Abstract
Multiple lipoxygenase sequence alignments and structural modeling of the enzyme/substrate interaction of the cucumber lipid body lipoxygenase suggested histidine 608 as the primary determinant of positional specificity. Replacement of this amino acid by a less-space-filling valine altered the positional specificity of this linoleate 13-lipoxygenase in favor of 9-lipoxygenation. These alterations may be explained by the fact that H608V mutation may demask the positively charged guanidino group of R758, which, in turn, may force an inverse head-to-tail orientation of the fatty acid substrate. The R758L+H608V double mutant exhibited a strongly reduced reaction rate and a random positional specificity. Trilinolein, which lacks free carboxylic groups, was oxygenated to the corresponding (13S)-hydro(pero)xy derivatives by both the wild-type enzyme and the linoleate 9-lipoxygenating H608V mutant. These data indicate the complete conversion of a linoleate 13-lipoxygenase to a 9-lipoxygenating species by a single point mutation. It is hypothesized that H608V exchange may alter the orientation of the substrate at the active site and/or its steric configuration in such a way that a stereospecific dioxygen insertion at C-9 may exclusively take place.
Keywords: Cucumis sativus L., active-site modeling, oxygenated lipids, product specificity
Lipoxgenases (LOXs; linoleate:oxygen oxidoreductase; EC 1.13.11.12) are widely distributed in the plant and animal kingdom (1, 2). They constitute a family of nonheme iron-containing dioxygenases that catalyze the regio- and stereoselective dioxygenation of polyenoic fatty acids forming hydroperoxy derivatives (3). In mammals, LOXs are classified according to their positional specificity of arachidonic acid oxygenation (2, 4). Because arachidonic acid either is not present in higher plants or is a minor constituent of cellular lipids, plant LOXs are classified into 9- and 13-LOXs with respect to their positional specificity of linoleic acid (LA) oxygenation (5). Recently, a more comprehensive classification of plant LOXs has been proposed based on the comparison of their primary structures (6).
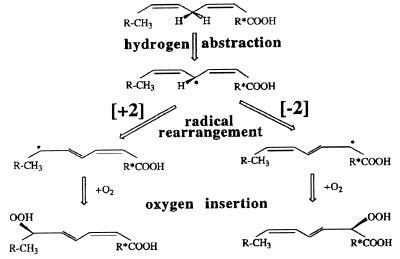
The positional specificity of LOXs is a result of two catalytic processes. (i) Regio- and stereospecific hydrogen removal; with substrate fatty acids containing several doubly allylic methylenes such as linolenic acid, arachidonic acid, or eicosapentaenoic acid hydrogen abstraction from two, three, or four doubly allylic methylenes, respectively, is possible. (ii) Regio- and stereospecific oxygen insertion: when hydrogen is abstracted from a certain doubly allylic methylene, molecular oxygen can be introduced either at the [+2] or at the [−2] position (Fig. 1). Thus, a fatty acid containing three doubly allylic methylenes such as arachidonic acid can be oxygenated by a LOX to six regioisomeric hydroperoxy derivatives (HPETEs), namely 15- and 11-HPETE (originating from C-13 hydrogen removal), 12- and 8-HPETE (C-10 hydrogen removal), and 9- and 5-HPETE (C-7 hydrogen removal). Experiments on mammalian 12- and 15-LOXs indicated that the site of hydrogen abstraction can be altered when critical amino acids are targeted by site-directed mutagenesis (7, 8). When the space-filling M419 or F353 of the human and/or rabbit reticulocyte-type 15-LOX is mutated to smaller residues, the substrate fatty acids are supposed to slide farther into the substrate-binding pocket approaching the doubly allylic carbon-10 of arachidonic acid, closer to the catalytically active nonheme iron (7, 8). In these experiments the site of hydrogen removal was altered, but the direction of radical rearrangement was not affected because formation of both 15- and 12-HPETE involves a [+2] radical rearrangement.
Figure 1.
Specificity of LOX reaction with substrates containing one doubly allylic methylene. In this case, the specificity of the LOX reaction solely depends on the direction of the radical rearrangement. [+2] Radical rearrangement indicates that dioxygen is inserted at the second carbon atom in the direction of the methyl terminus of the substrate counted from the site of hydrogen removal. [−2] Indicates an inverse direction of radical rearrangement.
