. Values were normalized to a UBQ3 control. Open diamonds, wild-type (WT) Wassilewskija ecotype; closed squares, cca1-1. The results from three independent experiments were averaged and are shown ±SEM.
DISCUSSION
We have identified a null mutation that results in the absence of CCA1, a potential component of the circadian regulatory pathway in plants. Circadian regulation of all the clock-controlled genes we examined was found to be affected in these lines. Our results clearly show that CCA1 is playing a central role in circadian control of gene expression in plants. The fact that circadian oscillations are not lost in the cca1-1 line suggests that many features of circadian control are still functional. It is possible that there is another protein partially able to compensate for the loss of CCA1. An obvious candidate is LHY, which has a very high degree of homology with CCA1 and strong similarities in the phenotypes of the overexpressing lines (4, 5). Furthermore, we have shown (Fig. 2 A and B) that there is a strong correlation between the period of oscillations of LHY and Lhcb1*1. However, the functions of CCA1 and LHY are not completely redundant, because the loss of CCA1 results in a clearly altered circadian gene expression phenotype. It remains to be seen whether LHY compensates for the loss of CCA1 in the cca1-1 plants, and it would be of interest to examine the effects of a CCA1-LHY double loss-of-function mutation on circadian rhythms.
The period of oscillations of LHY, CAT2, and Lhcb1*1 were clearly shorter in the cca1-1 line. However, the effect of the null mutation on CCR2 was less clear but may have involved a phase shift. If CCR2 is phase-shifted in the cca1-1 line, it would indicate that neither CCA1 nor LHY is controlling CCR2 oscillations directly and would suggest that there is at least one additional oscillator in the cell. The existence of multiple oscillators in plants has been suggested by physiological studies in bean and by an analysis of gene expression in very young tobacco seedlings (20–22). Alternatively, the pattern of CCR2 oscillations that we observed might be a result of the autoregulatory CCR2 oscillator winding down (16). However, it is also possible that, like the other clock-controlled genes we examined, CCR2 has an altered period in cca1-1 plants.
In addition, we have shown that CCA1 is involved directly in light regulation of gene expression, acting in the signal transduction pathway from phytochrome to Lhcb1*3. In this respect, CCA1 resembles the white collar genes of Neurospora whose products are involved in light-regulated transcriptional activation of gene expression and also play a role in the Neurospora circadian oscillator (23). These results are consistent with recent findings that circadian oscillations of a reporter gene driven by an Lhcb promoter are altered in phytochrome A and B mutants in Arabidopsis (24). It is possible that CCA1 provides a molecular link between phytochrome and the circadian oscillator in plant cells.
Although further experiments will be needed to elaborate fully the role of CCA1 and other proteins in circadian regulation, it is clear from the data presented here and from our studies on an overexpressing line (4) that CCA1 plays a significant role. We cannot eliminate the possibility that CCA1 may be part of an output from the oscillator that loops back to the input to the clock (25, 26) or acts directly as an input. However, our evidence is consistent with the idea that CCA1 may act as part of a central oscillator as illustrated in Fig. 4. In this model, a possible explanation for the shorter period of gene expression seen in the cca1-1 plants would be that, in wild-type plants, LHY and CCA1 may form heterodimers; however, in the absence of CCA1, LHY may form homodimers that can maintain oscillations and affect the feedback inhibition differently.
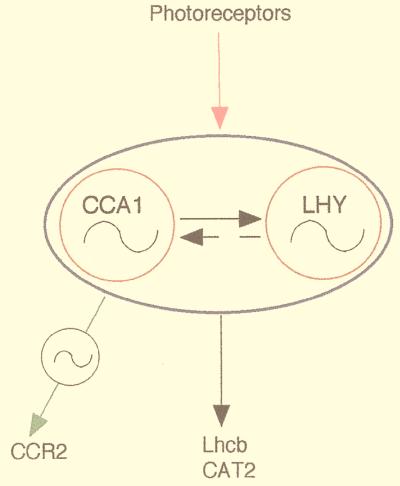
Figure 4.
