Abstract
Distant metastases of human breast cancers have been suggested to be more different from each other than from their respective primary tumors, based on expression profiling. The mechanism behind this lack of similarity between individual metastases is not known. We used cDNA microarrays to determine the expression profiles of pulmonary metastases and primary mammary tumors in two distinct transgenic models expressing either the Neu or the Wnt-1 oncogene from the mouse mammary tumor virus long terminal repeat (MMTV LTR). We found that pulmonary metastases are similar to each other and to their primary tumors within the same line. However, metastases arising in one transgenic mouse line are very different from either metastases or primary tumors arising in the other line. In addition, we found that, like their primary tumors, lung metastases in Wnt-1 transgenic mice harbor both epithelial and myoepithelial tumor cells and cells that express the putative progenitor cell marker keratin 6. Our data suggest that both gene expression profiles and cellular heterogeneity are preserved after breast cancer has spread to distant sites, and that metastases are similar to each other when their primary tumors were induced by the same oncogene and from the same subset of mammary cells.
Introduction
Breast cancer evolves from atypical ductal hyperplasia, to carcinoma in situ, then invasive carcinoma, and finally distant metastasis, which is usually the cause of death [1,2]. Expression array studies of a small number of human breast cancer metastases found that they were more similar to their corresponding primary tumors than to each other [3–6]. These results have been interpreted to suggest that expression profiles do not change significantly in the progression from primary lesions to distant metastases [7].
However, it is not clear why metastases are less similar to each other, especially because the local environment in the metastasis sites might induce a common set of genes in all of the metastases in question [4,8]. Because only a small number of paired samples have been analyzed, it is possible that the dissimilarity may have been caused by heterogeneity in causal mutations, differentiation stages of the cancer-originating cell, or genetic backgrounds, all of which have been reported to have an influence on gene expression patterns [7,9,10] (Bu et al., unpublished data).
Would the expression profiles of metastases still be more similar to those of their primary tumors than to each other, if the comparison could be made between tumors that were induced by the same oncogenic mutation from the same type of cell? Mouse models have the advantage of purer genetic background, known initiating genetic alterations, and better controlled cell of origin through the use of cell type-specific transgenic promoters; they have been widely used to provide insights in human cancer evolution [11]. In this report, we investigated these issues by comparing expression profiles and cellular composition of distant metastases and primary tumors in two distinct mouse models that express the Wnt or the Neu oncogene.
Materials and Methods
Tissue Samples
The MMTV-Neu transgenic mouse line (on the FVB background) carries a rat cDNA encoding the wild-type Neu (ErbB2/HER2) protein [12]. MMTV-Wnt-1 transgenic mice [13] were on a mixture of FVB (>75%), SJL, and C57BL/6 strains. Metastases used in the study were gross metastases dissected from lung tissues using scalpels. They included four from MMTV-Wnt-1 transgenic mice that were bearing a primary tumor or had had one primary tumor removed by survival surgery, six experimental metastases induced by injecting primary mammary tumor cells from MMTV-Wnt-1 transgenic mice into the tail vein of the same mice, three spontaneous metastases from tumor-bearing MMTV-Neu transgenic mice, and two experimental metastases induced by injecting primary MMTV-Neu-induced mammary tumor cells into the tail vein of nontransgenic mice. The 35 primary tumors—23 and 12 from MMTV-Wnt-1 and MMTV-Neu transgenic mice, respectively—have been described [14]. All tissue samples were snap-frozen in liquid nitrogen and kept at -80°C before use. After being ground into fine powder in liquid nitrogen, these tissues were subject to RNA extraction using Trizol (Invitrogen, Carlsbad, CA). The reference RNA was a mixture of ovarian RNA (cat no. 7824; Ambion, Austin, TX) and RNA extracted from tissues of liver, spleen, kidney, thymus, pancreas, lung, and normal lactating mammary gland of FVB mice of 6 months of age. All reference RNA used in this study and in our previous study [14] were from a single preparation, snapfrozen as aliquots (for single use), and stored at -80°C.
cDNA Microarray Hybridization, Data Extraction, and Statistical Analysis
All arrays used in this report were generated using the same batch of array prints (mouse 15k cDNA arrays, printed at the National Cancer Institute Microarray Facility) described previously [14]. Labeling, hybridization, scanning, and microarray analyses were performed for the 15 metastasis samples and the 35 primary tumors at the same time and as previously described [14]. The data for these primary tumors have been described in our previous report [14] and deposited into the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under the series number GSE2860. The 15 new arrays of pulmonary metastases have been deposited into the GEO database under the same series number.
