Abstract
TMPRSS2-ERG gene fusions are the predominant molecular subtype of prostate cancer. Here, we explored the role of TMPRSS2-ERG gene fusion product using in vitro and in vivo model systems. Transgenic mice expressing the ERG gene fusion product under androgen-regulation develop mouse prostatic intraepithelial neoplasia (PIN), a precursor lesion of prostate cancer. Introduction of the ERG gene fusion product into primary or immortalized benign prostate epithelial cells induced an invasion-associated transcriptional program but did not increase cellular proliferation or anchorage-independent growth. These results suggest that TMPRSS2-ERG may not be sufficient for transformation in the absence of secondary molecular lesions. Transcriptional profiling of ERG knockdown in the TMPPRSS2-ERG-positive prostate cancer cell line VCaP revealed decreased expression of genes over-expressed in prostate cancer versus PIN and genes overexpressed in ETS-positive versus -negative prostate cancers in addition to inhibiting invasion. ERG knockdown in VCaP cells also induced a transcriptional program consistent with prostate differentiation. Importantly, VCaP cells and benign prostate cells overexpressing ERG directly engage components of the plasminogen activation pathway to mediate cellular invasion, potentially representing a downstream ETS target susceptible to therapeutic intervention. Our results support previous work suggesting that TMPRSS2-ERG fusions mediate invasion, consistent with the defining histologic distinction between PIN and prostate cancer.
Introduction
Based on a bioinformatics strategy that nominated genes showing high expression in a subset of cancer cases, we identified fusions of the 5′-untranslated region of TMPRSS2 (21q22) to ERG (21q22), ETV1 (7p21), ETV4 (17q21), or ETV5 (3q27) in prostate cancer cases that over-expressed the respective ETS family member [1–3]. TMPRSS2-ERG fusions are the most predominant molecular subtype, with multiple studies showing that approximately 50% of prostate cancers from prostate-specific antigen (PSA) screened surgical cohorts are TMPRSS2-ERG fusion-positive, and greater than 90% of prostate cancers over-expressing ERG harbor TMPRSS2-ERG fusions [2,4–18].
As TMPRSS2 had previously been characterized as an androgen-regulated gene [19], and TMPRSS2 only contributes untranslated sequence to many TMPRSS2-ERG transcripts, we hypothesized that the androgen responsive regulatory elements of TMPRSS2 drive ERG over-expression in fusion-positive cases. In support of this hypothesis, we observed that treatment of the TMPRSS2-ERG-positive prostate cancer cell line VCaP with the synthetic androgen R1881 resulted in increased expression of the TMPRSS2-ERG [2,20] fusion product. Additionally, castration of mice with androgen-dependent TMPRSS2-ERG-positive xenografts resulted in decreased expression of ERG in the xenograft [21].
Following the identification of TMPRSS2 fusions to ERG, ETV1, and ETV4, we recently discovered additional 5′ fusion partners involved in ETV1 and ETV5 gene fusions, including the 5′ untranslated regions from SLC45A3, HERV-K_22q11.3, C15ORF21, and HNRPA2B1 [3,22]. Presently, these additional 5′ partners have only been identified in ETV1 and ETV5 fusions, and it is unknown if they can fuse with ERG (in rare TMPRSS2-ERG-negative cases with ERG outlier expression) or additional ETS family members. ETV1 and ETV5 gene fusions are relatively rare and account for only 2% to 8% of prostate cancers. Interestingly, in these recent studies, we observed that ETV1 or ETV5 over-expression induces a cell invasion program [3,22]. Furthermore, androgen regulation and over-expression of the ETV1 fusion product in the prostate induced mouse prostatic intraepithelial neoplasia (mPIN) in mice. Thus, whereas ETV1 and ETV5 are rare gene fusions in prostate cancer, it is unknown if the functional role of the most common aberration in prostate cancer, TMPRSS2-ERG, is similar. Here, we recapitulated TMPRSS2-ERG fusions in vivo and in vitro and used an integrative expression profiling strategy to determine functional roles for TMPRSS2-ERG in prostate cancer.