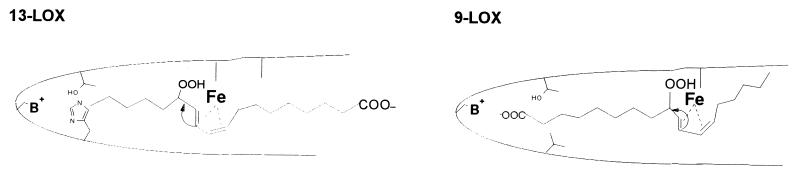
Attempts to alter the direction of radical rearrangement of the LOX reaction (e.g., conversion of a linoleate 13-LOX to a 9-LOX) by site-directed mutagenesis have not been successful so far. One possibility to achieve this goal (Fig. 2) would be to force an inverse head-to-tail substrate orientation (9–12). For linoleate 13-lipoxygenation the substrate appears to penetrate the active site, with its methyl terminus favoring a [+2] radical rearrangement (Fig. 2 Left). In contrast, when the substrate slides into the active site with its carboxyl terminus, a [−2] radical rearrangement would be favored (Fig. 2 Right). However, an inverse-substrate orientation should be strongly inhibited by a large energy barrier associated with burying the charged carboxylate group in the hydrophobic environment of the substrate-binding cage (13, 14). This energy barrier should be reduced strongly if a positively charged amino acid at the surface of the substrate-binding pocket were present or if such a charged residue that is normally buried behind a large hydrophobic side chain were demasked (Fig. 2).
Figure 2.
Straight- and inverse-substrate orientation at the active site of LOXs.
For a more comprehensive understanding of the mechanistic reasons for the positional specificity of LA oxygenation, we carried out structural modeling of enzyme/substrate interaction and site-directed mutagenesis on the lipid body LOX of cucumber seedlings and identified H608 as the primary determinant for the positional specificity. The data presented indicate the possibility of converting a plant 13-LOX catalyzing a [+2] radical rearrangement to a 9-lipoxygenating species.
EXPERIMENTAL PROCEDURES
Materials.
The chemicals used were from the following sources: standards of chiral and racemic hydroxy fatty acids were from Cayman Chemical (Ann Arbor, MI); trilinolein (TL) was from Sigma; methanol, hexane, 2-propanol (all HPLC grade) were from Baker; and restriction enzymes were purchased from New England Biolabs.
Site-Directed Mutagenesis and Protein Expression.
For bacterial expression of wild-type and mutant LOX species and for site-directed mutagenesis studies we used the plasmid pQE-30 (Qiagen) containing lipid body LOX cDNA from cucumber cotyledons (LOXpQE-30) as insert (15). Mutagenesis was carried out by using the QuikChange site-directed mutagenesis kit from Stratagene. Oligonucleotides containing the appropriate base changes were purchased from MWG-Biotec (Ebersberg, Germany). To analyze for the mutations, additional conservative base exchanges were introduced to construct new restriction sites or delete existing ones. In addition, all mutations were sequenced and at least three different bacterial colonies were expressed and used for analysis of enzymatic parameters. Expression of LOXpQE-30 and all mutations was performed as described (15). Cells from a 1-liter culture were resuspended in 5–7 ml lysis buffer and disrupted by using a sonifier tip with five pulses each of 30 s, cellular debris was pelleted, and affinity purification of His-tagged LOXs was performed as described (15).
Activity Assay and Sample Work-Up.
For product analysis, 0.9 ml cell lysates was incubated with 0.9 mM LA or 1.2 mM TL (final concentration) in 100 mM Tris buffer, pH 7.5, for 30 min at room temperature. Reactions were stopped by the addition of sodium borohydride. The samples were acidified to pH 3 and the lipids were extracted (16). The lower chloroform phase was recovered, the solvent was evaporated, the remaining lipids were reconstituted in 0.1 ml methanol, and aliquots were injected for HPLC analysis. For alkaline hydrolysis of the triacylglycerides, the lipid extracts were diluted with 0.4 ml methanol, 80 μl 40% (wt/vol) KOH was added, and the samples were hydrolyzed under argon atmosphere for 20 min at 60°C. After cooling down to room temperature the samples were acidified with glacial acetic acid and aliquots were analyzed by RP-HPLC.
Analytics.
