Abstract
It is increasingly clear that complex networks of relationships between genes and/or proteins govern neoplastic processes. Our understanding of these networks is expanded by the use of functional genomic and proteomic approaches in addition to computational modeling. Concurrently, whole-genome association scans and mutational screens of cancer genomes identify novel cancer genes. Together, these analyses have vastly increased our knowledge of cancer, in terms of both "part lists" and their functional associations. However, genetic interactions have hitherto only been studied in depth in model organisms and remain largely unknown for human systems. Here, we discuss the importance and potential benefits of identifying genetic interactions at the human genome level for creating a better understanding of cancer susceptibility and progression and developing novel effective anticancer therapies. We examine gene expression profiles in the presence and absence of co-amplification of the 8q24 and 20q13 chromosomal regions in breast tumors to illustrate the molecular consequences and complexity of genetic interactions and their role in tumorigenesis. Finally, we highlight current strategies for targeting tumor dependencies and outline potential matrix screening designs for uncovering molecular vulnerabilities in cancer cells.
Background
Most of the current knowledge of cancer susceptibility, progression and treatment has been generated by traditional approaches, in which small numbers of genes or proteins are characterized in depth to study the molecular mechanisms of neoplastic processes. With the advent of large-scale functional genomic and proteomic ("omic") methodologies, additional mechanistic insights into neoplasia have been uncovered. Whole-genome association studies for cancer risk variants and somatic mutation screening projects have completed their initial phases and will provide the "part lists" of cancer genes, both at the germline [1] and the somatic levels [2]. Transcript analyses have identified expression profiles that provide accurate prognoses for cancer patients [3]. Systematic mapping of protein-protein interactions is currently being carried out in what are referred to as 'interactome' mapping projects. This research will elucidate the wiring diagram of protein associations in cells [4,5]. These types of genes and/or protein (gene/protein) functional relationships can be modeled together to provide better understanding and predict molecular mechanisms of neoplasia [6-10].
Genetic interactions are identified when the action of one gene measured through a molecular, cellular or organism phenotype is modified by one or more other genes. They provide insight into biological processes that are complementary but which frequently do not overlap with other types of gene/protein associations [11]. To date, genetic interactions remain largely unknown on a large scale in human systems. Previous reviews and assays gave excellent descriptions of the role of genetic interactions in understanding phenotypic variability [12-14]. Here, we focus our discussion on the potential of studying human genetic interactions as a means of not only better understanding cancer susceptibility and progression but also, and most importantly, developing novel anticancer treatments.
Discussion
A step forward in cancer research
The use of 'model organisms' (i.e. species that meet the criteria of being representative of particular organisms or cellular behaviour) has played a prominent role in all aspects of biology. Since the first description of the complete sequence of an eukaryotic genome, the yeast Saccharomyces cerevisiae [15], the use of model organisms has grown with the development of high-throughput screenings for large-scale genomic and proteomic investigations [16]. Using these technologies and models, researchers have been able to acquire a greater understanding of how most, if not all, genes/proteins act coordinately to determine the properties of an organelle, cell or organism [8]. This "systems-level" research is also being used to decipher complex networks of molecular relationships in human systems, in healthy or normal conditions and their perturbation in disease or unconventional molecularly – or environmentally-modified conditions.
Of the possible gene-to-gene relationships in complex networks, genetic interactions were first mapped on a large scale in yeast [17]. These studies identified "synthetic lethal" interactions that occur when the combination of two gene deletions causes cell death whereas neither deletion is lethal by itself (i.e. non-essential genes). In addition to "synthesis", interactions can also be revealed by "epistasis", which is commonly used to define "genetic interactions" in statistical terms and describes the deviation from additivity for a quantitative phenotype by the effect of genetic variants or mutations at different loci [18-20]. Many models of interactions between different loci have been identified, including the combination of "aggravating" or agonistic and "alleviating" or buffering relationships [21,22].
Given the complexity of genetic interaction relationships, understanding the topology of genetic interaction networks is crucial for establishing genotype-phenotype correlations. This knowledge has clear implications in the study of certain aspects of common, "complex" or non-Mendelian diseases, such as the incidence of cancer in the general population, where the sum of minor gene effects contributes to the risk [23]. In addition to germline cancer risk, the mapping of genetic interactions in humans may also provide information on cancer progression and treatment, due to the large number of genes involved and the complexity of pathological phenotypes. Lessons learned from the study of genetic interactions in model organisms could be applied in defining hitherto unimaginable approaches for studying complex biological aspects of neoplastic processes.