Materials and Methods
Transgenic ERG Mice
cDNA of ERG (exon 2 to base 1533 of NM_182918.2), was amplified by reverse transcription-polymerase chain reaction (RT-PCR) from the VCaP cell line and TOPO cloned into the Gateway entry vector pCR8/GW/TOPO (Invitrogen, Carlsbad, CA), yielding pCR8-ERG. 3XFLAG-epitope-tagged construct was generated by PCR using pCR8-ERG as the template with the reverse primer encoding a triple FLAG tag before the stop codon. The product was TOPO cloned into pCR8, generating pCR8-3xFLAG-ERG. To generate a prostate-specific ERG transgenic construct, 3xFLAG-ERG was inserted into pBSII (Stratagene, La Jolla, CA) downstream of a modified small composite probasin promoter (ARR2PB) and upstream of a bovine growth hormone polyA site (PA-BGH) [23,24]. The ARR2Pb-ERG plasmid was linearized with PvuI/KpnI/SacII and microinjected into fertilized FVB mouse eggs and surgically transplanted into a pseudopregnant female by the University of Michigan Transgenic Animal Model Core. Transgenic founders were screened by PCR using genomic DNA isolated from tail snips. Multiple ARR2Pb-ERG transgenic founders were obtained and crossed with FVB mice, and transgene-positive male mice offspring were sacrificed at various time points.
Prostates from transgenic mice were dissected, stained with hematoxylin and eosin, and evaluated by three pathologists (R.M., M.A.R., and R.B.S.) as described earlier [22,25].
For immunohistochemical detection of Erg-FLAG, the basal cell marker p63, and smooth muscle actin, deparaffinized slides were subjected to microwave-citrate antigen retrieval and incubated with rabbit anti-FLAG polyclonal antibody (1:50 dilution, overnight incubation, #2368; Cell Signaling Technology, Danvers, MA), mouse monoclonal anti-p63 antibody (1:100 dilution, 45 minutes of incubation, MS1081P1; LabVision, Fremont, CA), and mouse monoclonal anti-smooth muscle actin antibody (1:50 dilution, 30 minutes of incubation, M0851; DakoAb, Carpinteria, CA), respectively. Visualization of p63 and SMA was performed using a standard biotin-avidin complex technique using M.O.M. Immunodetection kit (PK2200; Vector Laboratories, Burlingame, CA). FLAG was detected using Envision+System-HRP (DAB) kit (K4011; DakoCytomation, Carpinteria, CA).
Cell Lines and Samples
The benign immortalized prostate cell line RWPE was obtained from the American Type Culture Collection (Manassas, VA). Primary benign prostatic epithelial cells (PrEC) were obtained from Cambrex Bio Science (Walkersville, MD). VCaP was derived from a vertebral metastasis from a patient with hormone-refractory metastatic prostate cancer [26], and was provided by Kenneth Pienta (University of Michigan).
Prostate tissues were from the radical prostatectomy series at the University of Michigan and from the Rapid Autopsy Program, which are both part of the University of Michigan Prostate Cancer Specialized Program of Research Excellence Tissue Core. All samples were collected with informed consent of the patients and prior institutional review board approval. For all samples and cell lines, total RNA was isolated with Trizol (Invitrogen) according to the manufacturer's instructions.
In Vitro Over-expression of ERG
To generate adenoviral and lentiviral constructs, pCR8-ERG and a control entry clone (pENTR-GUS) were recombined with pAD/CMV/V5 (Invitrogen) and pLenti6/CMV/V5 (Invitrogen), respectively, using LR Clonase II (Invitrogen). Control pAD/CMV/LACZ clones were obtained from Invitrogen. Adenoviruses and lentiviruses were generated by the University of Michigan Vector Core. The benign immortalized prostate cell line RWPE was infected with lentiviruses expressing ERG or GUS, and stable clones were generated by selection with blasticidin (Invitrogen). Benign PrEC cells were infected with adenoviruses expressing ERG or LACZ. RWPE cells were also infected with ERG or LACZ adenoviruses for transient over-expression.
Immunoblot Analysis
Cell lysates transferred to polyvinylidene fluoride membranes were probed with rabbit polyclonal anti-ERG (sc-354; Santa Cruz Biotechnology, Santa Cruz, CA) at 1:500 dilution, mouse monoclonal anti-matrix metalloproteinase 3 (MMP3) (IM36L; Calbiochem, San Diego, CA) at 1:500 dilution, mouse monoclonal anti-uPA (IM13L, Calbiochem) at 1:500 dilution, and mouse anti-GAPDH antibody (Abcam, Cambridge, MA) at 1:30,000 dilution for loading control.
