Abstract
Association between Y chromosome haplotype variation and alcohol dependence and related personality traits was investigated in a large sample of psychiatrically diagnosed Finnish males. Haplotypes were constructed for 359 individuals using alleles at eight loci (seven microsatellite loci and a nucleotide substitution in the DYZ3 alphoid satellite locus). A cladogram linking the 102 observed haplotype configurations was constructed by using parsimony with a single-step mutation model. Then, a series of contingency tables nested according to the cladogram hierarchy were used to test for association between Y haplotype and alcohol dependence. Finally, using only alcohol-dependent subjects, we tested for association between Y haplotype and personality variables postulated to define subtypes of alcoholism—antisocial personality disorder, novelty seeking, harm avoidance, and reward dependence. Significant association with alcohol dependence was observed at three Y haplotype clades, with significance levels of P = 0.002, P = 0.020, and P = 0.010. Within alcohol-dependent subjects, no relationship was revealed between Y haplotype and antisocial personality disorder, novelty seeking, harm avoidance, or reward dependence. These results demonstrate, by using a fully objective association design, that differences among Y chromosomes contribute to variation in vulnerability to alcohol dependence. However, they do not demonstrate an association between Y haplotype and the personality variables thought to underlie the subtypes of alcoholism.
Although familial transmission of alcoholism is well established, the causation of this disease is complex, with both genetic and environmental components. Moreover, there is strong evidence for distinct clinical subtypes, which differ in their accompanying psychophysiological traits, clinical course, and genetic influences. Cloninger (1) has developed a conceptual framework of two principal but nonexclusive subtypes of alcoholism. Type I alcoholism, a late onset form, is characterized by a personality profile of low Novelty Seeking, high Harm Avoidance, and high Reward Dependence, whereas type II alcoholism is characterized by high Novelty Seeking, low Harm Avoidance, and low Reward Dependence. Type II alcoholics have an early age of onset and are more likely to have antisocial personality disorder (ASPD) (2, 3). Both types of alcoholism are more common in men than women, but Type II alcoholism is almost exclusively limited to males. Type I and Type II alcoholism are not recognized diagnoses, however, ASPD is a well defined clinical phenotype and strong evidence for cotransmission of ASPD and vulnerability to alcoholism has been shown in adoption studies from Sweden (4) and the United States (5).
It is well known that the Y chromosome affects the expression of many traits, but the extent to which differences among Y chromosomes account for phenotypic differences among males is largely unknown. With the exception of a few traits, e.g., azoospermia and possibly gonadoblastoma, pure holandric inheritance in humans is unknown. It has been difficult to demonstrate any influence of polymorphic Y-linked genes for several reasons. First, there do not appear to be many coding sequences on the Y chromosome. Second, much of the Y chromosome is nonrecombining, which makes classical linkage analysis based on meiotic recombination impossible. In addition, the best known Y linked genes regulate the expression of autosomal genes, so that it is difficult to isolate the genetic origin of individual differences in sex-associated traits. Until recently, Y chromosomal genetic markers have provided insufficient resolving power.
Compared with the autosomes, the Y chromosome contains a limited number of sequence polymorphisms. However, the polymorphisms that have been described now include multiple microsatellites and several unique sequence variants found on the nonrecombining portion of the chromosome (6). The lack of recombination for the Y chromosome-specific polymorphisms and the haploid status of this chromosome in males provides for straightforward construction of definitive haplotypes. In addition, highly informative paternal lineages can be evaluated by constructing compound haplotypes using the different types of available polymorphisms (7, 8).
The purpose of this study was to analyze the effects of differences between Y chromosomes on the vulnerability to alcoholism, ASPD, and related personality dimensions by using cladistic association analysis (9). To assess Y chromosome differences, seven Y chromosome-specific microsatellite loci and a restriction site at the DYZ3 alphoid satellite DNA locus (10) were used to construct haplotypes from a large sample of psychiatrically evaluated and unrelated Finnish males (11–13). Males from Finland provide an ideal study population because of the country’s relative isolation since its founding some 80 generations ago and the availability of excellent health care and family records (14–17). The founding event and subsequent isolation has been purported to be responsible for the high prevalence of over 30 rare genetic diseases in Finland (14–16). In fact, Finnish Y chromosome diversity is significantly lower than the diversity in other European populations (18) but it is similar to the diversity in several Native American populations that have undergone severe population bottlenecks after European contact (19).
In isolated populations, the evolutionary history of Y chromosome haplotypes can be reliably inferred and represented in an evolutionary tree, or cladogram. The cladogram of Finnish Y chromosomes forms a hierarchy depicting ancestor–descendant relationships and groups of haplotypes (clades) that share a common ancestor. We used this hierarchy as an inherent nested design (9) to investigate whether any particular Y chromosome or group of related Y chromosomes was associated with alcoholism. Then, by using only alcoholics, we applied the same strategy to test for association between Y chromosomes and the personality characteristics postulated to define alcoholism subtypes (1). Alcoholism was defined by the DSM-III-R alcohol-dependence criteria (20). The alcoholism-related personality traits tested were ASPD defined by DSM-III-R criteria, and the three principal axes of the Tridimensional Personality Questionnaire (TPQ) (21, 22): Harm Avoidance, Novelty Seeking, and Reward Dependence.