HPLC analysis was carried out on a Hewlett–Packard 1100 HPLC system coupled to a diode array detector. RP-HPLC of the free fatty acid derivatives was carried out on a Nucleosil C-18 column (250 × 4 mm, 5-μm particle size; Macherey & Nagel) with a solvent system of methanol/water/acetic acid (85:15:0.1, by vol) and a flow rate of 1 ml/min. The absorbances at 234 nm (conjugated diene system of the hydroxy fatty acids) and 210 nm (polyenoic fatty acids) were recorded simultaneously. Oxygenated LA containing triacylglycerides was separated on a Nucleosil C-18 column (250 × 4 mm, 5-μm particle size; Macherey & Nagel) using a binary gradient system: solvent A, methanol/water/acetic acid (90:10:0.1, by vol); solvent B, methanol/acetic acid (100:0.1, by vol); and the following gradient program: 10 min at 100% solvent A, then 20 min with a linear increase of solvent B to 100% solvent B followed by an isocratic postrun of 50 min at 100% B. The absorbance at 234 nm was recorded. Straight-phase HPLC (SP-HPLC) of hydroxy fatty acid isomers was carried out on a Zorbax SIL column (250 × 4.6 mm, 5-μm particle size; Hewlett–Packard) with a solvent system of n-hexane/2-propanol/acetic acid (100:2:0.1, by vol) and a flow rate of 1 ml/min. The enantiomer composition of the hydroxy fatty acids was analyzed by chiral-phase HPLC on a Chiralcel OD column (250 × 4.6 mm, 5-μm particle size; Diacel Chemical Industries, distributed by Baker) with a solvent system of hexane/2-propanol/acetic acid (100:5:0.1, by vol) and a flow rate of 1 ml/min (17).
Structural Modeling.
Because of the high degree of homology between the soybean LOX-1 and the cucumber enzyme, the x-ray coordinates for the former LOX (18) were used for modeling the enzyme–substrate interaction. The alignment of the substrate fatty acid (LA) at the active site of the enzyme was estimated assuming a rigid-probe/rigid-receptor model. Thus, the docking problem of LA in the substrate-binding pocket essentially was reduced to a six-dimensional search over translational and rotational degrees of freedom in some selected regions around the substrate-binding site. Initially, the orientation of LA was calculated with the manual docking method by using functions involving steric complementarity and distance geometry. Then, the geometry of the interaction between LA and the side chains of the amino acids was optimized by using the molecular mechanics force-field method MM+ of the hyperchem software package. For H608V mutation the amino acid side chains were exchanged on the computer and the energetically most favorable position of the new side chains was calculated.
RESULTS
Structural Modeling of Enzyme/Substrate Interaction.
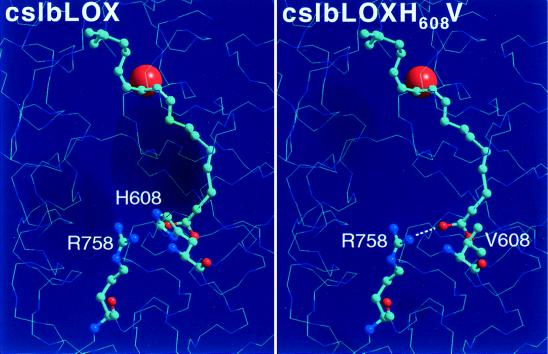
Site-directed mutagenesis studies on mammalian 12/15-LOXs suggested that the substrate alignment at the active site appears to be of major importance for the positional specificity of the LOX reaction (7, 8, 13). For better understanding of the mechanism of LA oxygenation by the lipid body 13-LOX, we carried out structural modeling of the enzyme/substrate complex. Because the LOX shares a high degree of amino acid similarity (70%) with the soybean seed LOX-1 (19), the x-ray coordinates of this enzyme were used (18, 20). These modeling studies and earlier literature reports (20) indicated that R758 is localized in the vicinity of the putative substrate-binding pocket but that its positive charge may be shielded by the rather bulky imidazole ring of H608 (Fig. 3, cslbLOX). Thus, the positive charge appears to buried inside the protein and may not disturb hydrophobic milieu inside the substrate-binding pocket. Multiple amino acid sequence alignments of various plant LOXs (13-LOX: lipid body LOX, soybean seed LOX-1, potato LOX-H1, Arabidopsis LOX-2; 9-LOX: potato tuber LOX, tobacco elicitor-induced LOX, barley LOX-A) exhibiting different positional specificities indicated that all enzymes contain an arginine at that position that aligns with R758 of the lipid body LOX. The data suggest that this amino acid alone may not be critical for the positional specificity. However, there was an obvious conserved difference between 9- and 13-LOXs. For all linoleate 13-LOXs the positions that align with H608 of the lipid body LOX are occupied by space-filling amino acids (phenylalanine or histidine). In contrast, a rather space-saving amino acid (valine) is located at this position in all plant 9-LOXs cloned so far (Table 1). Interestingly, H608 of the lipid body LOX aligns with the M418 of the human reticulocyte-type 15-LOX, which was identified as the primary determinant of the positional specificity of arachidonic acid oxygenation (8). In addition, for the rabbit 15-LOX it has been reported that F353, which aligns with S491 of soybean LOX-1 and V542 of lipid body LOX, constitutes another primary determinant of the positional specificity (7). However, sequence alignments of plant LOXs did not reveal conserved differences at this position between linoleate 13- or 9-LOXs.
Figure 3.