Proliferation Assay
Cell counts were estimated by trypsinizing the cells and, analysis was done using a Coulter counter (Beckman Coulter, Fullerton, CA) at the indicated time points in triplicate.
FACS Cell Cycle Analysis
Propidium iodide-stained RWPE-ERG and RWPE-GUS cells were analyzed on a LSR II flow cytometer (BD Biosciences, San Jose, CA) running FACSDivia, and cell cycle phases were calculated using ModFit LT (Verity Software House, Topsham, ME).
Soft Agar Assay
A 0.6% (wt./vol.) bottom layer of low melting point agarose in normal medium was prepared in six-well culture plates. On top, a layer of 0.3% agarose containing 1 × 104 RWPE-GUS, RWPE-ERG, or DU145 (positive control) cells was placed. After 12 days, foci were stained with crystal violet and counted.
Invasion Assays
Invasion assays were performed using PrEC and RWPE-ERG and -LACZ cells (48 hours after infection with adenoviruses), stable RWPE-ERG and -GUS cells, or VCaP cells as described earlier [22].
For inhibitor studies, amiloride (20 µM; EMD Biosciences, San Diego, CA), MMP3 inhibitor (10 µM; EMD Biosciences), MMP2/9 inhibitor (10 µM; EMD Biosciences), MMP8 inhibitor (10 µM; EMD Biosciences), the pan MMP inhibitor GM 6001 (10 µM; EMD Biosciences), the EWS:FLI inhibitor cytosine arabinoside (250 nM) [27], or vehicle control was added to VCaP and stable RWPE-ERG or -GUS cells for 24 hours, before trypsinization and seeding for invasion assays. For PAI-1, VCaP and stable RWPE-ERG or -GUS cells were trypsinized and treated with the indicated amount of recombinant PAI-1 (EMD Biosciences) for 15 minutes at indicated concentrations, before seeding.
ERG, PLAU, and PLAT Knockdown
For short interfering RNA (siRNA) knockdown of ERG, PLAT, or PLAU, the individual siRNA composed of the Dharmacon SMART-pool against ERG (MQ-003886-01; Lafayette, CO), PLAT (LQ-005999-00), or PLAU (LQ-006000-00), were tested for knockdown by quantitative polymerase chain reaction (qPCR), and the most effective single siRNA (ERG, D-003886-01; PLAT, J-005999-05; PLAU, J-006000-07) was used for further experiments. siCONTROL Non-Targeting siRNA #1 (D-001210-01) or siRNA against ERG, PLAT, or PLAU was transfected into VCaP or RWPE-ERG cells as indicated using Oligofectamine (Invitrogen). After 24 hours, we carried out a second identical transfection and cells were harvested 24 hours later for RNA isolation, invasion assays, or proliferation assays.
Expression Profiling
Expression profiling was performed using the Agilent Whole Human Genome Oligo Microarray (Santa Clara, CA) according to the manufacturer's protocol [22]. For all hybridizations involving ERG over-expression by adenovirus or lentivirus, the reference was the same cell line expressing LACZ or GUS, respectively. For profiling of ERG knockdown in VCaP, the reference was VCaP treated with nontargeting siRNA. All hybridizations were performed in duplicate with duplicate dye flips, for a total of four arrays, except for transiently expressing RWPE-ERG, which consisted of duplicate hybridizations and a single dye flip. Over- and under-expressed signatures were generated by filtering to include only features with significant differential expression (Log ratio, P < .01) in all hybridizations and two-fold average over- or under-expression (Log ratio) after correction for the dye flip. For VCaP profiling, all features with significant differential expression (Log ratio, P < .01) and Cy5/Cy3 ratios of > or < 1 in all hybridizations were included in the over- and under-expressed signatures, respectively.
Molecular Concepts Analysis
All expression signatures were uploaded into the Oncomine Concepts Map (OCM, www.oncomine.org) [28] as molecular concepts, using all features on the Agilent Whole Human Genome Oligo Microarray as the null set. For the assessment of prostate-specific gene expression, the expO (GSE2109) and Shyamsundar normal tissue [29] data sets were accessed using the Oncomine database. Cancer and normal tissue types are defined in Table W3. For the assessment of prostate cell type expression, the Prostate cell-specific expression Affymetrix data set of Oudes et al. [30] was downloaded from the Gene Expression Omnibus (GSE3998). Data are reported as RMA-normalized fluorescent intensities.