For this study, the Y chromosome cladogram was estimated from alleles at seven microsatellite loci and a DYZ3 restriction site. Although a functional relationship between these polymorphisms and behavioral differences is unlikely, we can expect that functional polymorphisms will be embedded within the same cladogram, because this region of the Y chromosome is nonrecombining and clonally inherited. We present here data that suggest an association between evolutionarily related Y chromosome haplotypes and male-associated behavioral traits.
MATERIALS AND METHODS
Subjects and Psychiatric Diagnoses.
DNA was acquired from 359 unrelated adult males born in Finland. All subjects had Finnish surnames and volunteered during the past 10 years for a long-term psychiatric genetics study in Helsinki, Finland. Subjects were ascertained throughout Finland either as criminal probands undergoing court-ordered forensic psychiatric examinations (n = 154), relatives of criminal probands (unrelated to probands analyzed here) (n = 53), or a population sample (n = 152). The Structured Clinical Interview for DSM-III-R was administered by a psychiatrist (M.E.) to 104 criminal probands, 26 relatives of other criminal probands, and 69 Finnish volunteers. Diagnoses were made from the interviews by a trained clinical social worker under the supervision of a research psychiatrist, both of whom were blind to the previous histories of the subjects. One hundred thirty-two males were diagnosed alcohol dependent (DSM-III-R 303.90) and among these, 66 were also diagnosed as having antisocial personality disorder (DSM-III-R 301.7). Of the alcohol-dependent subjects, 15 were from the population volunteer sample, and 7 of these population volunteers also had ASPD. Eighty-nine of the alcohol-dependent males were scored by using the Tridimensional Personality Questionnaire (TPQ). The TPQ assesses four dimensions, of which three were used in this study: Novelty Seeking, Harm Avoidance, and Reward Dependence. A written informed consent for genetic studies on psychiatric disorders and alcoholism was obtained from all subjects. The human studies protocol was approved by the Institutional Review Board of the National Institutes of Health and the National Institute of Mental Health, by the Office for Protection from Research Risks, by the University of Helsinki Department of Psychiatry Institutional Review Board, and by the University of Helsinki Central Hospital Institutional Review Board.
Polymorphism Typing.
Genotyping was performed blind to phenotypic information. Seven microsatellite loci specific to the nonrecombining portion of the Y chromosome were amplified by using PCR. Primer sequences for DYS388, DYS389, DYS390, DYS391, DYS392, DYS393, and DYS394 were identified from the Genome DataBase and then tested for Y chromosome specificity. Primers for the DYS389 locus amplified two products, a smaller (240–260 bp) and a larger (370–400 bp) product. Only the smaller product was used for the analyses. For each PCR reaction, 50 ng of genomic DNA was added to 200 μM of dNTPs, 10 mM Tris⋅HCl (pH = 8.3), 50 mM KCl, 1.0–2.0 mM MgCl2, 0.6 units of AmpliTaq polymerase (Perkin–Elmer), and 0.33 μM primers. The forward primers were fluorescently labeled. The PCR cycling conditions consisted of 93°C for 3 min, 10 cycles at 94°C for 15 s, 55°C for 15 s, and 72°C for 30 s, followed by 20 cycles at 89°C for 15 s, 55°C for 15 s, and 72°C for 30 s. Finally, the extension cycle was 72°C for 10 min. The PCR products were then pooled in the presence of a size standard (Genescan 500 GS) and electrophoresed by using an Applied Biosystem 373A DNA sequencer. Alleles at each locus were determined by using the gs analysis and genotyper programs (Applied Biosystems). Corrections were made for individual gel shifts by using the computer program bioautograph (written by J.C.L. and M. Ross), and discrete size categories were assigned to the PCR products.
A PCR-based assay was used to type the DYZ3 locus (23, 24). Under a slight modification of Santos et al. (10), 40 ng of genomic DNA was amplified by using 200 μM dNTPs, 10 mM Tris⋅HCl (pH = 8.3), 50 mM KCl, 1.0–2.0 mM MgCl2, 0.6 units of AmpliTaq polymerase, and 0.33 μM forward and reverse primers designated U972 (5′-TCTGAGACACTTCTTTGTGGTA-3′) and L1214 (5′-CGCTCAAAATATCCACTTTCAC-3′). The PCR conditions were as follows; 94°C for 3 min, and then 30 cycles at 94°C for 30 s, 65°C for 30 s, and 72°C for 1 min. After amplification, the PCR products were visualized on a 3.5% agarose gel. The presence or absence of the restriction site polymorphism was detected by using 5 μl of PCR mixture, 20 units of the restriction enzyme HindIII (New England Biolabs), and the recommended buffer incubated overnight at 37°C.
Cladogram Construction.
All eight loci were used to construct Y chromosome haplotypes. The method of phylogenetic inference was maximum parsimony under the single stepwise mutation model. The single stepwise mutation model, after the mutation/drift model of Kimura and Ohta (25), has been proposed as a model for microsatellite mutations (26–28). The model assumes that an allele in state i will mutate to an allele either in state i + 1 or i − 1 with equal probability. The most parsimonious tree was determined by using the program paup (Phylogenetic Analysis Using Parsimony) Version 3.1.1 (29). More than 100,000 trees were evaluated by using extensive branch swapping. To resolve tree topology ambiguities, equal weights of one were used for the microsatellite characters, and a weight of three was used for the less variable DYZ3 HindIII site. The cladogram provided the design for the nestings by using clades of related haplotypes.