Microarray images were analyzed and normalized to generate logarithmic ratios of test-to-reference. Corresponding quality measurements in each experiment were calculated using the ArraySuite software based on the Scanalytics IPlab platform (Scanalytics, Fairfax, VA) and as described [15]. Arrays of poor quality were excluded from further analysis. Genes with a low average quality (<0.5) across all arrays were excluded from subsequent data analyses. Unsupervised average linkage hierarchical clustering analysis and multidimensional scaling (MDS) analysis were performed as described [14] to assess the relationship among different sample groups. A two-sample permutation t test, as described previously [14], was used to identify genes that were significantly different between two groups of samples (P < .001).
Immunohistochemical Staining
Tissues were freshly collected, fixed in 10% neutral formalin overnight, and processed as previously described [16] to obtain paraffin sections of 4 µm thickness. Immunohistochemical staining was performed using Vector ABC kits and the Nova-Red substrate (Vector Laboratories, Burlingame, CA) following the manufacturer's recommendations and as described [14]. Primary antibodies used include rabbit IgGs against keratin 6 (Covance, Philadelphia, PA), mouse monoclonal antibodies against α-smooth muscle actin (α-SMA; Dako, Philadelphia, PA), and rat anti-keratin 8 [17,18].
Results
Comparing Metastases and Primary Tumors in Either the MMTV-Neu or the MMTV-Wnt-1 Model
Neu (HER2/ErbB2) encodes a member of the epidermal growth factor receptor family of receptor tyrosine kinases [19], and is amplified in approximately 25% of human breast cancers [20]. Transgenic mice (MMTV-Neu) expressing Neu from the promoter in the mouse mammary tumor virus long terminal repeat (MMTV LTR), which is active primarily in more differentiated mammary epithelial cells as well as in some undefined mammary progenitor cells [21–23], develop mammary tumors at a median age of 7 months [12]. Microscopic lesions are detected in the lung in 75% of cases, but gross metastases are rare [12]. Members of the Wnt gene family encode an extracellular protein that binds to membrane coreceptors Frizzled and low-density lipoprotein receptor-related protein (LRP) 5/6, leading to stabilization of β-catenin and activation of the mammalian target of rapamycin (mTOR) pathway [24,25]. Wnt signaling is implicated in tumorigenesis of the breast and many other tissue types [25]. MMTV-Wnt-1 transgenic mice develop mammary tumors with a median latency of six months [13]. Microscopic lung metastases are found in approximately 35% of tumor-bearing mice on euthanasia (Podsypanina, unpublished data). Gross metastases are infrequent as in MMTV-Neu transgenic mice, but can be enhanced after removal of the primary lesion through survival surgery [13] (Podsypanina, unpublished data).
One of the major obstacles in profiling metastatic tumors is obtaining sufficient amounts of tumor samples for RNA extraction. In this study, we examined a large cohort of mice and/or employed survival surgery to remove the initial primary tumor to enhance the growth of metastases (see Material and Methods section for detail). We were able to collect three spontaneous pulmonary metastases from MMTV-Neu transgenic mice, and four pulmonary metastases from MMTV-Wnt-1 transgenic mice through survival surgery to remove primary tumors. In addition, we generated two and six experimental metastases from tail vein injection of cell suspensions made from primary tumors from MMTV-Neu and MMTV-Wnt-1 transgenic mice, respectively.