Quantitative PCR
Quantitative PCR was performed using Power SYBR GreenMastermix (Applied Biosystems, Foster City, CA) on an Applied Biosystems 7300 Real Time PCR system as described [1,2]. All oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA) and are listed in Table W2. All reactions were performed in duplicate unless otherwise indicated.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to published protocols using anti-ERG (sc-354x; Santa Cruz) or rabbit anti-IgG (sc-2027; Santa Cruz) antibodies [31]. For PCR analysis of enrichment of target gene promoters, 2 µl each of input DNA, ERG-enriched, or IgG-enriched DNA were subjected to PCR using Platinum PCR Supermix (Invitrogen) and primers specific for target gene promoters (Table W2).
Results
Transgenic Expression of ERG in the Mouse Prostate Induces mPIN
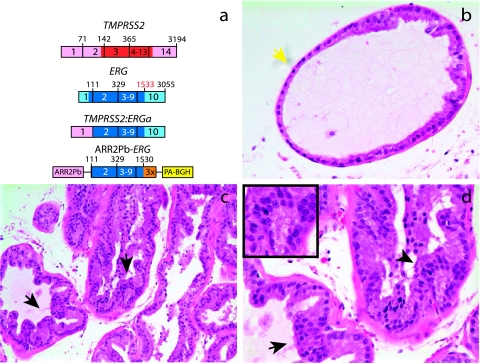
Fusion transcripts juxtaposing exon 1 of TMPRSS2 (NM_005656.2) to exon 2 of ERG isoform 1 (NM_182918.2; identical to exon 4 of ERG isoform 2, NM_004449.3) are the most commonly detected transcripts in TMPRSS2-ERG-positive cases (TMPRSS2-ERGa) [2,5,9]. Because exon 1 of TMPRSS2 is entirely noncoding, this fusion transcript likely results in a truncated ERG protein product. Thus, we generated transgenic mice expressing the truncated ERG product from TMPRSS2-ERGa (beginning at exon 2 through the reported stop codon (base 1533) of NM_182918.2, C-terminal FLAG-tagged) under the control of the modified probasin promoter (ARR2Pb-ERG) (Figure 1a), which drives androgen-regulated transgene expression exclusively in the prostate [23,24]. This transgene is functionally analogous to the TMPRSS2-ERGa fusion product. We obtained multiple ARR2Pb-ERG founders and lines were expanded for phenotypic analysis. By 12 to 14 weeks of age, three of eight (37.5%) ARR2Pb-ERG mice developed mPIN (Table W1 and Figure 1), the candidate precursor lesion of prostate cancer [25].
Figure 1.
Transgenic mice recapitulating TMPRSS2-ERG in the prostate develop mPIN. (a) To recapitulate TMPRSS2-ERG in vivo, we generated transgenic mice over-expressing the ERG gene fusion product (exons 2 through the reported stop codon; 1533 of NM_182918.2, C-terminal 3X-FLAG epitope tag) with a bovine growth hormone polyA signal (PA-BGH) under the control of the enhanced probasin promoter (ARR2Pb). Mice were sacrificed at 12 to 14 weeks or >20 weeks, and mouse prostatic intraepithelial neoplasia (mPIN) was observed in 4 of 11 ARR2Pb-ERG mice as described in Table W1. Benign epithelia and areas of mPIN are indicated by yellow and black arrows, respectively. (b–d) Hematoxylin and eosin staining of ARR2Pb-ERG prostates for morphologic assessment. Consistent with the focality of mPIN, (b) benign glands and (c and d) mPIN were observed in the ventral prostate (VP) of ARR2Pb-ERG mice. Original magnification: (b) ×400, (c) ×200, and (d) inset showing area of mPIN with macronucleoli, ×400.