Model showing inverse-substrate binding with respect to the proposed mode of binding of the lipid body LOX and its H608V mutant. (Left) Active-site model of the lipid body LOX. Here, the methyl terminus of LA is nearby the side chain of H608. The charged R758 is shielded by H608. (Right) Model for the H608V mutant. When oriented inversely, the negatively charged carboxylic group of LA (red) may form a salt bridge (dotted line) with positively charged nitrogens (blue) of R758.
Table 1.
Alignment of amino acid residues possibly determining the positional specificity of plant LOXs
| Enzyme | Accession no. | Position of amino acid residues | Amino acid residues |
|---|---|---|---|
| 13-LOX | |||
| Cucumber lipid body LOX | X92890 | 607/608 | Thr/His |
| Soybean seed LOX-1 | P08170 | 556/557 | Thr/Phe |
| Potato LOX-H1 | X96405 | 614/615 | Ser/Phe |
| Arabidopsis LOX-2 | P38418 | 611/612 | Cys/Phe |
| 9-LOX | |||
| Potato tuber LOX | P37831 | 579/580 | Thr/Val |
| Tobacco elicitor-induced LOX | X84040 | 580/581 | Thr/Val |
| Barley grain LOX-A | L35931 | 574/575 | Thr/Val |
In the case of the wild-type lipid body LOX, H608 may shield the charged guanidino group of R758, making it functionally silent. However, if a less space-filling amino acid would be introduced at this position the shielding may not be as effective. Thus, the positive charge may be exposed to the interior of the substrate-binding pocket and may favor an inverse head-to-tail substrate orientation by forming a salt bridge to the carboxyl terminus of LA (Fig. 3, cslbLOXH608V, dotted line). If these theoretical considerations were correct one might be able to convert a linoleate 13-LOX to a 9-lipoxygenating species mutating H608 to a less space-filling residue.
Site-Directed Mutagenesis Studies.
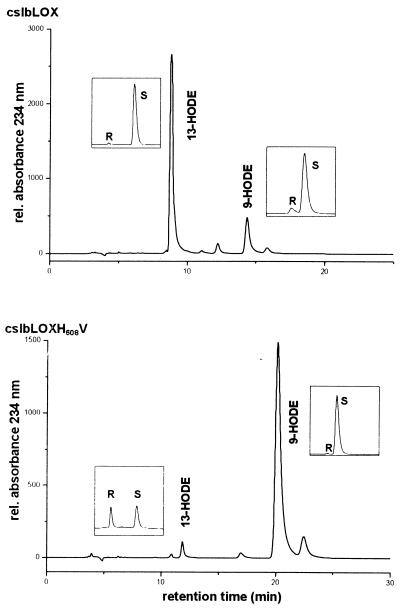
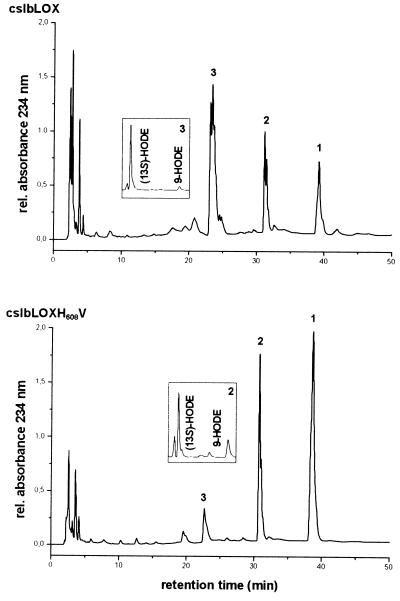
As indicated above, H608 of the lipid body 13-LOX aligns with V580 of the potato tuber 9-LOX. To test our working hypothesis, H608 of the lipid body LOX was mutated to a valine (H608V mutation). The wild-type and mutant enzymes were overexpressed as His-tagged fusion proteins and purified on a nickel-Sepharose column. As expected, HPLC analysis of the LA oxygenation products indicated (13S,9Z,11E)-13-hydro(pero)xy-9,11-octadecadienoic acid [13-H(P)ODE] as the major product of the wild-type enzyme (Fig. 4). However, for the H608V mutant, (9S,10E,12Z)-9-hydro(pero)xy-10,12-octadecadienoic acid [9-H(P)ODE] was identified as the main product. To find out whether mutation of H608 to an amino acid residue that is more space filling than valine but smaller than histidine would have a similar effect, the H608M mutant was tested. Here again, we found a strong preponderance of 9-H(P)ODE formation. Basic kinetic characterization of the 13-lipoxygenating wild type and the 9-lipoxygenating H608M mutant indicated that mutation resulted in a strongly impaired substrate affinity and a decrease in reaction rate. For the wild-type enzyme a Km of 114.9 μM and LA turnover under Vmax conditions (substrate saturation) of 12 s−1 were determined (23-point measurement between 100 μM and 250 μM LA concentration). In contrast, we calculated a Vmax of 2 s−1 and a Km of 1333.3 μM for the H608M mutant (21-point measurement between 300 μM and 1.4 mM LA concentration). These data suggest that substrate binding by the mutant enzyme may be hindered energetically and more substrate is required to reach Vmax.