Statistical Analyses.
Association between Y haplotype and alcohol dependence was evaluated by using a series of nested two (affected and unaffected) × ni likelihood ratio χ2 contingency analyses, where ni is the number of clades in nesting category i. Then, using only alcohol-dependent subjects, the contingency table analyses were repeated for ASPD, and nested ANOVA was used to test for association between Y haplotype and the three TPQ personality scales. The nested design was determined from the Y cladogram by using the algorithm of Templeton et al. (9). Starting at the terminal ends of branches, we defined larger branches (clades) of evolutionary related haplotypes in the cladogram by single mutational steps until all mutations between haplotypes were incorporated into the nested design. For example, 0-step clades were the actual haplotypes and groups of 0-step clades related by a single mutational step comprise 1-step clades, groups of 1-step clades related by a single mutational step comprise 2-step clades, etc., until the entire cladogram was incorporated into a step-clade. Likelihood ratio χ2 statistics and general linear model ANOVAs were calculated by using SAS Version 6.0 software. When significant association was detected within a nesting category that had many related haplotypes, a series of tests were performed separately for all of the clades within the nesting category to localize the specific evolutionarily related haplotypes that contributed to the phenotype–haplotype association.
P values for statistically significant comparisons were confirmed by using the following randomization procedure. First, phenotypes were permuted at random among individuals. Second, the cladistic association analysis was performed. Third, steps 1 and 2 were repeated 10,000 times, and the results were sorted and tabulated. Fourth, the permuted test statistic value above the observed test statistic value is the empirical 100α% significance level.
To evaluate the sensitivity of our results to the possibility of non-Finnish admixture, DNA was acquired from 45 unrelated Swedish subjects living in Lund, Sweden, and 32 unrelated subjects of European heritage living in the Washington, D.C. metropolitan area. After genotyping, these samples were used to identify portions of the Finnish Y chromosome cladogram that are shared by non-Finns. Our analyses were then repeated by using only those Finnish subjects whose Y chromosomes belong to the major Finn-exclusive clade.
RESULTS
Y Chromosome Haplotypes.
Among the 359 unrelated males, 102 distinct eight-locus haplotypes were observed. Genic diversity (30), the probability that two randomly sampled alleles from the population are different, averaged across all loci was 0.479 ± 0.051. The number of alleles per locus ranged from two to six (Table 1). The HindIII polymorphism at the DYZ3 subunit consists of an A → G transition, and thus is less variable than the microsatellite loci. The HindIII site at the DYZ3 locus was present in 89% of the Y chromosomes. The number of observed haplotypes was thus substantially lower than the number expected on the basis of random allelic association. The most common haplotype was found in 82 males (23%), whereas 66 haplotypes were observed only once. Haplotype diversity calculated according to Nei (30) was 0.926 ± 0.005, which indicated that the ability of these markers to resolve Y chromosomes was near optimal, with cladistic analysis providing the means to make objective groupings as described below.
Table 1.
Allele frequencies and genic diversity
| Locus | No. of alleles | Size, bp/frequency | Genic diversity (h) | |||||
|---|---|---|---|---|---|---|---|---|
| DYS388 | 6 | 123/0.006 | 126/0.039 | 129/0.713 | 132/0.006 | 135/0.240 | 138/0.012 | 0.441 |
| DYS389 | 3 | 247/0.281 | 251/0.291 | 255/0.429 | 0.653 | |||
| DYS390 | 5 | 199/0.006 | 203/0.033 | 207/0.415 | 211/0.429 | 215/0.117 | 0.629 | |
| DYS391 | 4 | 283/0.398 | 287/0.593 | 291/0.006 | 295/0.003 | 0.493 | ||
| DYS392 | 5 | 248/0.357 | 251/0.003 | 254/0.039 | 257/0.599 | 260/0.003 | 0.513 | |
| DYS393 | 4 | 120/0.006 | 124/0.418 | 128/0.549 | 132/0.028 | 0.523 | ||
| DYS394 | 6 | 243/0.025 | 247/0.774 | 251/0.086 | 255/0.031 | 259/0.070 | 263/0.014 | 0.386 |
| DYZ3 | 2 | +/0.890 | −/0.110 | 0.196 | ||||
| Mean | 4.4 | 0.479 ± 0.051 | ||||||
Genic diversity (h) was calculated as h = n(1 − Σx12)/(n − 1) where n = number of individuals and x1 = allele frequency.
Y Chromosome Cladogram.