We observed normal glands in the prostates of ARR2Pb-ERG mice containing focal proliferative lesions displaying nuclear atypia, including stratification, hyperchromasia, and macronucleoli (Figure 1, b–d), consistent with the definition of mPIN [25]. In 12- to 14-week-old ARR2Pb-ERG mice, foci of mPIN were observed exclusively in the ventral lobe (Table W1). Immunohistochemistry in ARR2Pb-ERG mice demonstrated strong ERG-FLAG expression primarily in mPIN foci and not benign glands (Figure W1, a and b), and qPCR confirmed that transgene expression was limited to the prostate (data not shown).
All lesions were confirmed to be in situ by the presence of an intact fibromuscular layer, as demonstrated by contiguous smooth muscle actin staining (Figure W1, c and d). However, immunohistochemistry with the basal cell marker p63 demonstrated loss of the circumferential basal epithelial layer in ARR2Pb-ERG mPIN compared to benign glands (Figure W1, e and f), indicating the disruption of the basal cell layer. Because loss of the basal layer is a hallmark of prostate carcinoma development in both mice and humans [32], ARR2Pb-ERG mice will be closely monitored for the development of invasive carcinoma at later time points. Whereas we have not observed progression to invasive carcinoma in ARR2Pb-ERG mice, we have only characterized three mice at greater than 20 weeks of age, one of which (33.3%) also had mPIN in both the ventral and dorsolateral lobes (Table W1). These results demonstrate that, although ERG induces a neoplastic phenotype in the mouse prostate, providing support for an oncogenic role in human prostate cancer, it is not sufficient for the development of prostate cancer in mice.
ERG Over-expression Induces an Invasion Program In Vitro
Next, we determined the effects of ERG over-expression in vitro, by generating adenoviruses and lentiviruses that express the same truncated ERG product from TMPRSS2-ERGa as in the ARR2Pb-ERG mice (Figure 2a). We infected the benign immortalized prostate epithelial cell line RWPE with lentivirus expressing ERG and selected for stable RWPE-ERG cells, and transiently over-expressed ERG in primary benign prostate epithelial cells (PrEC) by infection with adenovirus expressing ERG. By immunoblot analysis, we confirmed the expression of a protein product recognized by a commercial anti-ERG antibody in both RWPE and PrEC (Figure W2).
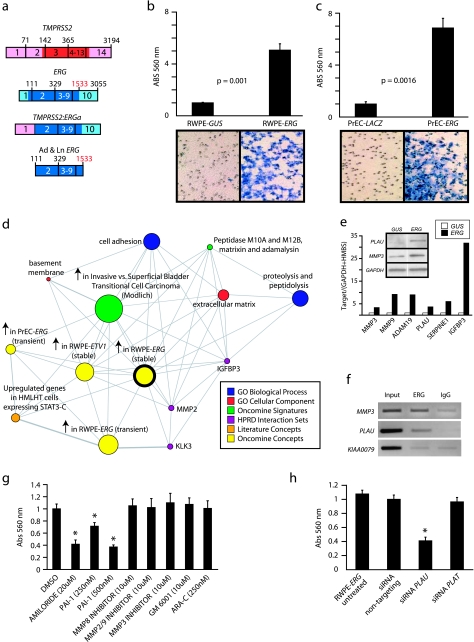
Figure 2.