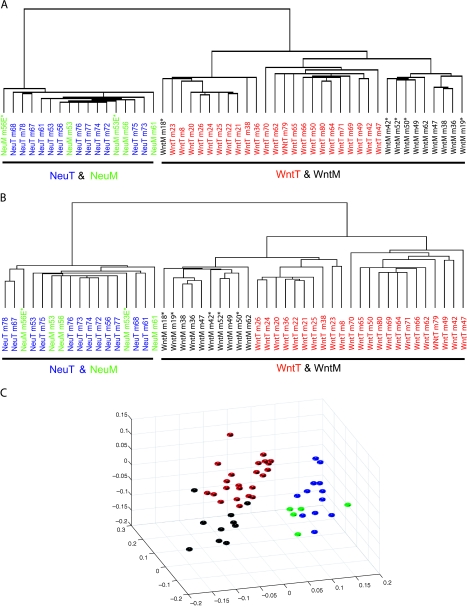
Using mouse 15k cDNA arrays, we determined the expression profiles of these metastases (5 from MMTV-Neu transgenic mice and 10 from MMTV-Wnt-1 transgenic mice) and their primary tumors in both models (12 from MMTV-Neu transgenic mice and 23 from MMTV-Wnt-1 transgenic mice). Using unsupervised average linkage hierarchical clustering analysis and MDS [14] to analyze these arrays, we identified two main clusters: one comprises metastases and primary tumors from MMTV-Neu transgenic mice, and the other contains metastases and primary tumors from MMTV-Wnt-1 transgenic mice (Figure 1A and data not shown).
Figure 1.
Comparison of gene expression between primary tumors and metastases in MMTV-Wnt-1 and MMTV-Neu transgenic models. Average linkage hierarchical clustering analysis was performed on arrays of lung metastases (WntM or NeuM) and primary tumors (WntT or NeuT) from MMTV-Wnt-1 and MMTV-Neu transgenic mice using 13,162 genes (all the genes on the array except those with low quality across all samples) (A), or the 5704 genes that are not differentially expressed between primary tumors of MMTV-Wnt-1 and MMTV-Neu transgenic mice (see Material and Methods section for more detail) (B). Paired primary tumors and lung metastases have the same number. Experimental metastases are noted by an asterisk. These arrays were also analyzed by MDS using the 5704 genes (C). The color of each tumor spot is coded as in A and B.
This segregation based on the model may not be surprising, since besides the initiating oncogene, these two models are also different in histopathologic features, cellular composition, estrogen receptor (ER) status, and possibly the cell of origin—whereas tumors in the MMTV-Neu model appear to arise from more differentiated mammary epithelial cells, lack histologic differentiation or ER expression, and contain predominantly epithelial tumor cells; tumors arising in MMTV-Wnt-1 transgenic mice appear to arise from progenitor cells, display a more differentiated histopathology, are usually ER+, and harbor mixed cell lineages including epithelial cells, myoepithelial cells, and cells that express markers (Sca-1, keratin 6, and CD24/CD29) putatively associated with progenitor cells [18,26–31]. In addition, cyclin D1 is required for carcinogenesis in the MMTV-Neu model, but is dispensable in the MMTV-Wnt-1 model [32]. Furthermore, there is a slight difference in the genetic background between these two models.
To determine whether eliminating the genes associated with these other differences might change the dendrogram structure and reveal similarities between metastases in the two models, we filtered the 1296 genes that we have reported to be differentially expressed between primary tumors in these two models (P < .001) [14], and an additional 1162 genes that had a P value between .001 and .05 to ensure a more stringent analysis. Of the remaining 12,361 genes, 6657 genes were excluded for either low quality or near identical expression across all the samples. In the end, 5704 genes were included for hierarchical clustering and MDS analyses. Again, the two models separated into two clusters, with the metastases grouping with primary tumors in each model (Figure 1, B and C).
Within the MMTV-Neu transgenic model, the expression profiles of the metastases were highly similar to each other, and also to those of the primary tumors (Figure 1). In accord, 96% of the genes (1207 of 1263) that we have previously identified as differentially expressed between primary tumors from MMTV-Neu transgenic mice and virgin mammary glands from nontransgenic mice [14] were not significantly differentially expressed between metastases and primary tumors in this model (P > .001). For three of these five metastases, matching primary tumors from the same mouse were included in this analysis. However, these metastases did not appear to have a closer similarity to primary tumors in the same mouse than to either metastases or primary tumors in other mice (Figure 1, A and B). These observations suggest that, in the MMTV-Neu transgenic model of breast cancer, expression profiles change very little either between metastases or between metastases and primary tumors.