Figure 4.
HPLC analysis of hydroxy fatty acids formed from LA by the lipid body LOX and its H608V mutant. Equal amounts of LOX protein were incubated with 0.9 mM LA at room temperature for 30 min. After reduction of lipids with sodium borohydride, the reaction mixture was acidified and the lipids were extracted. Oxygenated fatty acid derivatives were isolated by RP-HPLC, and positional isomers were analyzed by SP-HPLC. Ratios of S and R were determined by CP-HPLC (Insets).
For the human 15-LOX it has been shown that amino acids 418 and 419, which align with T607 and H608, respectively, of the lipid body LOX form the bottom of the substrate-binding pocket (8). To test whether T607 and H608 also work together, we created a mutant in which the polar threonine was exchanged to an isoleucine that is similar in size but does not contain a polar hydroxy group. This mutant was catalytically active (comparable to the wild-type enzyme) but exhibited a random positional specificity (Table 2). The mechanistic reasons for this effect remain unclear, but it may be possible that the polar hydroxy group is required for lege artis catalysis. Interestingly, the double mutations T607IH608V and T607IH608M exhibited a high degree of positional specificity strongly favoring linoleate 9-lipoxygenation. These data suggest that the effect caused by T607I exchange is suppressed when H608 is mutated to a less space-filling amino acid. In this case, other amino acids may be of major importance for the positional specificity of LA oxygenation.
Table 2.
Products formed from the reaction of cslbLOX and of cslbLOXH608V with either LA or TL
| Mutants | Products obtained with LA
|
Products obtained with TL
|
|
|---|---|---|---|
| Positional isomers 13-HODE:9HODE and major enantiomer | Amount of TL oxygenation and molar ratios of oxygenation products, TL-3HODE : TL-2HODE : TL-1HODE | Positional isomers 13-HOME:9-HODE and major enantiomer | |
| cslbLOX | 84 : 16 | 100% | 90 : 10 |
| S : S | 44 : 30 : 36 | S : rac | |
| H608V | 5 : 95 | 42% | 65 : 35 |
| rac : S | 2 : 26 : 72 | S : rac | |
| H608M | 21 : 79 | 71% | 83 : 17 |
| S : S | 25 : 34 : 41 | S : rac | |
| T607I | 45 : 55 | 74% | 85 : 15 |
| S : S | 19 : 29 : 52 | S : rac | |
| T607IH608V | 11 : 89 | 88% | 72 : 28 |
| s : S | 18 : 30 : 52 | S : rac | |
| T607IH608M | 18 : 82 | 90% | 80 : 20 |
| S : S | 18 : 33 : 49 | S : rac | |
| R758LH608V | 60 : 40 | 20% | 50 : 50 |
| rac : rac | 5 : 29 : 66 | rac : rac | |
| R758LT607IH608V | 52 : 48 | 17% | 50 : 50 |
| rac : rac | 2 : 31 : 67 | rac : rac | |
S > 80% S-enantiomer; s = 70–80% S-enantiomer; rac = 50–70% S-enantiomer, racemic. Oxygenated fatty acid derivatives were isolated by RP-HPLC (shown for TL in Fig. 4). Positional isomers of hydroxylinoleic acid (HODE) were given as molar ratios determined by SP-HPLC (Fig. 3). Optical isomers were determined by CP-HPLC (Fig. 3 Insets).
Structural modeling suggested that the H608V and H608M mutations may demask R758, and, thus, a positive charge may be exposed in the substrate-binding pocket that favors an inverse substrate orientation. Consequently, mutation of this arginine to a neutral residue in connection with the H608V or T607I+H608V exchanges should not lead to 9-lipoxygenating species. In fact, the R758L+H608V double mutant and R758L+T607IH608V triple mutant exhibited a random positional specificity and a reduced reaction rate (5% residual activity), suggesting the importance of R758 for linoleate 9-lipoxygenation.
Reaction Specificity with TL.
Earlier studies on the substrate specificity of the lipid body LOX indicated the enzyme’s capability to oxygenate esterified polyenoic fatty acids (15, 17). Because triacylglycerols do not contain free carboxylic groups, no major differences were expected when the patterns of oxygenation products of the wild-type enzyme and of the linoleate 9-lipoxygenating mutants were compared. Indeed, we found that the wild-type enzyme and all 9-lipoxygenating mutants exhibited a trilinoleate 13-LOX activity (Table 2). However, the rates of TL oxygenation by the 9-lipoxygenating mutants were only about 50% of that measured for the wild-type enzyme. Moreover, TL oxygenation by the mutant enzymes mainly led to triacylglycerol species in which one LA moiety was oxygenated. In contrast, with the wild-type enzyme, all three linoleate residues were oxygenated (Fig. 5).