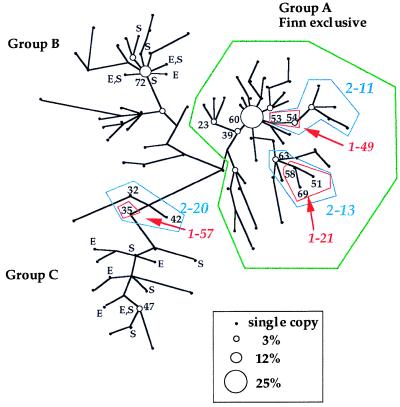
The evolutionary relationships of the eight-locus haplotypes are depicted by the cladogram in Fig. 1. Resamplings (10,000 bootstraps) and repeated cladogram constructions yielded high branch and clade support (>92%) throughout the cladogram. The DYZ3 locus helped distinguish three major lineages (haplogroups) of Y chromosomes. Y chromosome haplogroups A and B (89% of the sample) are HindIII (+), whereas Y chromosome lineage C is HindIII (−). Recently, we have shown that haplogroup A is a monophyletic clade of Y chromosome haplotypes not found in other European populations (31). Many of the haplotypes in haplogroups B and C also are observed in Swedish and European-American males (Fig. 1). Within each major lineage, there are several clusters of star-like extensions that are indicative of a recent expansion of the male population of Finland. Both the frequencies and the distributions of haplotypes within the tree suggest that it is an accurate portrayal of the mutational history. For instance, haplotypes 60 (n = 89), 72 (n = 44), 23 (n = 13), 54 (n = 12), 63 (n = 10), and 39 (n = 10) were positioned at internal nodes of the tree. Consistent with coalescence theory (32–34), these more frequent haplotypes (>10 copies) are placed at interior nodes within the cladogram, whereas the haplotypes represented by the terminal nodes were found only as single copies.
Figure 1.
Unrooted Y chromosome cladogram. Phylogeny of 102 Y chromosome haplotypes inferred by using maximum parsimony. Haplotype relationships are displayed as an unrooted phylogram with branch lengths proportional to the number of mutational steps between haplotypes. Total number of steps in the tree is 130. Observed haplotypes are depicted as circles (filled or open) whose sizes reflect haplotype frequency. Numbers in black are haplotypes noted in the text. Haplotypes found on branches A and B possessed the DYZ3 HindIII site, whereas haplotypes on branch C lacked the site. Haplotypes observed in Swedish and European American males are labeled S and E, respectively. Two-step clades 2-11, 2-13, and 2-20 (outlined in blue) were significantly associated with alcohol dependence, P = 0.002, P = 0.020, and P = 0.010, respectively. The association was attributable to 1-step clades 1-49, 1-21 and 1-57 (outlined in red). Every individual in clade 1-57 (haplotype 35) was also diagnosed with ASPD.
Clades connected by differing numbers of steps were identified by using the algorithm of Templeton et al. (9). The number of 0-step clades is simply equal to the number of haplotypes, in this case 102. At the next level, 0-step clades are embedded in a total of 61 1-step clades—45 representing external branches and 16 found internally. Proceeding inward from the “1-step-terminal” clades, we found 25 2-step clades, 8 3-step clades, 3 4-step clades, and 2 5-step clades. These step clades created the nested categories in which to test for phenotype associations.
Localization of Alcohol Dependence-Associated Clades.
Each step clade represents an independent partition of the data in which to test for Y haplotype–phenotype associations. The nested analyses provide immediate insight into which Y chromosomes, or clades of Y chromosomes, contribute to phenotypic differences. The psychiatric diagnosis of alcohol dependence represents a discrete categorization and was evaluated by using nested likelihood ratio χ2 statistics for each level of nesting. Contingency tests at each level of nesting are mutually independent (35). Furthermore, contingency tests performed within the same step clade also are independent, because the subsets of data do not overlap (36). Significant-likelihood ratio χ2 values for alcohol dependence were detected both at the 0-step (haplotype) and at the 1-step levels of the cladogram. A partition of the χ2 values reveals that all of the 0-step clade (haplotype) effects can be explained by the effects at the 1-step clade level. No overall significant effect was detected at the 2-step or higher clades. Table 2 shows the significant Y chromosome effects found at the 1-step level of the hierarchy. The alcohol-dependence associations are because of the 34 individuals who comprised 1-step clades contained within 2-step clades 2-11, 2-13, and 2-20 (highlighted in blue in Fig. 1), with χ2 P values of 0.001, 0.020, and 0.010, respectively. The P values were closely matched by the randomization procedure. The phylogenetic relationships of Y haplotypes in the three alcohol dependence-related clades were supported by bootstrap confidence values of 100%. Also, because the contingency analysis used only clinically diagnosed individuals (n = 199), cladogram construction was repeated using only the haplotypes from diagnosed individuals. The results of the second cladistic analysis were nearly identical to those found using the full data set (data not shown).
Table 2.
Nested contingency analysis for alcohol dependence
| Source | χ2 | Observed P | Empirical P |
|---|---|---|---|
| Full data set | |||
| 5-Step clades | 0.11 | 0.737 | n.s. |
| 4-Step clades | 2.45 | 0.293 | n.s. |
| 3-Step clades | 4.44 | 0.727 | n.s. |
| 2-Step clades | 21.13 | 0.451 | n.s. |
| 1-Step clades | 84.40 | 0.002 | 0.002 |
| Within 2-11 | 13.69 | 0.001 | 0.001 |
| Within 2-13 | 5.66 | 0.017 | 0.020 |
| Within 2-20 | 6.73 | 0.009 | 0.010 |
| 0-Step clades | 107.19 | 0.006 | 0.020 |
| Finn-exclusive clade A | |||
| 3-Step clades | 1.27 | 0.258 | n.s. |
| 2-Step clades | 12.23 | 0.201 | n.s. |
| 1-Step clades | 47.49 | 0.003 | 0.003 |
| 0-Step clades | 61.63 | 0.005 | 0.004 |
Likelihood ratio χ2 values were calculated for each nesting category of 132 alcohol-dependent men and 67 psychiatrically interviewed controls. Empirical P values were determined after 10,000 randomizations. n.s., P > 0.05.