Within the MMTV-Wnt-1 transgenic model, two clusters were detected (Figure 1, A and B). The metastases (with the exception of one in Figure 1A) formed a distinct subcluster within one of these two clusters, suggesting an insignificant change in expression patterns in the transition from primary site to the lung. In accord, 83% of the genes (322 of 388) that we previously reported to be differentially expressed between primary tumors and hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice [14] were not differentially expressed between metastases and primary tumors in this model. For 7 of 10 metastases, matching primary tumors from the same mouse were included in this analysis. However, metastases did not have closer relations with their matching primary tumors than with other metastases or other primary tumors (Figure 1, A and B). This observation and the results from the MMTV-Neu model suggest that expression profiles change very little either between metastases or between metastases and primary tumors when the same oncogene is targeted to the same subset of mammary cells. However, analyses of a larger number of metastases may reveal more subtle differences between metastases and/or between metastases and primary tumors within each model, especially because our collection of metastases included eight experimental metastases. Of note, experimental metastases in both models did not segregate from the corresponding spontaneous metastases, suggesting that these experimental metastases probably did not affect the clustering hierarchy of these arrays.
Pulmonary Metastases Preserve the Cellular Heterogeneity of Primary Tumors in MMTV-Wnt-1 Transgenic Mice
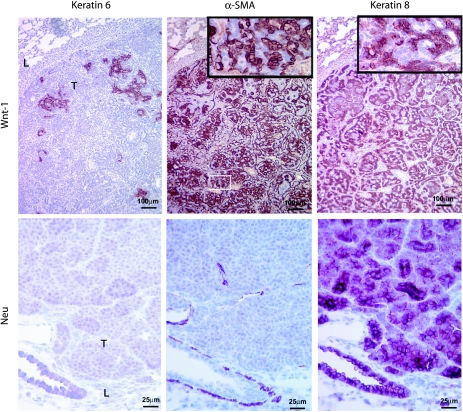
It has not been demonstrated whether breast cancer metastases preserve the cellular composition of their primary tumors, though histologic features may be shared [6]. Using the permutation t test, we found that pulmonary metastases from MMTV-Wnt-1 transgenic mice expressed, at similar levels as their primary tumors, the epithelial markers E-cadherin and β-catenin, the myoepithelial cell markers calponin1 and 2, and a few markers that have been reported [27,33,34] to be associated with stem and/or progenitor cells, such as keratin 6, Sca1, CD44, CD24, integrin β1, and integrin α6 (data not shown). Using immunohistochemical staining of paraffin sections of metastases from MMTV-Wnt-1 transgenic mice, we found positive staining for the epithelial cell marker keratin 8, the myoepithelial cell marker α-SMA, and the potential progenitor cell marker keratin 6 (Figure 2), as in primary tumors in the same transgenic mice [18,26,28,30]. In addition, the ratio (0.98 ± 0.29) of epithelial cells to myoepithelial cells in metastases was similar to that (1.20 ± 0.39) in primary tumors (P = .47). In contrast, only keratin 8 was detected in metastases from MMTV-Neu transgenic mice (Figure 2), as has been reported in their primary tumors [18,28,30]. These data suggest that the cellular composition of primary tumors is preserved in distant metastases.
Figure 2.
Expression of keratin 6, α-SMA, and keratin 8 in lung metastases from MMTV-Wnt-1 and MMTV-Neu transgenic mice. Consecutive paraffin sections were stained by immunohistochemistry for the protein indicated at the top. The transgenic oncogenes are indicated at the left. The inserts are four times the views of the boxed areas. Lung (L) and tumor mass (T) are indicated. Scale bars are as shown.
Discussion
The expression profiling in two distinct mouse models of breast cancer suggests that, at the expression level, individual metastases from the same transgenic model are very similar to each other and to their primary tumors (Figure 1). The high degree of similarity between metastases and primary tumors in the same model suggests that changes in gene expression are probably small as primary tumors in these models disseminate to distant locations, consistent with the observation in humans that metastases closely recapitulate their corresponding primary tumors at the expression level [3–6]. Further supporting the close similarity between metastases and primary tumors, tumors arising from orthotopic transplantation of metastases from MMTV-Wnt-1 transgenic mice did not have improved potential to metastasize (Podsypanina, unpublished data), as has been reported for comparisons between cells isolated from metastatic sites versus from primary sites [35]. However, our observations, as well as the observations in human metastases, do not exclude the possibility that, in the progression from primary tumors to metastases, additional mutations occur, which promote metastasis but do not significantly perturb global expression patterns [7,36,37].