Lessons from model organisms
The study of genetic interactions has been typically used in the annotation of signaling or metabolic pathways and protein complexes as a means of inferring the logical order of their components and examining pathway cross-talk [24]. A remarkable finding made in early systematic deletion analysis studies of yeast indicated that most eukaryotic genes are dispensable for viability. Only ~20% of Saccharomyces cerevisiae genes are essential for haploid cells grown in standard laboratory conditions [25,26].
Topological analysis of the yeast genetic interaction network has revealed the importance of gene interactions in phenotype modelling. More than 30 interactions on average were identified for non-essential genes and five-fold more for essential genes [17,27]. Accordingly, a recent estimate predicts ~200,000 synthetic lethal interactions in the global yeast genetic network [12]. When extrapolated to humans, these figures result in a vast number of genetic interactions with a specific phenotype potentially influenced by hundreds of different gene combinations. This estimate does not include combinations of more than two genes, which remain difficult to calculate at this stage. These predictions for phenotype determinants in humans clearly reveal the need for large-scale genetic interaction projects to provide more in-depth knowledge of complex diseases. Beyond the examination of single or small numbers of genes, systems-level analyses of model organisms have also uncovered the complex structure of genetic interaction networks. Epistasis extends beyond individual genes to clusters or modules of functionally related genes [28]. At this systems-level, perturbing one functional module can have aggravating or alleviating consequences on another.
Further advances could potentially be made through computational prediction of human genetic interactions using different types of experimentally generated molecular interactions. However, the modest overlap observed between experimentally identified genetic interactions and protein-protein interactions in yeast suggests that any predictions will render largely incomplete data [11,29-31]. Furthermore, given a pair of genes and their corresponding gene products, calculations in yeast give approximately four times more genetic interactions than protein-protein interactions. Finally, using orthologs to transfer genetic interactions across species may show even lower confidence than for protein-protein interologs [32], due to the observed complexity of the relationships between apparently molecularly unrelated signaling or metabolic pathways [28,33]. Taken together, computational predictions of human genetic interactions may provide lists of candidate gene pairs but, until further advances are made, they will require extensive experimental validation.
Systematic identification of genetic interactions in a multicellular organism has been described in a recent study by Lehner et al. [34]. The authors identified genetic interactions in Caenorhabditis elegans using the RNA interference (RNAi) method, which entails analyzing hypomorphic alleles rather than complete transcript depletion, which is the preferred methodology in the majority of yeast studies. Lehner et al. [34] highlighted the global relevance to phenotype modeling of genetic interactions between components of the same pathway, rather than interactions between components of different pathways. This study also anticipated the methodological problems and interpretational caveats that may appear when performing large-scale screening of genetic interactions in humans. Human studies require vast amounts of effort, the prevention of false positives in large-scale RNAi screens [35], and the incorporation of multiple designs to account for different cellular models and conditions [14]. Nevertheless, we believe that they can have an enormous impact on our knowledge of cancer.
Genetic interactions and cancer susceptibility
Projects such as the Cancer Genetic Markers of Susceptibility (CGEMS) initiative and the work carried out by Cancer Research UK within an international consortium are currently providing the preliminary partial lists of low-penetrance genetic variants for risk of different cancer types in the general population [36,37]. Initial analyses of this data have used a whole-genome approach to identify the main effects of individual variants. However, earlier studies based on candidate-gene approaches had already highlighted the existence of interactions between variants in different genes influencing cancer susceptibility [38]. It is now thought that the analysis of large series of individuals, together with the development of novel statistical approaches, will facilitate the assessment of millions of variant combinations which will, in turn, enable us to elucidate the impact of genetic interactions in cancer susceptibility [39]. Promising analyses based on simulations and empirical data have demonstrated that the likelihood of detecting significant associations increases when epistasis is taken into account [39-41].
The drawback of analyses taking into account epistasis is the trade-off between statistical power and false discovery rate. Current whole genome association studies consider over 500,000 single nucleotide polymorphisms, and P values need to be under 10-7 to account for multiple comparisons. Since low-penetrance alleles provide modest increases in risk, in the range of 20–30%, the sample sizes needed to detect main effects are in the range of 2,000 to 5,000 cases and a similar number of controls. When the aim is to detect gene interactions, the required number of tests increases to 2.5 × 1011 and the P value should be corrected to 2 × 10-13. The sample sizes needed for these significance levels are larger, but currently feasible – in the range of 6,000 to 15,000 cases for variants with high frequencies (20–40%) [41,42]. Novel statistical approaches have been proposed which reduce the dimensionality of the problem of searching for relevant gene interactions. The multifactor dimensionality reduction (MDR) method has been successfully applied to limited-scale scenarios [38,43,44]. Other promising proposals include the restricted partition method (RPM) [45] and combinatorial searching methods (CSM) [46]. Nevertheless, given the large number of expected false positive genetic interactions that will be proposed following analysis of whole-genome association studies, it will be necessary to carry out extensive validation in multiple populations and biological models based on the experimental identification of genetic interactions [47].