To localize within the cladogram the association of Y chromosome haplotypes to alcohol dependence, 25 additional contingency tests were performed; i.e., affected vs. not affected for each 2-step clade. Fig. 1 identifies the location of Y chromosome effects on alcohol dependence at three 1-step clades (1-49, 1-21, and 1-57, which are embedded within 2-11, 2-13, and 2-20, respectively). Within these 1-step clades of related haplotypes, there were 16 unrelated individuals and all (100%) were alcohol-dependent.
To overcome the problem of ethnic stratification, we again restricted the cladistic association analysis, this time to the Finn-exclusive clade (haplogroup A). This restricted data set revealed quite similar levels of significance for alcohol dependence (Table 2).
Analysis of Personality Traits Related to Alcoholism Subtypes.
To determine whether Y chromosome haplotype variation underlies the type 1/type 2 alcoholism subtypes postulated by Cloninger (1), we performed the cladistic association analysis on ASPD (DSM-III-R 301.7) and the novelty seeking (NS), harm avoidance (HA), and reward dependence (RD) scales of the TPQ (21, 22), using only the alcohol-dependent subjects in our sample. Within our clinical sample, every male with ASPD was also alcohol-dependent; however, not all alcohol dependents were diagnosed with ASPD. To determine whether ASPD was related to Y chromosome haplotype, we calculated likelihood ratio χ2 values for the clinical diagnosis of ASPD within all alcoholics in the cladogram (Table 3). Within the alcohol dependence-associated clade 1-57 is haplotype 35, possessed by four individuals, all of whom have a comorbid ASPD diagnosis. Although this effect was nominally significant by theχ2 P value (P = 0.015), the randomization procedure failed to confirm the finding (P = 0.131). Thus, we cannot show a Y chromosome effect for ASPD that is independent of alcoholism alone.
Table 3.
Nested contingency analysis for aspd among alcoholics
| Source | χ2 | Observed P | Empirical P |
|---|---|---|---|
| Full data set | |||
| 5-Step clades | 1.77 | 0.183 | n.s. |
| 4-Step clades | 3.02 | 0.221 | n.s. |
| 3-Step clades | 11.13 | 0.133 | n.s. |
| 2-Step clades | 29.07 | 0.065 | n.s. |
| 1-Step clades | 56.41 | 0.035 | 0.248 |
| 0-Step clades | 80.06 | 0.015 | 0.131 |
| Finn exclusive clade A | |||
| 3-Step clades | 1.67 | 0.196 | n.s. |
| 2-Step clades | 8.36 | 0.301 | n.s. |
| 1-Step clades | 19.99 | 0.333 | n.s. |
| 0-Step clades | 28.22 | 0.348 | n.s. |
Likelihood ratio χ2 values were calculated for each nesting category of 66 individuals with the DSM-III-R diagnoses of alcohol dependence and ASPD vs. 66 individuals with alcohol dependence only. Empirical P values were determined after 10,000 randomizations. n.s., P > 0.05.
A nested ANOVA was used to test for association between particular clades within the cladogram and the three quantitative dimensions (NS, HA, and RD) of the TPQ (21, 22). As shown in Table 4, no significant association between a personality dimension and Y haplotype was seen at any level of the cladogram. Overall P values incorporating all five levels of the cladogram hierarchy were only 0.735, 0.531, and 0.141 for NS, HA, and RD, respectively.
Table 4.
Nested analysis of variance for TPQ personality dimensions
| Dimension | df | ss | F | P |
|---|---|---|---|---|
| Novelty-seeking | ||||
| 5-Step clades | 1 | 2.49 | 0.05 | 0.821 |
| 4-Step clades | 1 | 1019.00 | 0.02 | 0.875 |
| 3-Step clades | 5 | 319.99 | 1.33 | 0.269 |
| 2-Step clades | 10 | 324.57 | 0.67 | 0.743 |
| 1-Step clades | 15 | 501.15 | 0.69 | 0.778 |
| 0-Step clades | 9 | 479.43 | 1.11 | 0.378 |
| Overall | 41 | 1628.84 | 39.73 | 0.735 |
| Harm avoidance | ||||
| 5-Step clades | 1 | 21.67 | 0.84 | 0.364 |
| 4-Step clades | 1 | 11.44 | 0.44 | 0.509 |
| 3-Step clades | 5 | 15.05 | 0.12 | 0.988 |
| 2-Step clades | 10 | 364.52 | 1.41 | 0.205 |
| 1-Step clades | 15 | 412.72 | 1.06 | 0.412 |
| 0-Step clades | 9 | 207.19 | 0.89 | 0.540 |
| Overall | 41 | 1032.61 | 0.97 | 0.531 |
| Reward dependence | ||||
| 5-Step clades | 1 | 7.25 | 0.36 | 0.553 |
| 4-Step clades | 1 | 10.09 | 0.49 | 0.485 |
| 3-Step clades | 5 | 35.24 | 1.73 | 0.146 |
| 2-Step clades | 10 | 33.32 | 1.63 | 0.126 |
| 1-Step clades | 15 | 25.20 | 1.24 | 0.280 |
| 0-Step clades | 9 | 27.94 | 1.37 | 0.228 |
| Overall | 41 | 1156.4 | 1.38 | 0.141 |
DISCUSSION
It is known that alcoholism is strongly associated with additional psychiatric disorders, the most common of which are ASPD and depression (37, 38). Significant heritability for ASPD has been detected in numerous adoption (5) and twin (39) studies, as has a high cotransmission of alcoholism and ASPD (38). ASPD is more often diagnosed (comorbid) in alcoholic men, whereas depression is more often diagnosed in alcoholic women (40, 41). It has been suggested that one possible origin of the higher prevalence of alcoholism with ASPD in men than in women may reside on the Y chromosome (41). Less obvious and more controversial is that variation in the Y chromosome may be implicated in interindividual (male-to-male) variation in behavioral traits. Early studies on the Y chromosome and behavior were flawed by availability of relatively crude genetic markers (i.e., Y chromosome aneuploidy and length) and subjective phenotypes such as “aggression” and “criminality”(39–42). Later, studies on the role of the Y chromosome in mouse aggressive behavior were able to relate strain of origin of Y chromosome to a well defined phenotype (intermale attack behavior) (43–45). The precise relevance of murine attack behavior to human behavior is unknown.