The high similarity between individual metastases in each model suggest that expression patterns vary only slightly among individual metastases when their primary tumors are induced using the same oncogene in the same subset of mammary cells. This is interesting because differences in expression profiles [14,28,38], cell composition [28], and susceptibility to mTOR inhibitors (Li, unpublished data) are notable among primary tumors within the MMTV-Wnt-1 model. These data also imply that secondary genetic alterations play only minor roles in the expression profiles when the Neu or Wnt oncogenic pathway is the initiating event. Approximately 70% of the tumors exhibit point mutations, small deletions, or insertions in the transgenic Neu protooncogene in MMTV-Neu transgenic mice [39]; 50% of tumors suffer an activating mutation in the H-Ras locus [40,41] in MMTV-Wnt-1 transgenic mice. Because tumors in both models are presumably clonal [16,39,40], a secondary mutation in either Ras or the Neu transgene itself likely exists at the same frequency in its respective metastasis group, and has no significant influence on the expression profile of these metastases, as in the primary lesions [14,38].
Although highly similar to each other within the same model, expression patterns are quite different between MMTV-Neu-induced metastases and those induced by MMTV-Wnt-1 (Figure 1A). This difference may be attributed to several sources including the initiating oncogene and the cell of origin, both of which have an influence on gene expression profiles of primary mammary tumors [9,28] (Bu and Li, unpublished data). The addition of metastases from models that share an initiating oncogenic pathway and/or a cell of origin may reveal more similarities among metastases from different models. However, even after genes that accounted for differences in primary tumors between the MMTV-Neu and MMTV-Wnt-1 models were excluded, large expression differences persisted between these two sets of metastases (Figure 1, B and C).
The high expression pattern similarity between individual metastases in each model as well as the obvious expression dissimilarity in metastases between the two models suggest that variations in the cancerinitiating genetic alterations and possibly in the cell of cancer origin between patients may be partly responsible for the lack of close expression similarity of distant metastases seen in the smaller number of breast cancer metastases that have previously been analyzed [3–6]. Analysis of more human samples with similar sets of genetic alternations and/or with similar histopathology features will help provide a more conclusive answer to this possibility.
Multiple genetic mutations are usually required to cause human malignancies [42,43]. The reported overall lack of similarity among human breast metastases [3–6] might also be attributed to variations in secondary oncogenic mutations, although secondary oncogenic mutations do not seem to greatly influence the expression profiles of primary tumors in several models [14,28,38,44]. Furthermore, variation of the genetic background in the human population may also be a contributing factor to the differences in metastases between individual patients. In addition, genomic instability is much higher in human breast cancers than in these two and most other mouse models [45,46]. Such genomic heterogeneity may also add to the more unique expression profiles of human metastases. Analysis of metastatic tumors from genomically unstable models such as p53-deficient mice [46] may help measure the contribution of genomic instability to gene expression profile differences.
Of note, most humans breast cancers arise from a few mutated cells in a field of normal cells. However, the two transgenic models used in this study and most other models of breast cancer were generated by expressing oncogenes in essentially all mammary epithelial cells, and this widespread transgenic expression frequently impairs the development of the mammary glands [46]. This abnormal environment in transgenic models may have a different effect on the expression profiles of both primary tumors and metastases than the usual environment of normal cells. Expression profiling of breast cancer models that initiate oncogenic mutations after the gland has developed and/or, in only a small number of cells [46–48], may elucidate more realistically mechanisms underlying breast cancer development and metastasis.
Interestingly, metastases in the MMTV-Wnt-1 transgenic model were found to have high cellular heterogeneity (Figure 2), as do their primary tumors [14,18,38]. Because these metastases are usually focal, it is likely that they had a clonal origin, as has been suggested for metastases of several tissue types [49–52]. If so, these metastases may arise from spread and colonization of cancer stem cells [35,53], which appear to exist in this model [18,26,27]. Nevertheless, this heterogeneity may also result from dissemination and colonization of heterogeneous tumor cells that traveled as a group.