Genetic interactions and cancer progression
Cancer arises from the consecutive acquisition of genetic alterations that, in general, produce the loss of function or transcriptional down-regulation of tumor suppressor genes and the activation or transcriptional up-regulation of oncogenes [48]. Downstream of the genetic alterations in tumorigenesis we find expression changes in many genes, which lead to abnormal regulation of biological processes such as the cell cycle and apoptosis [9,49]. Consequently, it is thought that genetic and molecular alterations promote tumorigenesis in the context of highly connected and regulated gene/protein networks [7,8,50]. Thus, the progression from normal epithelium to benign dysplasia to metastases is relatively well characterized for some cancer types and highlights a specific molecular program [51]. The importance of studying genetic interactions between tumor suppressor genes, such as the retinoblastoma RB1 gene, and oncogenes, such as the RAS family, was established in earlier works that provided fundamental insights into the mechanisms of tumorigenesis and metastasis [52-54].
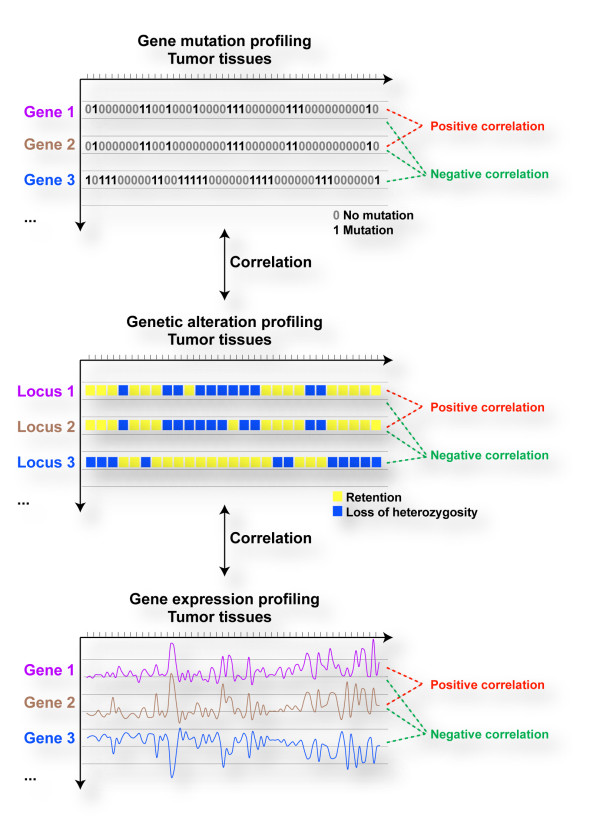
Knowledge of the sequence of genetic alterations that contribute to the neoplastic process could be used to target cancer cells according to the concept of synthetic lethality. Thus, it may be possible to reveal cancer genetic alterations useful for identifying genetic interactions by examining the profiles of somatic mutations, genomic alterations or gene expression changes in large series of tumors (Fig. 1). Under this hypothesis, the examination of highly correlated profiles may help to detect strong gene/protein dependencies in cancer cells. A greater understanding of these dependencies may illuminate mechanisms of carcinogenesis with insights into prognosis. Differences in biological meanings could also be extrapolated from positive versus negative correlations, implying molecular dependencies in different directions. Research into this hypothesis will be aided by the characterization at different genomic levels of large series of tumors. The Cancer Genome Project at the Sanger Institute has sequenced hundreds of human genes in tumors, tissues and cell lines [55,56]. Recent results obtained by other groups have demonstrated that oncogene activation in tumors is usually mutually exclusive. However, it has also been shown that some co-occurring mutations may reflect dependencies that are critical for neoplasia [57]. The identification of these types of neoplastic-specific functional dependencies could improve prognosis prediction and tumor classification, while targeting these dependencies may enhance therapeutic efficacy.
Figure 1.
Correlation of genetic and genomic alterations and transcript profiles across hundreds of tumors may reveal gene/protein functional dependencies that are useful for predicting genetic interactions of interest for cancer progression and anticancer therapies.