We have shown, using cladistic association analysis, that differences in Y chromosomes are associated with the alcohol-dependence diagnosis. Because alcohol dependence is a heterogeneous disorder, we analyzed the cladogram with respect to several variables that are implicated to define subtypes (1–3). With regard to the three TPQ dimensions, none showed evidence for association. Our findings with respect to ASPD, an almost male sex-limited diagnosis, were more interesting. Although two clades contained subjects comorbid with alcohol dependence and ASPD, we were unable to show an association between Y haplotype and ASPD that was independent of alcohol dependence alone. This is perhaps unsurprising, because an autosomal gene basis for ASPD is suggested by an increased risk in female relatives of ASPDs for somatization disorder, a nearly female-limited trait. Regarding both TPQ and ASPD, we should remember that very large sample sizes may be necessary to dissect the individual component traits underlying the subtypes of alcoholism.
Using Haplotype Phylogeny to Design Association Studies.
Cladistic association analysis provides several advantages for population-based association studies. First, it provides objective criteria for the grouping of haplotypes. Haplotypes that may contribute to phenotypic variation are grouped according to evolutionary history. In reconstructing the Y haplotype phylogeny microsatellite convergence was not a problem for several reasons. First, valuable information for phylogeny construction was gained by incorporating the single stepwise mutation model for the microsatellite alleles (based on the number of repeat units). By ordering the characters (alleles), we effectively optimized our inferred haplotype mutational history in light of the convergence of allele states. Second, because the haplotypes were sampled from a recently expanded small population, the haplotypes were closely related and coalesce to ancestral haplotypes still present within the population. Finally, the point mutation at the DYZ3 locus firmly identified two of the three Y chromosome lineages in the phylogeny. We presented our unrooted network as a phylogram whose branch lengths explicitly illustrate the number of mutational steps that separate each haplotype. Branch lengths are important because the identification of the step-clade hierarchy required information on the exact number of mutational steps separating haplotypes.
Finnish Population Heterogeneity.
Like other methods for detecting association, cladistic association analysis can provide spurious findings if a heterogeneous population has been sampled. Therefore, it is recommended (46) that such studies be performed on relatively homogeneous populations. Although Finland is often portrayed by geneticists as a relatively homogeneous genetic isolate (46, 47), a separate haplotype analysis using geographic origin of samples revealed differences between eastern and western Finland (48). In fact, two major groups of Finnish Y chromosomes exist in Finland: one is of Asian origin (haplogroup A) and is not found in other European populations, whereas the other (haplogroup B) is common in Europe. Because Finland may have been influenced by Swedish immigration throughout history, we tested the validity of the association by using only the Y chromosomes exclusive to Finland (haplogroup A). A sample of these closely related Finn-exclusive haplotypes were significantly associated with alcohol dependence. Thus, the pattern of association we have observed is opposite that which would be created by population stratification.
Unfortunately, other precautions against erroneous associations, such as by using untransmitted alleles methods (49) are impossible for the Y-chromosome because it is clonally inherited. In fact, pedigrees would not be as informative as samples of unrelated males. Family-based data would hinder this type of association analysis because of the high levels of gene identity and allele sharing at possible contributing autosomal and X-linked loci. It would be difficult to extricate Y chromosome effects from those owing to autosomal and X-linked sources. However, a viable and potentially powerful utilization of pedigrees would be to ascertain different groups of distantly related males who share by descent, particular Y chromosomes, for the purpose of phenotypic comparisons across haplotypes. This is because autosomal genetic identity decays by a factor of two with each meiosis, but the Y chromosome component is undiluted. By using this approach one can obtain as many representatives of a haplotype as required.