In conclusion, expression profiling of metastases and primary tumors in two distinct mouse models of breast cancer suggests that pulmonary metastases that share the same cancer-initiating oncogene and cellular origin are similar in expression profiles to each other and to their primary tumors, and that breast cancer metastases preserve the cellular heterogeneity found in primary tumors.
Acknowledgments
We thank Harold Varmus for advice and resources for generating the array data reported in this study, Jeffrey Green and David Petersen for the array slides, Susan Hilsenbeck for advice on statistical analyses, and Gary Chamness for assistance in the preparation of the manuscript.
Abbreviations
- α-SMA
alpha-smooth muscle actin
- ER
estrogen receptor
- LRP
low-density lipoprotein receptor-related protein
- MDS
multidimensional scaling
- MMTV
mouse mammary tumor virus
- mTOR
mammalian target of rapamycin
Footnotes
This work was supported in part by funds from the National Cancer Institute (R01CA113869-01 to Y.L.).
References
- 1.Allred DC, Brown P, Medina D. The origins of estrogen receptor alpha-positive and estrogen receptor-negative human breast cancer. Breast Cancer Res. 2004;6:240–245. doi: 10.1186/bcr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van't Veer LJ. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci USA. 2003;100:15901–15905. doi: 10.1073/pnas.2634067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, Glas AM, Perou CM, van't Veer LJ. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65:9155–9158. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 6.Weigelt B, Wessels LF, Bosma AJ, Glas AM, Nuyten DS, He YD, Dai H, Peterse JL, van't Veer LJ. No common denominator for breast cancer lymph node metastasis. Br J Cancer. 2005;93:924–932. doi: 10.1038/sj.bjc.6602794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weigelt B, Peterse JL, van't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 8.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 9.Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, Dickson R, Furth P, Hunter K, Kucherlapati R, et al. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci USA. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford NP, Hunter KW. New perspectives on hereditary influences in metastatic progression. Trends Genet. 2006;22:555–561. doi: 10.1016/j.tig.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–144. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- 12.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 14.Huang S, Li Y, Chen Y, Podsypanina K, Chamorro M, Olshen AB, Desai KV, Tann A, Petersen D, Green JE, et al. Changes in gene expression during the development of mammary tumors in MMTV-Wnt-1 transgenic mice. Genome Biol. 2005;6:R84. doi: 10.1186/gb-2005-6-10-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Kamat V, Dougherty ER, Bittner ML, Meltzer PS, Trent JM. Ratio statistics of gene expression levels and applications to microarray data analysis. Bioinformatics. 2002;18:1207–1215. doi: 10.1093/bioinformatics/18.9.1207. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Podsypanina K, Liu X, Crane A, Tan LK, Parsons R, Varmus HE. Deficiency of Pten accelerates mammary oncogenesis in MMTV-Wnt-1 transgenic mice. BMC Mol Biol. 2001;2:2. doi: 10.1186/1471-2199-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemler R, Brulet P, Schnebelen MT, Gaillard J, Jacob F. Reactivity of monoclonal antibodies against intermediate filament proteins during embryonic development. J Embryol Exp Morphol. 1981;64:45–60. [PubMed] [Google Scholar]
- 18.Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 20.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 21.Munoz B, Bolander FF., Jr Prolactin regulation of mouse mammary tumor virus (MMTV) expression in normal mouse mammary epithelium. Mol Cell Endocrinol. 1989;62:23–29. doi: 10.1016/0303-7207(89)90109-3. [DOI] [PubMed] [Google Scholar]
- 22.Gunther EJ, Belka GK, Wertheim GB, Wang J, Hartman JL, Boxer RB, Chodosh LA. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 2002;16:283–292. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 23.Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- 24.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Lindvall C, Bu W, Williams BO, Li Y. Wnt signaling, stem cells, and the cellular origin of breast cancer. Stem Cell Rev. 2007;3:157–168. doi: 10.1007/s12015-007-0025-3. [DOI] [PubMed] [Google Scholar]