Empirical analysis of genetic interactions acting on tumor phenotypes
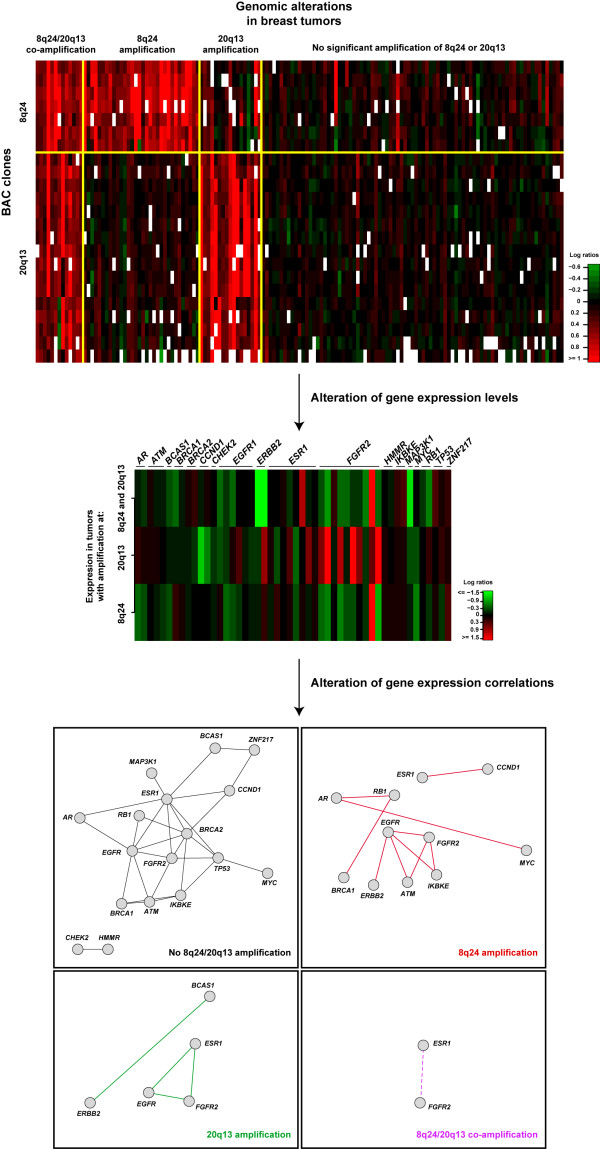
The integrative analysis of cancer molecular data sheds light on the mechanisms of neoplasia [6,10,58-60]. As discussed above, as large series of tumors and cell lines are characterized we will gradually be able to detect molecular dependencies and the ways in which they influence cancer progression. Co-amplification of specific genomic regions in breast tumors has been suggested and may represent cancer gene interactions between oncogenes that promote neoplasia. Specifically, co-amplification of the 8q24 and 20q13 regions may target the c-MYC and ZNF217 or BCAS1 proto-oncogenes, respectively [60].
To assess the 8q24/20q13 interaction, we evaluated molecular phenotypes such as transcript levels and co-expression between known breast cancer tumor suppressors and oncogenes (Fig. 2). In the absence of emergent genetic interactions, one may expect co-amplification of 8q24/20q13 to produce no relevant differences in gene expression between breast tumors. However, differences were observed beyond the simple additivity of expression levels for many genes when comparing tumors showing co-amplification with tumors showing single region amplification or no amplification; for example, ERBB2 showed marked down-regulation in tumors with co-amplification while the other tumors showed higher expression levels (Fig. 2, middle panel). Molecular phenotypic alterations also produced increases or decreases in correlation values of gene expression profiles, which are comparable to the concepts of alleviating or aggravating interactions; for example, a cluster of significant correlations between ATM,EGFR, ERBB2, FGFR2 and IKBKE appeared to be stronger in tumors with only 8q24 amplifications (Fig. 2, bottom panel). Portraits of expression correlations were noticeably different between tumors with different genomic alterations, to the extent that in co-amplification there were no significant correlations. These observations suggest differences in the molecular mechanisms that promote cancer progression determined by genetic interactions between cancer genes. This analysis is limited by the sample size series and could be extended by using multivariate analyses. Nevertheless, it illustrates the potential usefulness of genetic interactions in understanding tumor phenotypes.
Figure 2.
Empirical analysis of genetic interactions acting on tumorigenesis. Pre-processed and normalized data were taken from Chin et al. [60]. White squares represent missing data. Log expression ratios are relative to tumors that do not show amplification at 8q24 or 20q13. Average expression values are shown for each gene probe (not detailed) in each tumor set (8q24/20q13, 8q24 or 20q13 amplification). Transcript correlations were calculated using the Pearson correlation coefficient and adjusted P values with a false discovery rate of 5%; only significant correlations with Q values < 0.05 are shown, except for the 8q24/20q13 co-amplification set (dashed line; Q ~0.07).