Diagram of a possible role of CCA1 in circadian rhythms. The solid and dashed arrows represent potential interactions between CCA1 and LHY. Constitutive expression of CCA1 represses the expression of the LHY gene (4); the effect of constitutive expression of LHY on CCA1 has not yet been established. The wavy lines represent potential oscillators, and the solid line around CCA1 and LHY indicates possible redundant functioning of CCA1 and LHY in regulating rhythms of target genes.
Acknowledgments
We thank members of the Tobin laboratory, J. Brusslan, D. S. Greenberg, S. Jacobsen, C. Lin, and K. Singh, for their critical reading of this manuscript; C. Lin for making the DNA pools available; Z.-Y. Wang for carrying out the Western analysis; and J. Kreps and G. Coupland for providing cDNA clones for CCR2 and LHY. This work was supported by National Institutes of Health RO1GM23167.
References
- 1.Dunlap J C. Curr Opin Genet Dev. 1998;8:400–406. doi: 10.1016/s0959-437x(98)80109-3. [DOI] [PubMed] [Google Scholar]
- 2.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson C R, Tanabe A, Golden S S, Johnson C H, Kondo T. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 3.Millar A J, Carré I A, Strayer C A, Chua N-H, Kay S A. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z-Y, Tobin E M. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 5.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré I A, Coupland G. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 6.Krysan P J, Young J C, Tax F, Sussman M R. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z-Y, Kenigsbuch D, Sun L, Harel E, Ong M, Tobin E. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brusslan J A, Tobin E M. Proc Natl Acad Sci USA. 1992;89:7791–7795. doi: 10.1073/pnas.89.16.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevalier C, Yamaguchi J, McCourt P. Plant Physiol. 1992;99:1726–1728. doi: 10.1104/pp.99.4.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenigsbuch D, Tobin E M. Plant Physiol. 1995;108:1023–1027. doi: 10.1104/pp.108.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Doxsee R A, Harel E, Tobin E M. Plant Cell. 1993;5:109–121. doi: 10.1105/tpc.5.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson S L, Teakle G R, Martino-Catt S J, Kay S A. Plant J. 1994;6:457–470. doi: 10.1046/j.1365-313x.1994.6040457.x. [DOI] [PubMed] [Google Scholar]
- 13.Zagotta M T, Hicks K A, Jacobs C I, Young J C, Hangarter R P, Meeks-Wagner D R. Plant J. 1996;10:691–702. doi: 10.1046/j.1365-313x.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- 14.Hicks K A, Millar A J, Carré I A, Somers D E, Straume M, Meeks-Wagner D R, Kay S A. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- 15.Zhong H H, McClung C R. Mol Gen Genet. 1996;251:196–203. doi: 10.1007/BF02172918. [DOI] [PubMed] [Google Scholar]
- 16.Heintzen C, Nater M, Apel K, Staiger D. Proc Natl Acad Sci USA. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter C D, Kreps J A, Simon A E. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson S L, Kay S A. Trends Plant Sci. 1996;1:51–57. [Google Scholar]
- 19.Zhong H H, Painter J E, Salomé P A, Straume M, McClung C R. Plant Cell. 1998;10:2005–2018. doi: 10.1105/tpc.10.12.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennessey T L, Field C B. J Biol Rhythms. 1992;7:105–113. doi: 10.1177/074873049200700202. [DOI] [PubMed] [Google Scholar]
- 21.Kolar C, Fejes E, Ádám E, Schãfer E, Kay S, Nagy F. Plant J. 1998;13:563–569. doi: 10.1046/j.1365-313x.1998.00048.x. [DOI] [PubMed] [Google Scholar]
- 22.Kolar C, Ádám E, Schãbfer E, Nagy F. Proc Natl Acad Sci USA. 1995;92:2174–2178. doi: 10.1073/pnas.92.6.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crosthwaite S K, Dunlap J C, Loros J J. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 24.Somers D E, Devlin P F, Kay S A. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 25.Millar A. Ann Bot (London) 1998;81:581–587. [Google Scholar]
- 26.Roenneberg T, Merrow M. J Biol Rhythms. 1998;13:167–179. doi: 10.1177/074873098129000011. [DOI] [PubMed] [Google Scholar]