Our results indicate that the risk of alcoholism in Finnish males is influenced by differences in Y chromosomes. Risk ratios suggest that males within clades 1-49, 1-21, and 1-57 were 1.5 times more likely to be alcoholic than males with other Y haplotypes, and the risk for alcohol dependence with ASPD was increased 2-fold within clade 1-57. However, the majority of the risk of alcoholism in these Finnish males is not Y chromosome-associated, and in fact, alcohol dependence is observed with Y haplotypes distributed throughout the cladogram. Twin studies suggest that alcoholism has a heritability of ≈50% (50, 51). Using this figure and data from our population sample, we estimate that Y chromosome variability may account for ≈7% of the total variance and 15% of the genetic variance of alcoholism in these Finnish males. These values are consistent with our present understanding of an etiology of alcoholism that encompasses contributions from many environmental factors and multiple genetic loci.
Acknowledgments
We thank Elisa Moore, Longina Akhtar, and Betsy Davis for their excellent technical support. We also thank Charles F. Sing for comments on an earlier draft and discussions with Jaakko Lappalainen, Raymond Peterson, and Margrit Urbanek.
ABBREVIATIONS
- ASPD
antisocial persoality disorder
- TPQ
Tridimensional Personality Questionnaire
References
- 1.Cloninger C R. Science. 1987;236:410–416. doi: 10.1126/science.2882604. [DOI] [PubMed] [Google Scholar]
- 2.Cloninger C R, Bohman M, Sigvardsson S. Arch Gen Psychiatry. 1981;38:861–868. doi: 10.1001/archpsyc.1981.01780330019001. [DOI] [PubMed] [Google Scholar]
- 3.Cloninger C R. Nat Med. 1995;1:623–625. doi: 10.1038/nm0795-623. [DOI] [PubMed] [Google Scholar]
- 4.Bohman M, Sigvardsson S, Cloninger C R. Arch Gen Psychiatry. 1981;38:965–969. doi: 10.1001/archpsyc.1981.01780340017001. [DOI] [PubMed] [Google Scholar]
- 5.Cadoret R J, Troughton E, O’Gorman T W, Heywood E. Arch Gen Psychiatry. 1986;43:1131–1136. doi: 10.1001/archpsyc.1986.01800120017004. [DOI] [PubMed] [Google Scholar]
- 6.Jobling M, Tyler-Smith C. Trends Genet. 1995;11:449–456. doi: 10.1016/s0168-9525(00)89144-1. [DOI] [PubMed] [Google Scholar]
- 7.Oakley R, Tyler-Smith C. Genomics. 1990;7:325–330. doi: 10.1016/0888-7543(90)90165-q. [DOI] [PubMed] [Google Scholar]
- 8.Mathias N, Bayes M, Tyler-Smith C. Hum Mol Genet. 1994;3:115–123. doi: 10.1093/hmg/3.1.115. [DOI] [PubMed] [Google Scholar]
- 9.Templeton A R, Boerwinkle E, Sing C F. Genetics. 1987;117:343–351. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos F, Pena S, Tyler-Smith C. Gene. 1995;165:191–198. doi: 10.1016/0378-1119(95)00501-v. [DOI] [PubMed] [Google Scholar]
- 11.Linnoila M, De Jong J, Virkkunen M. Arch Gen Psychiatry. 1989;46:613–616. doi: 10.1001/archpsyc.1989.01810070039006. [DOI] [PubMed] [Google Scholar]
- 12.Virkkunen M, Rawlings R, Tokola R, Poland R E, Guidotti A, Nemeroff C, Bissette G, Kalogeras K, Karonen S-L, Linnoila M. Arch Gen Psychiatry. 1994;51:20–27. doi: 10.1001/archpsyc.1994.03950010020003. [DOI] [PubMed] [Google Scholar]
- 13.Virkkunen M, Kallio E, Rawlings R, Tokola R, Poland R E, Guidotti A, Nemeroff C, Bissette G, Kalogeras K, Karonen S-L, Linnoila M. Arch Gen Psychiatry. 1994;51:28–33. doi: 10.1001/archpsyc.1994.03950010028004. [DOI] [PubMed] [Google Scholar]
- 14.Nevanlinna H R. Hereditas. 1972;71:195–236. doi: 10.1111/j.1601-5223.1972.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 15.Norio R. In: Biocultural Aspects of Disease. Rothschild H, editor. New York: Academic; 1981. pp. 359–415. [Google Scholar]
- 16.de la Chapelle A. J Med Genet. 1993;30:857–865. doi: 10.1136/jmg.30.10.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urbanek M, Goldman D, Long J C. Mol Biol Evol. 1996;13:943–953. doi: 10.1093/oxfordjournals.molbev.a025662. [DOI] [PubMed] [Google Scholar]
- 18.Sajantila A, Salem A, Savolainen P, Bauer K, Gierig C, Paabo S. Proc Natl Acad Sci USA. 1996;93:12035–12039. doi: 10.1073/pnas.93.21.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kittles R A, Bergen A W, Virkkunen M, Linnoila M, Urbanek M, Goldman D, Long J C. Am J Phys Anthropol. 1999;108:881–900. doi: 10.1002/(SICI)1096-8644(199904)108:4<381::AID-AJPA1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 20.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, Revised Third Edition. Washington, DC: Am. Psychiatric Assoc.; 1987. [Google Scholar]