Genetic interactions and cancer treatment
Robustness, which describes the ability of cells or organisms to maintain viability and functionality despite (multiple) molecular perturbations, is a fundamental principle in many biological systems [50,61-63]. Tumors are robust systems par excellence and it is thought that robustness is sustained by functional redundancy maintained by cell heterogeneity and feedback transcriptional control mechanisms [50]. However, cancer robustness might be offset by the extreme fragility of molecular networks. This idea is the basis for the hypothesis that uncommon perturbations of regular neoplastic processes would have dramatic effects on cancer cells [64].
Information on the acquisition of germline and somatic genetic alterations that contribute to neoplasia could then be systematically analyzed to find the Achilles' heel of cancer cells. The authors of a landmark study that paved the way for this type of strategy discovered that cells deficient in the breast cancer tumor suppressors BRCA1 and BRCA2 are completely dependent on the normal activity of the poly (ADP-ribose) polymerase family member 1 (PARP1) [65,66]. Inherited germline mutations in BRCA1 or BRCA2 genes confer high risk of breast and ovarian cancer, always accompanied by the somatic inactivation of the remaining wild-type allele [67-69]. Consequently, BRCA1/BRCA2-deficient tumor cells show genetic and genomic instability due in part to impaired DNA damage repair [70]. Against this background, depletion of PARP1 causes synthetic lethality due to severe defective homologous recombination necessary for regular DNA repair activity. Clinical trials using PARP1 inhibitors for breast cancer, and other malignancies characterized by DNA repair defects, now explore the potential of these interventions.
The BRCA/PARP1 paradigm could be applied in different terms to the observed tumor resistance to anticancer therapies. An example of this would be the use of trastuzumab for treating breast cancer with ERBB2 amplifications, which represent ~25% of all cases [71]. Although these tumors show great dependence on the HER-2 receptor for growth and, as a result, frequently respond to trastuzumab treatment, they almost always develop resistance. Genetic interaction screens performed in the presence of trastuzumab, or blocking HER-2, may reveal those genes whose inactivation leads to synthetic lethality or sickness pattern in breast cancer cells. Similar observations could be made using imatinib and KIT gene mutations in gastrointestinal stromal tumors [72] or the Philadelphia chromosome in chronic myeloid leukaemia [73]. A landmark study has recently demonstrated the usefulness of the concept of synthetic lethality in identifying genes that, when depleted, increase the sensitivity of cancer cells to paclitaxel [74]. Another recent study has revealed the co-activation of receptor tyrosine kinases as a mechanism that allows cancer cells to survive single compound-based treatments [75]. These observations further support the use of combined target therapies based on the depletion of selected genes to achieve superior therapeutic efficacy.
Genetic interactions screen designs
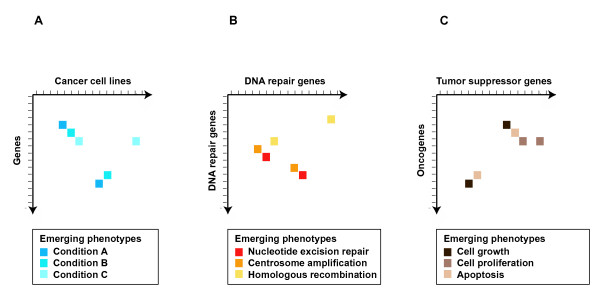
The fundamental aim of whole-genome systematic screens of human genetic interactions is to reveal multiple new anticancer therapy alternatives. Among various possible designs, these screens could be performed with a matrix format and standard cancer cell lines and by depleting genes using siRNAs or viral-packed shRNA constructs [76,77] (Fig. 3A). This approach would uncover a cancer genetic interaction network without a priori hypotheses concerning gene function, although current knowledge of genetic alterations in cancer cell lines should help to identify promising targets. This design could be extended following the suggestion made by Whitehurst et al. [74], in which sublethal concentrations of anticancer agents are used.
Figure 3.
Examples of matrix screens of genetic interactions of interest in studying cancer susceptibility, progression and treatment: (A) Interactions are identified in standard cancer cell lines by depleting one gene at a time and through quantitation of emerging phenotypes; (B) Interactions are identified between genes involved in the DNA repair processes, with quantitation of emerging phenotypes highlighting repair subcategories; and (C) Interactions are revealed between known tumor suppressor genes and oncogenes, with quantitation of emerging phenotypes highlighting different properties of the neoplastic process.
