Abstract
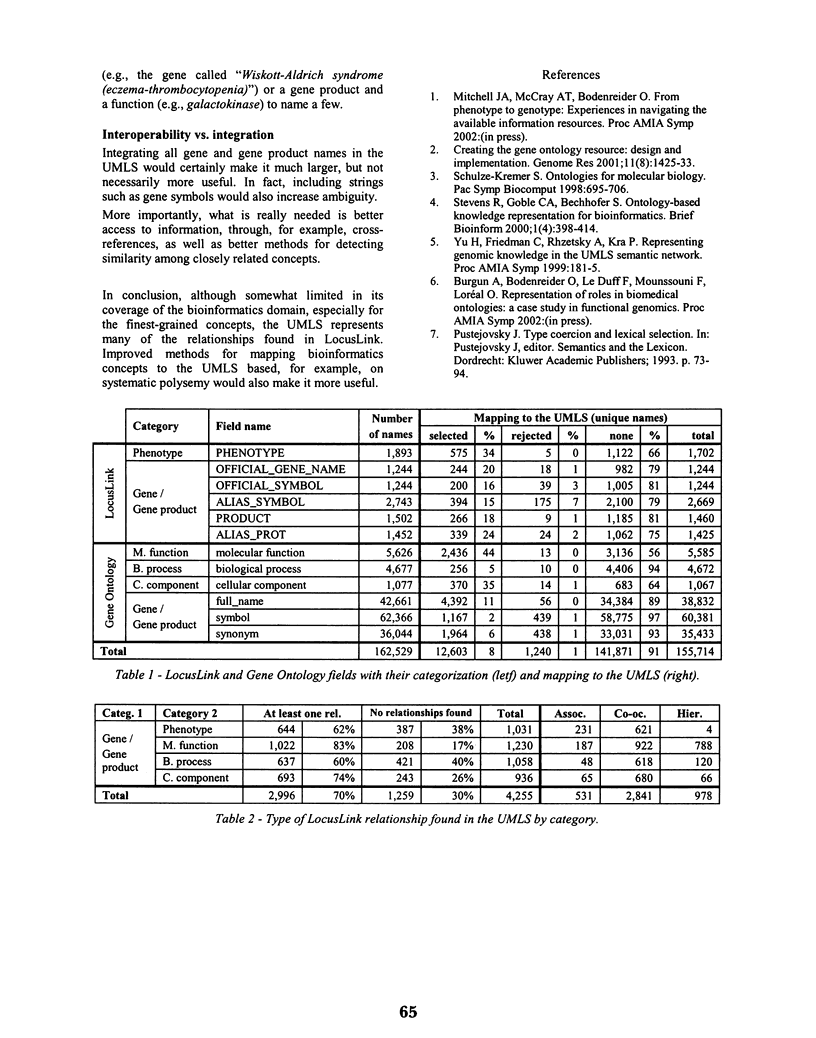
Objectives: Terminology and knowledge resources are essential components of interoperability among disparate systems. This paper evaluates whether names and relationships needed in biomedical informatics are present in the UMLS. METHODS: Terms for five broad categories of concepts were extracted from LocusLink and mapped to the UMLS Metathesausus. Relationships between gene products and the other four categories (phenotype, molecular function, biological process, and cellular component) were searched for in the Metathesaurus. All gene products in the Gene Ontology database were also mapped to the UMLS in order to evaluate its global coverage of the domain. RESULTS: The coverage of concepts ranged from 2% (gene product symbols) to 44% (molecular functions). The coverage of relationships ranged from 60% for Gene product-Biological process to 83% for Gene product-Molecular function. DISCUSSION: Terminology and ontology issues are discussed, as well as the need for integrating additional resources to the UMLS.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gene Ontology Consortium Creating the gene ontology resource: design and implementation. Genome Res. 2001 Aug;11(8):1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Kremer S. Ontologies for molecular biology. Pac Symp Biocomput. 1998:695–706. [PubMed] [Google Scholar]
- Stevens R., Goble C. A., Bechhofer S. Ontology-based knowledge representation for bioinformatics. Brief Bioinform. 2000 Nov;1(4):398–414. doi: 10.1093/bib/1.4.398. [DOI] [PubMed] [Google Scholar]
- Yu H., Friedman C., Rhzetsky A., Kra P. Representing genomic knowledge in the UMLS semantic network. Proc AMIA Symp. 1999:181–185. [PMC free article] [PubMed] [Google Scholar]