Figure 5.
HPLC analysis of oxidized TL derivatives formed by the lipid body LOX and its H608V mutant. Equal amounts of LOX protein were incubated with an emulsion of 1.2 mM TL for 30 min. Lipids were reduced with sodium borohydride, and the reaction mixture was acidified. After extraction the lipids were analyzed by RP-HPLC. A representative chromatogram is shown. Numbers mark the resulting LOX-derived products: 1, TL derivative containing one oxygenated fatty acid; 2, doubly oxygenated TL isomers; and 3, triple-oxygenated TL. For analysis of positional isomers of the LA moieties, the free fatty acid derivatives were obtained by alkaline hydrolysis and isolated by RP-HPLC. Positional isomers of hydroxy LA (HODE) were given as molar ratios determined by SP-HPLC as shown in the Insets. Optical isomers were determined by CP-HPLC.
DISCUSSION
The structural determinants for the positional specificity of mammalian LOXs have been studied for a long time. In 1991 it was reported that mutation of rather bulky amino acids at positions 418 and 419 of the human reticulocyte-type 15-LOX to smaller residues led to a 12-lipoxygenating species (8). Later, similar mutation strategies converted the porcine leukocyte 12-LOX (21) and the human platelet 12-LOX to a 15-lipoxygenating species (22). However, attempts failed to convert the murine macrophage 12-LOX (23) and the rat pineal gland 12-LOX (24) to a 15-LOX. Formation of LOX chimera of the rabbit 15-LOX and the murine macrophage 12-LOX led to the identification of F353 of the rabbit 15-LOX as a primary determinant of the positional specificity (7). Mutation of this bulky amino acid to a smaller valine altered the positional specificity to 12-lipoxygenation. These data and the x-ray coordinates for two plant LOXs (18, 20, 25) and for the rabbit 15-LOX inhibitor/complex (13) led to the hypothesis that the volume of the substrate-binding pocket may be decisive for the positional specificity of the LOX reaction. 15-LOXs have a small cavity and, because fatty acids may penetrate the active site with their methyl terminus first, they are dioxygenated close to this end of the molecule. In contrast, more space appears to be available in the substrate-binding cleft of 12- and 5-LOXs. In these cases, the fatty acid substrates may slide farther into the substrate-binding pocket and, thus, are oxygenated closer to the carboxyl terminus. However, there may be an alternative hypothesis explaining 5-lipoxygenation. This theory suggests that, for 5-lipoxygenation, the substrate is bound inversely at the active site, so the carboxylate group may penetrate into the substrate-binding pocket (9–12). According to this inverse-orientation hypothesis, the stereochemistry of linoleate 9-lipoxygenation is straightforward and there is no need to postulate conformational changes of either substrate or enzyme. However, a major drawback of this hypothesis is that burying the charged carboxylic group in the hydrophobic environment of the putative substrate-binding pocket should be hindered strongly by a large energy barrier (14). This energy barrier can be reduced if a charged residue that is masked in wild-type LOXs was exposed to the inner surface of the substrate-binding cage. Our search for potential candidates of charged amino acids located in close proximity to the putative substrate-binding pocket identified R758 as a possible candidate for the lipid body LOX. Interestingly, in all plant LOXs this residue is highly conserved, and, in the case of linoleate 13-LOXs, this charged residue appears to be shielded by a space-filling amino acid (histidine or phenylalanine, Table 1) at position 608. In contrast, plant 9-LOXs contain a rather small amino acid (valine, Table 1) at this position, and, thus, the positively charged guanidino group of R758 may be exposed at the surface of the substrate-binding cage, where it may interact with the negatively charged carboxylic group of the fatty acid substrate. In this context the data obtained here appear to be supportive for the hypothesis of a head-to-tail inverse-substrate orientation at the active site of 9-LOXs.
It should, however, be stressed that an inverse head-to-tail substrate orientation is not the only explanation for an effective conversion of a [+2] LOX to a [−2] lipoxygenating species. Even if a conserved singular orientation of LOX substrates is assumed (methyl end of the fatty acid penetrates the active site), the configuration of the substrate at the active site may vary depending on the side-chain geometry of the amino acids lining the substrate-binding pocket. Fatty acids are rather flexible molecules and may adopt several thermodynamically favored configurations when binding at the active site, and these different substrate configurations may influence the position of dioxygen insertion.