Different interpretations of genetic interactions and their applications will depend on whether the studied cell lines or models represent dominant or recessive inherited cancer. Genetic interactions in dominant inherited cancer syndromes should specifically target tumor cells that, for instance, lack both autosomal copies of a tumor suppressor gene. When targeting these tumors with synthetic lethal interventions, normal tissue and cells should remain unaffected unless dose effects appear due to the inactivation of a single allele in all cells. In recessive inherited cancer syndromes such as Fanconi anemia, in which all cells are inactivated for a specific gene, therapeutic targeting with synthetic lethal disruption could potentially act systemically, depending on cell-type specificities.
Designs for matrix screens of genetic interactions can also be based on the current knowledge of common molecular alterations in neoplasia (Fig. 3B). Many human genes encoding for proteins involved in DNA damage repair processes are mutated or epigenetically altered in cancer cells [78]. Currently, there are ~150 human genes annotated with DNA repair-related Gene Ontology terms [79]. Mapping interactions between these genes, including somatic or germline mutations such as the mismatch defects observed in colorectal tumors [80], is therefore expected to be beneficial in different cancer types. Again, the suggestion of Whitehurst et al. [74] could be applied to the identification of interactions between DNA repair genes by administering DNA-damaging agents at sublethal concentrations. In addition, many of these genes have orthologs in model organisms that could be used to study the conservation of synthetic lethality relationships and their role in altering the cellular response to DNA damage.
Another possible matrix strategy that could be used to identify new treatments consists of mapping interactions between known tumor suppressor genes and oncogenes (Fig. 3C). Unlike "passenger alterations", genetic alterations that contribute to neoplasia occur in a specific combination and order between tumor suppressor genes and oncogenes [48,49]. This observation, otherwise fragility of cancer genetic networks, could be useful in the identification of therapeutic targets. This matrix design combines the over-expression of tumor suppressors and the down-regulation or depletion of oncogenes. The experimental designs presented here are intended to stimulate interest in the utility and importance of this type of gene relationship in different areas of cancer research. We are confident that future research can and will lead to more sophisticated experimental designs for mapping genetic interactions in human models, which will greatly enhance our understanding of the molecular mechanisms of cancer.
Conclusion
Interactions between human genes are largely unknown. From studies in model organisms, it is clear that genetic interactions at the human genome-scale need to be identified in order to better understand common human diseases, with cancer being the paradigm. The detection of these interactions will be invaluable to our understanding of cancer risk, by suggesting hypotheses concerning the molecular mechanisms of susceptibility; of cancer progression, by revealing gene/protein functional dependencies; and of cancer treatment, by providing the knowledge required to develop new, efficient anticancer strategies.
Authors' contributions
XS, VM and MAP designed and performed the empirical data analysis. VM performed the simulation of association data. CAM and MAP drafted the manuscript. VM, LG, PH, AU helped to draft the manuscript. MAP conceived the article. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This study was supported by the Catalan Institute of Oncology, the Fundació la Caixa (BM05-254-00) and the Instituto de Salud Carlos III (FIS-PI060545). CAM is supported by a Beatriu de Pinós fellowship from the Agència de Gestió d'Ajuts Universitaris i de Recerca. MAP is a Ramón y Cajal Researcher with the Spanish Ministry of Education and Science.
Contributor Information
Christopher A Maxwell, Email: cmaxwell@iconcologia.net.
Víctor Moreno, Email: v.moreno@iconcologia.net.
Xavier Solé, Email: x.sole@iconcologia.net.
Laia Gómez, Email: lgomez@iconcologia.net.
Pilar Hernández, Email: phgutierrez@iconcologia.net.
Ander Urruticoechea, Email: anderu@iconcologia.net.
Miguel Angel Pujana, Email: mapujana@iconcologia.net.