- 26.Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci USA. 2004;101:4158–4163. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 28.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Podsypanina K, Huang S, Mohsin SK, Chamness GC, Hatsell S, Cowin P, Schiff R, Li Y. Estrogen receptor positivity in mammary tumors of Wnt-1 transgenic mice is influenced by collaborating oncogenic mutations. Oncogene. 2005;24:4220–4231. doi: 10.1038/sj.onc.1208597. [DOI] [PubMed] [Google Scholar]
- 30.Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR, MacLeod CL, Shyamala G, Gillgrass AE, Cardiff RD. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol. 2002;161:1087–1097. doi: 10.1016/S0002-9440(10)64269-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui XS, Donehower LA. Differential gene expression in mouse mammary adenocarcinomas in the presence and absence of wild type p53. Oncogene. 2000;19:5988–5996. doi: 10.1038/sj.onc.1203993. [DOI] [PubMed] [Google Scholar]
- 32.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 33.Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 34.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 35.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells—an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 36.Gangnus R, Langer S, Breit E, Pantel K, Speicher MR. Genomic profiling of viable and proliferative micrometastatic cells from early-stage breast cancer patients. Clin Cancer Res. 2004;10:3457–3464. doi: 10.1158/1078-0432.CCR-03-0818. [DOI] [PubMed] [Google Scholar]
- 37.Meng S, Tripathy D, Shete S, Ashfaq R, Haley B, Perkins S, Beitsch P, Khan A, Euhus D, Osborne C, et al. HER-2 gene amplification can be acquired as breast cancer progresses. Proc Natl Acad Sci USA. 2004;101:9393–9398. doi: 10.1073/pnas.0402993101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, Podsypanina K, Chen Y, Cai W, Tsimelzon A, Hilsenbeck S, Li Y. Wnt-1 is dominant over neu in specifying mammary tumor expression profiles. Technol Cancer Res Treat. 2006;5:565–571. doi: 10.1177/153303460600500603. [DOI] [PubMed] [Google Scholar]
- 39.Siegel PM, Dankort DL, Hardy WR, Muller WJ. Novel activating mutations in the neu proto-oncogene involved in induction of mammary tumors. Mol Cell Biol. 1994;14:7068–7077. doi: 10.1128/mcb.14.11.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Podsypanina K, Li Y, Varmus H. Evolution of somatic mutations in mammary tumors in transgenic mice is influenced by the inherited genotype. BMC Med. 2004;2:24. doi: 10.1186/1741-7015-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang JW, Boxer RB, Chodosh LA. Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Mol Cell Biol. 2006;26:8109–8121. doi: 10.1128/MCB.00404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 43.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu ML, Von Lintig FC, Liyanage M, Shibata MA, Jorcyk CL, Ried T, Boss GR, Green JE. Amplification of Ki-ras and elevation of MAP kinase activity during mammary tumor progression in C3(1)/SV40 Tag transgenic mice. Oncogene. 1998;17:2403–2411. doi: 10.1038/sj.onc.1202456. [DOI] [PubMed] [Google Scholar]
- 45.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 46.Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. 2007;7:659–672. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- 47.Du Z, Li Y. RCAS-TVA in the mammary gland: an in vivo oncogene screen and a high fidelity model for breast transformation? Cell Cycle. 2007;6:823–826. doi: 10.4161/cc.6.7.4074. [DOI] [PubMed] [Google Scholar]
- 48.Du Z, Podsypanina K, Huang H, McGrath A, Toneff MJ, Bogoslovskaia E, Zhang X, Moraes RC, Fluck MM, Allred DC, et al. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc Natl Acad Sci USA. 2006;103:17396–17401. doi: 10.1073/pnas.0608607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talmadge JE, Wolman SR, Fidler IJ. Evidence for the clonal origin of spontaneous metastases. Science. 1982;217:361–363. doi: 10.1126/science.6953592. [DOI] [PubMed] [Google Scholar]
- 50.Fidler IJ. The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 51.Kerbel RS, Waghorne C, Korczak B, Lagarde A, Breitman ML. Clonal dominance of primary tumours by metastatic cells: genetic analysis and biological implications. Cancer Surv. 1988;7:597–629. [PubMed] [Google Scholar]
- 52.Moffett BF, Baban D, Bao L, Tarin D. Fate of clonal lineages during neoplasia and metastasis studied with an incorporated genetic marker. Cancer Res. 1992;52:1737–1743. [PubMed] [Google Scholar]
- 53.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1886–1895. [DOI] [PubMed] [Google Scholar]