The detailed mechanism of the LOX reaction remains a matter of discussion. Although there is convincing evidence for a radical mechanism (26), the formation of organo-iron intermediates either at C-13 or C-9 still might be possible (27). However, there are no sufficient data to support this hypothesis, and, thus, the radical-based mechanism generally is accepted. On the other hand, this radical theory has several cavities and there appear to be two problems of particular interest: (i) the chemical nature of the radical intermediates and (ii) the mechanism of dioxygen insertion.
The classical mechanism of the LOX reaction involves the formation of a pentadienyl radical formed during the initial hydrogen abstraction. In this radical intermediate the electron density may be delocalized over the entire pentadienyl system (28), which would require a more or less planar structure of the intermediate. If an electron-drawing amino acid residue would be localized close to C-13 of the LA molecule, the electron density may be highest at this carbon atom ([+2] radical rearrangement), and, thus, oxygen may be inserted at C-13. In contrast, if an electrophilic residue is localized close to C-9, the electron density at C-9 should be higher and C-9 oxygenation may be favored. However, more recent structural data (20) suggest that the formation of a planar pentadienyl radical may be hindered sterically because of the bend configuration of the substrate fatty acid. Thus, for 13-lipoxygenation by the soybean LOX-1, the formation of a dioxygen-bridged [9,10,11]-allyl radical was proposed in which the radical electron is delocalized between C-9 and C-11. Such a rearrangement may not require a planar structure of the radical intermediate and, thus, appears to be sterically favored (29). This mechanism rationalizes the stereospecificity of the oxygen insertion and does not contradict an inverse head-to-tail substrate orientation. More recent spectroscopic studies on the soybean LOX-1 suggested the formation of such a dioxygen-bridged allyl radical (29). However, for now, it is not clear which steric factors will favor a [9,10,11]-allyl radical leading to 13-HPODE formation and which will favor a [11,12,13]-allyl radical (9-HPODE formation). It might be possible that the H608V exchange reported here may alter the overall substrate configuration in such a way that the formation of a [11,12,13]-allyl radical becomes sterically favored, which would lead to 9-HPODE formation.
The second point of particular interest is the mechanism of dioxygen insertion. According to the current status of knowledge there is no evidence for a specific dioxygen-binding site at the active site of LOXs. Although the ferrous nonheme iron should be capable of binding dioxygen, there are no experimental data to prove this. Instead, it is generally believed that dioxygen insertion may be diffusion-controlled. Originally, it was suggested that atmospheric dioxygen may diffuse from the medium to the active site through an 18-Å long hydrophobic tunnel that narrows to a diameter of 2.5 Å just before reaching the iron (18). In contrast, more recent x-ray data suggested that dioxygen movement through this tunnel may not be possible without substantial rearrangement of several residues lining the tunnel (20). Instead, it may be possible that dioxygen may pass through the fatty acid-binding cavity, or through a 20-Å path starting from H248 and R533 at the surface, passing between P834 and I839, and ending in the vicinity of H690. We carried out structural modeling to find out whether H608V or the H608M mutation would have any impact on either of the putative dioxygen paths but were not able to come up with a satisfactory explanation. Thus, further analyses are needed to explain the observed alteration of the positional specificity of LA dioxygenation.
Acknowledgments
We thank S. Barth and M. Anton for excellent technical assistance. This paper is dedicated to Prof. Dr. E. Heinz on the occasion of his 60th birthday. This work was supported by the Bundesministerium für Landwirtschaft, 96NR173, and, in part, by a research grant from the Deutsche Forschungsgemeinschaft to H.K. (Ku 961/2-3) and by the European Community (BMH4-CT-98-3191).