References
- Hunter DJ, Thomas G, Hoover RN, Chanock SJ. Scanning the horizon: what is the future of genome-wide association studies in accelerating discoveries in cancer etiology and prevention? Cancer Causes Control. 2007;18:479–484. doi: 10.1007/s10552-007-0118-y. [DOI] [PubMed] [Google Scholar]
- Kim SY, Hahn WC. Cancer genomics: integrating form and function. Carcinogenesis. 2007;28:1387–1392. doi: 10.1093/carcin/bgm086. [DOI] [PubMed] [Google Scholar]
- Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van't Veer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat Genet. 2005;37 Suppl:S31–7. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- Khalil IG, Hill C. Systems biology for cancer. Curr Opin Oncol. 2005;17:44–48. doi: 10.1097/01.cco.0000150951.38222.16. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Hernandez P, Huerta-Cepas J, Montaner D, Al-Shahrour F, Valls J, Gomez L, Capella G, Dopazo J, Pujana MA. Evidence for systems-level molecular mechanisms of tumorigenesis. BMC Genomics. 2007;8:185. doi: 10.1186/1471-2164-8-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, Assmann V, Elshamy WM, Rual JF, Levine D, Rozek LS, Gelman RS, Gunsalus KC, Greenberg RA, Sobhian B, Bertin N, Venkatesan K, Ayivi-Guedehoussou N, Sole X, Hernandez P, Lazaro C, Nathanson KL, Weber BL, Cusick ME, Hill DE, Offit K, Livingston DM, Gruber SB, Parvin JD, Vidal M. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Hartman JL, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Lehner B. Modelling genotype-phenotype relationships and human disease with genetic interaction networks. J Exp Biol. 2007;210:1559–1566. doi: 10.1242/jeb.002311. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:546, 563–7. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Robinson MD, Mitsakakis N, Johnston M. The promise of functional genomics: completing the encyclopedia of a cell. Curr Opin Microbiol. 2004;7:546–554. doi: 10.1016/j.mib.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinb. 1918;52:399–433. [Google Scholar]
- Bateson W. Mendel's Principles of Heredity. Cambridge University Press, Cambridge. 1909.
- Waddington CH. Canalization of development and the inheritance of acquired characters. 1942. pp. 563–565.
- Drees BL, Thorsson V, Carter GW, Rives AW, Raymond MZ, Avila-Campillo I, Shannon P, Galitski T. Derivation of genetic interaction networks from quantitative phenotype data. Genome Biol. 2005;6:R38. doi: 10.1186/gb-2005-6-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Avery L, Wasserman S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Davierwala AP, Haynes J, Li Z, Brost RL, Robinson MD, Yu L, Mnaimneh S, Ding H, Zhu H, Chen Y, Cheng X, Brown GW, Boone C, Andrews BJ, Hughes TR. The synthetic genetic interaction spectrum of essential genes. Nat Genet. 2005;37:1147–1152. doi: 10.1038/ng1640. [DOI] [PubMed] [Google Scholar]
- Segre D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- Myers CL, Robson D, Wible A, Hibbs MA, Chiriac C, Theesfeld CL, Dolinski K, Troyanskaya OG. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6:R114. doi: 10.1186/gb-2005-6-13-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SL, Zhang LV, Tong AH, Li Z, Goldberg DS, King OD, Lesage G, Vidal M, Andrews B, Bussey H, Boone C, Roth FP. Combining biological networks to predict genetic interactions. Proc Natl Acad Sci U S A. 2004;101:15682–15687. doi: 10.1073/pnas.0406614101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- Mika S, Rost B. Protein-protein interactions more conserved within species than across species. PLoS Comput Biol. 2006;2:e79. doi: 10.1371/journal.pcbi.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, Uetz P, Sittler T, Karp RM, Ideker T. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci U S A. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr., Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PDP, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Brinton L, Chanock S, Lissowska J, Peplonska B, Nevanlinna H, Fagerholm R, Eerola H, Kang D, Yoo KY, Noh DY, Ahn SH, Hunter DJ, Hankinson SE, Cox DG, Hall P, Wedren S, Liu J, Low YL, Bogdanova N, Schurmann P, Dork T, Tollenaar RAEM, Jacobi CE, Devilee P, Klijn JGM, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MWR, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den Ouweland A, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X, Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BAJ. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu JS. Bayesian inference of epistatic interactions in case-control studies. Nat Genet. 2007;39:1167–1173. doi: 10.1038/ng2110. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nat Rev Genet. 2004;5:89–100. doi: 10.1038/nrg1270. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhao H. Am J Epidemiol. 2003/10/31. Vol. 158. 2003. Sample size needed to detect gene-gene interactions using association designs; pp. 899–914. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Am J Epidemiol. 2002/02/28. Vol. 155. 2002. Sample size requirements for association studies of gene-gene interaction; pp. 478–484. [DOI] [PubMed] [Google Scholar]