- 21.Cloninger C R. Psychiatr Dev. 1986;3:167–226. [PubMed] [Google Scholar]
- 22.Cloninger C R. The Tridimensional Personality Questionaire, Version IV. St. Louis, MO: Washington Univ. School of Med.; 1987. [Google Scholar]
- 23.Tyler-Smith C, Brown W. J Mol Biol. 1987;195:457–470. doi: 10.1016/0022-2836(87)90175-6. [DOI] [PubMed] [Google Scholar]
- 24.Tyler-Smith C, Oakley R T, Larin Z, Fisher R, Crocker M, Affara N A, Ferguson-Smith M A, Muenke M, Zuffardi O, Jobling M A. Nat Genet. 1993;5:368–375. doi: 10.1038/ng1293-368. [DOI] [PubMed] [Google Scholar]
- 25.Kimura M, Ohta T. Proc Natl Acad Sci USA. 1978;75:2868–2872. doi: 10.1073/pnas.75.6.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valdes A, Slatkin M, Freimer N. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiRienzo A, Peterson A, Garza J, Valdes A, Slatkin M, Freimer N. Proc Natl Acad Sci USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shriver M, Jin L, Boerwinkle E, Deka R, Ferrell R, Chakraborty R. Mol Biol Evol. 1993;12:914–920. doi: 10.1093/oxfordjournals.molbev.a040268. [DOI] [PubMed] [Google Scholar]
- 29.Swofford D L. paup. Washington, DC: Smithsonian Institute; 1993. ,Version 3.1.1. [Google Scholar]
- 30.Nei M. Mol. Evol. Genet. New York: Columbia Univ. Press; 1993. [Google Scholar]
- 31.Kittles R A, Perola M, Peltonen L, Bergen A W, Aragon R A, Virkkunen M, Linnoila M, Goldman D, Long J C. Am J Hum Genet. 1998;62:1171–1179. doi: 10.1086/301831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castelloe J, Templeton A R. Mol Phylogenet Evol. 1994;3:102–113. doi: 10.1006/mpev.1994.1013. [DOI] [PubMed] [Google Scholar]
- 33.Crandall K, Templeton A R. Genetics. 1993;134:959–969. doi: 10.1093/genetics/134.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donnelly P. Variation in the Human Genome. New York: Wiley; 1996. [Google Scholar]
- 35.Prum B, Guilloud-Bataille M, Clerget-Darpoux F. Ann Hum Genet. 1990;54:315–320. doi: 10.1111/j.1469-1809.1990.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 36.Templeton A R. Genetics. 1995;140:403–409. doi: 10.1093/genetics/140.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin R, Cloninger C R, Guze S. J Clin Psychiatry. 1982;43:400–403. [PubMed] [Google Scholar]
- 38.Cadoret R J, O’Gorman T W, Troughton E, Heywood E. Arch Gen Psychiatry. 1985;42:161–167. doi: 10.1001/archpsyc.1985.01790250055007. [DOI] [PubMed] [Google Scholar]
- 39.Carey G. J Abnorm Psychol. 1992;101:18–25. doi: 10.1037//0021-843x.101.1.18. [DOI] [PubMed] [Google Scholar]
- 40.Hesselbrock M N. J Subst Abuse. 1991;3:205–519. doi: 10.1016/s0899-3289(05)80037-9. [DOI] [PubMed] [Google Scholar]
- 41.Cloninger C R, Christiansen K O, Reich T, Gottesman I I. Arch Gen Psychiatry. 1978;35:941–951. doi: 10.1001/archpsyc.1978.01770320035002. [DOI] [PubMed] [Google Scholar]
- 42.Hook E B. Science. 1973;179:139–150. doi: 10.1126/science.179.4069.139. [DOI] [PubMed] [Google Scholar]
- 43.Witkin H A, Mednick S A, Schulsinger F, Bakkestrom E, Christiansen K O, Goodenough D R, Hirschhorn K, Lundsteen C, Owen D R, Philip J, et al. Science. 1976;193:547–555. doi: 10.1126/science.959813. [DOI] [PubMed] [Google Scholar]
- 44.Urdal T, Brogger A. Lancet. 1974;i:626–627. doi: 10.1016/s0140-6736(74)92685-3. [DOI] [PubMed] [Google Scholar]
- 45.Nielson J, Henrikson F. Acta Psychiatr Scand. 1972;48:82–102. [Google Scholar]
- 46.Guillot P-V, Carlier M, Maxson S C, Roubertoux P L. Behav Genet. 1995;25:357–360. doi: 10.1007/BF02197285. [DOI] [PubMed] [Google Scholar]
- 47.Carlier M, Roubertoux P, Kottler M, Degrelle H. Behav Genet. 1990;20:137–154. doi: 10.1007/BF01070750. [DOI] [PubMed] [Google Scholar]
- 48.Maxson S C, Ginsburg B E, Trattner A. Behav Genet. 1979;9:219–226. doi: 10.1007/BF01071302. [DOI] [PubMed] [Google Scholar]
- 49.Lander E S, Schork N J. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 50.Hastbacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E. Nat Genet. 1992;2:204–211. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- 51.Spielman R S, Ewens W J. Am J Hum Genet. 1996;59:983–989. [PMC free article] [PubMed] [Google Scholar]
- 52.Pickens R W, Svikis D S, McGue M, Lykken D T, Heston L L, Clayton P J. Arch Gen Psychiatry. 1991;48:19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- 53.Kendler K S, Heath A C, Neale M C, Kessler R C, Eaves L J. J Am Med Assoc. 1992;14:1877–1882. [PubMed] [Google Scholar]