ABBREVIATIONS
- CP-HPLC
chiral-phase HPLC
- SP-HPLC
straight-phase HPLC
- 9-H(P)ODE
(9S,10E,12Z)-9-hydro(pero)xy-10,12-octadecadienoic acid
- 13-H(P)ODE
(13S,9Z,11E)-13-hydro(pero)xy-9,11-octadecadienoic acid
- LA
linoleic acid
- LOX
lipoxygenase
- TL
trilinolein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Siedow J N. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:145–188. [Google Scholar]
- 2.Yamamoto S. Biochim Biophys Acta. 1992;1128:117–131. doi: 10.1016/0005-2760(92)90297-9. [DOI] [PubMed] [Google Scholar]
- 3.Rosahl S. Z Naturforsch. 1996;51c:123–138. doi: 10.1515/znc-1996-3-401. [DOI] [PubMed] [Google Scholar]
- 4.Schewe T, Rapaport S M, Kühn H. Adv Enzymol Mol Biol. 1986;58:191–272. doi: 10.1002/9780470123041.ch6. [DOI] [PubMed] [Google Scholar]
- 5.Gardner H W. Biochim Biophys Acta. 1991;1084:221–239. doi: 10.1016/0005-2760(91)90063-n. [DOI] [PubMed] [Google Scholar]
- 6.Shibata D, Axelrod B. J Lipid Mediators Cell Signal. 1995;12:213–228. doi: 10.1016/0929-7855(95)00020-q. [DOI] [PubMed] [Google Scholar]
- 7.Borngräber S, Kuban R J, Anton M, Kühn H. J Mol Biol. 1996;264:1145–1153. doi: 10.1006/jmbi.1996.0702. [DOI] [PubMed] [Google Scholar]
- 8.Sloane D L, Leung R, Craik C S, Sigal E. Nature (London) 1991;354:149–152. doi: 10.1038/354149a0. [DOI] [PubMed] [Google Scholar]
- 9.Kühn H, Schewe T, Rapoport S M. Adv Enzymol Mol Biol. 1986;58:273–311. doi: 10.1002/9780470123041.ch7. [DOI] [PubMed] [Google Scholar]
- 10.Prigge S T, Gaffney B J, Amzel L M. Nat Struct Biol. 1998;5:178–179. doi: 10.1038/nsb0398-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egmond M R, Veldink G A, Vliegenthart J F G, Boldingh J. Biochem Biophys Res Commun. 1973;54:1178–1184. doi: 10.1016/0006-291x(73)90816-4. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann W D. Free Radical Biol Med. 1994;16:241–253. doi: 10.1016/0891-5849(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 13.Gillmor S A, Villasenor A, Fletterick R, Sigal E, Browner M F. Nat Struct Biol. 1998;4:1005–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 14.Browner M F, Gillmor S A, Fletterick R. Nat Struct Biol. 1998;5:179. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 15.Feussner I, Bachmann A, Höhne M, Kindl H. FEBS Lett. 1998;431:433–436. doi: 10.1016/s0014-5793(98)00808-4. [DOI] [PubMed] [Google Scholar]
- 16.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 17.Feussner I, Balkenhohl T J, Porzel A, Kühn H, Wasternack C. J Biol Chem. 1997;272:21635–21641. doi: 10.1074/jbc.272.34.21635. [DOI] [PubMed] [Google Scholar]
- 18.Boyington J C, Gaffney B J, Amzel L M. Biochem Soc Trans. 1993;21:744–748. doi: 10.1042/bst0210744. [DOI] [PubMed] [Google Scholar]
- 19.Höhne M, Nellen A, Schwennesen K, Kindl H. Eur J Biochem. 1996;241:6–11. doi: 10.1111/j.1432-1033.1996.0006t.x. [DOI] [PubMed] [Google Scholar]
- 20.Minor W, Steczko J, Stec B, Otwinowski Z, Bolin J T, Walter R, Axelrod B. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki H, Kishimoto K, Yoshimoto T, Yamamoto S, Kanai F, Ebina Y, Miyatake A, Tanabe T. Biochem Biophys Acta. 1994;1210:308–316. doi: 10.1016/0005-2760(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 22.Chen X S, Funk C D. FASEB J. 1993;7:694–701. doi: 10.1096/fasebj.7.8.8500694. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe T, Haeggstrom J Z. Biochem Biophys Res Commun. 1993;192:1023–1029. doi: 10.1006/bbrc.1993.1519. [DOI] [PubMed] [Google Scholar]
- 24.Hada T, Hagiya H, Suzuki H, Arakawa T, Nakamura M, Matsuda S, Yoshimoto T, Yamamoto S, Azekawa T, Morita Y, Ishimura K, Kim H Y. Biochem Biophys Acta. 1994;1211:221–228. doi: 10.1016/0005-2760(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 25.Skrzypczak-Jankun E, Amzel L M, Kroa B A, Funk M O. Proteins Struct Funct Genet. 1997;29:15–31. [PubMed] [Google Scholar]
- 26.Nelson M J, Seitz S P. Curr Opin Struct Biol. 1994;4:878–884. doi: 10.1016/0959-440x(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 27.Corey E J, Nagata R. J Am Chem Soc. 1987;109:8107–8108. [Google Scholar]
- 28.De Groot J J M C, Veldink G A, Vliegenthart J F G, Boldingh J, Wever R, Van Gelder B F. Biochim Biophys Acta. 1975;377:71–79. doi: 10.1016/0005-2744(75)90287-9. [DOI] [PubMed] [Google Scholar]
- 29.Nelson M J, Cowling R A, Seitz S P. Biochemistry. 1994;33:4966–4973. doi: 10.1021/bi00182a027. [DOI] [PubMed] [Google Scholar]