- Hahn LW, Ritchie MD, Moore JH. Bioinformatics. 2003/02/14. Vol. 19. 2003. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions; pp. 376–382. [DOI] [PubMed] [Google Scholar]
- Manuguerra M, Matullo G, Veglia F, Autrup H, Dunning AM, Garte S, Gormally E, Malaveille C, Guarrera S, Polidoro S, Saletta F, Peluso M, Airoldi L, Overvad K, Raaschou-Nielsen O, Clavel-Chapelon F, Linseisen J, Boeing H, Trichopoulos D, Kalandidi A, Palli D, Krogh V, Tumino R, Panico S, Bueno-De-Mesquita HB, Peeters PH, Lund E, Pera G, Martinez C, Amiano P, Barricarte A, Tormo MJ, Quiros JR, Berglund G, Janzon L, Jarvholm B, Day NE, Allen NE, Saracci R, Kaaks R, Ferrari P, Riboli E, Vineis P. Carcinogenesis. 2006/09/08. Vol. 28. 2007. Multi-factor dimensionality reduction applied to a large prospective investigation on gene-gene and gene-environment interactions; pp. 414–422. [DOI] [PubMed] [Google Scholar]
- Culverhouse R, Klein T, Shannon W. Genet Epidemiol. 2004/08/12. Vol. 27. 2004. Detecting epistatic interactions contributing to quantitative traits; pp. 141–152. [DOI] [PubMed] [Google Scholar]
- Sha Q, Zhu X, Zuo Y, Cooper R, Zhang S. Ann Hum Genet. 2006/08/16. Vol. 70. 2006. A combinatorial searching method for detecting a set of interacting loci associated with complex traits; pp. 677–692. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P. Drinking from the fire hose--statistical issues in genomewide association studies. N Engl J Med. 2007;357:436–439. doi: 10.1056/NEJMp078120. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/S1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- Kitano H. Cancer as a robust system: implications for anticancer therapy. Nat Rev Cancer. 2004;4:227–235. doi: 10.1038/nrc1300. [DOI] [PubMed] [Google Scholar]
- Sancho E, Batlle E, Clevers H. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Behrendt N, Chen Z, Noda T, Hino O, Cordon-Cardo C, Pandolfi PP. Genetic analysis of Pten and Tsc2 functional interactions in the mouse reveals asymmetrical haploinsufficiency in tumor suppression. Genes Dev. 2005;19:1779–1786. doi: 10.1101/gad.1314405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC, Deng CX. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- Takahashi C, Ewen ME. Genetic interaction between Rb and N-ras: differentiation control and metastasis. Cancer Res. 2006;66:9345–9348. doi: 10.1158/0008-5472.CAN-06-1250. [DOI] [PubMed] [Google Scholar]
- Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, Flanagan A, Teague J, Wooster R, Futreal PA, Stratton MR. Cosmic 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, Lee JC, Nicoletti R, Hatton C, Goyette M, Girard L, Majmudar K, Ziaugra L, Wong KK, Gabriel S, Beroukhim R, Peyton M, Barretina J, Dutt A, Emery C, Greulich H, Shah K, Sasaki H, Gazdar A, Minna J, Armstrong SA, Mellinghoff IK, Hodi FS, Dranoff G, Mischel PS, Cloughesy TF, Nelson SF, Liau LM, Mertz K, Rubin MA, Moch H, Loda M, Catalona W, Fletcher J, Signoretti S, Kaye F, Anderson KC, Demetri GD, Dummer R, Wagner S, Herlyn M, Sellers WR, Meyerson M, Garraway LA. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, Greulich H, Stewart CJ, Mulvey LA, Shen RR, Ambrogio L, Hirozane-Kishikawa T, Hill DE, Vidal M, Meyerson M, Grenier JK, Hinkle G, Root DE, Roberts TM, Lander ES, Polyak K, Hahn WC. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Kitano H. Cancer robustness: tumour tactics. Nature. 2003;426:125. doi: 10.1038/426125a. [DOI] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Csete ME, Doyle JC. Reverse engineering of biological complexity. Science. 2002;295:1664–1669. doi: 10.1126/science.1069981. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Deng CX, Wang RH. Roles of BRCA1 in DNA damage repair: a link between development and cancer. Hum Mol Genet. 2003;12 Spec No 1:R113–23. doi: 10.1093/hmg/ddg082. [DOI] [PubMed] [Google Scholar]
- Hudis CA. Trastuzumab--mechanism of action and use in clinical practice. N Engl J Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Heinrich MC, Corless CL. Gastrointestinal stromal tumour. Lancet. 2007;369:1731–1741. doi: 10.1016/S0140-6736(07)60780-6. [DOI] [PubMed] [Google Scholar]
- Deininger MW. Optimizing therapy of chronic myeloid leukemia. Exp Hematol. 2007;35:144–154. doi: 10.1016/j.exphem.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, Xie XJ, White MA. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, Chin L, Depinho RA. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007 doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Sachidanandam R, McCombie WR, Elledge SJ, Hannon GJ. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner SM, Kloor M, von Knebel Doeberitz M, Gebert JF. Microsatellite instability in the development of DNA mismatch repair deficient tumors. Cancer Biomark. 2006;2:69–86. doi: 10.3233/cbm-2006-21-208. [DOI] [PubMed] [Google Scholar]