Over-expression of ERG in RWPE cells increases invasion through the plasminogen activator pathway. (a) To recapitulate TMPRSS2-ERG in vitro, we generated adenoviruses and lentiviruses expressing the ERG gene fusion product (exons 2 through the reported stop codon). (b and c) Infected (b) RWPE and (c) PrEC cells as indicated were assayed for invasion through a modified basement membrane. Photomicrographs of invaded cells are shown below. (d) RWPE-ERG and RWPE-GUS (control vector) cells were profiled on Agilent Whole Genome microarrays and expression signatures were loaded into the Oncomine Concept Map. Molecular concept map analysis of the over-expressed in RWPE-ERG compared to RWPE-GUS signature (ringed yellow node). Each node represents a molecular concept, or set of biologically related genes. The node size is proportional to the number of genes in the concept. The concept color indicates the concept type according to the legend. Each edge represents a significant enrichment (P < .005). (e) qPCR confirmation of increased expression of genes involved in invasion. The amount of the indicated gene (normalized to the average of GAPDH and HMBS) in RWPE-GUS (white) and RWPE-ERG (black) is shown. Inset shows immunoblot confirmation of increased expression of PLAU and MMP3 in RWPE-ERG cells. (f) Chromatin immunoprecipitation shows enrichment of ERG binding to the proximal promoters of PLAU and MMP3 compared to IgG control. The promoter of KIAA0089 was used as a negative control. (g) RWPE-ERG cells were treated with PLAU inhibitors amiloride or ectopic PAI-1, MMP inhibitors (including the pan-MMP inhibitor GM-6001), or the EWS:FLI inhibitor ARA-C (EWS:FLI inhibitor) as indicated and assayed for invasion as in c. (h) RWPE-ERG cells were treated with transfection reagent alone (untreated), or transfected with nontargeting, PLAU or PLAT siRNA as indicated and assayed for invasion through a modified basement membrane. For all invasion assays, mean (n = 3) ± SEM are shown; *P < .05.31
In both RWPE and PrEC cells, over-expression of ERG did not increase proliferation (Figure W2), and ERG did not affect the percentage of RWPE cells in S phase by cell cycle analysis (Figure W2c). Additionally, soft agar transformation assays showed that ERG over-expression was not sufficient to transform RWPE cells (Figure W2d). Finally, orthotopic xenograft assays using RWPE-ERG cells did not result in tumor formation (data not shown). However, ERG over-expression markedly increased invasion in a modified basement membrane invasion assay in both RWPE (5-fold, P = .001) (Figure 2b) and PrEC cells (6.9-fold, P = .0016) (Figure 2c). Transient over-expression of ERG in RWPE using ERG adenovirus similarly increased invasion (Figure W3). These results are similar to over-expression of ETV1 and ETV5, which we have previously shown to increase invasion in PrEC and RWPE cells [3,22].
To investigate the transcriptional program regulated by ERG, we profiled stable RWPE-ERG and transiently expressing RWPE-ERG and PrEC-ERG cells using Agilent Whole Genome Oligo Expression Arrays, and identified 865, 854, and 221 features that were over-expressed in the respective cell lines (as described in the Materials and Methods section). We have recently developed a resource termed the Oncomine Concepts Map (OCM, www.oncomine.org) to look for associations between more than 20,000 biologically related gene sets by disproportionate overlap [28,33]. Thus, we uploaded these expression signatures into the OCM to identify transcriptional programs induced by ERG.We began by seeding the OCM analysis with the over-expressed in stable RWPE-ERG signature. OCM analysis identified the most significantly enriched concept as our previous over-expressed in stable RWPE-ETV1 signature [22] [odds ratio (OR) = 59.43, P = 1 × 10−100] (Figure 2d), consistent with their similar phenotypes and supporting the functional redundancy of these ETS family members in gene fusions.
The stable RWPE-ERG signature also shared significant enrichment with the over-expressed in transient RWPE-ETV5 (OR = 3.9, P = 1.2 × 10−9), over-expressed in transient RWPE-ERG (OR = 19.43, P = 1.1 × 10-100), and transient PrEC-ERG (OR = 5.77, P = 3.1 × 10−10) signatures, demonstrating similarities in these transcriptional programs, as well as several molecular concepts related to invasion. These concepts include the Interpro concept of gene products containing Peptidase M10A and M12B, matrixin or adamalysin domains (OR = 5.27, P = .002), which includes MMPs and a disintegrin and metalloproteinase domains (ADAM), and a signature of genes over-expressed in benign breast epithelial cells (HMLHT) over-expressing the STAT3-C oncogene (OR = 4.04, P = 6.3 × 10−5). In this system, STAT3-C over-expression did not increase proliferation, but increased invasion in an MMP9- dependent manner [34].
ERG-Mediated Induction of the Plasminogen Activator Pathway
We identified several genes over-expressed in RWPE-ERG that were present in multiple concepts in this enrichment network and have been directly implicated in the invasion in multiple cancers and models, including the metalloproteinases MMP3, MMP9, and ADAM19, the urokinase plasminogen activator (PLAU), and the plasminogen activator inhibitor type 1 (SERPINE1, also known as PAI-1) [35,36]. Both MMPs and the urokinase plasminogen pathway have been reported to be direct targets of ETS transcription factors [35–37]. By qPCR, we confirmed the over-expression of these genes, as well as the MMP cleavage target IGFBP3 in RWPE-ERG cells (Figure 2e).
By immunoblot analysis, we confirmed the over-expression of PLAU and MMP3 in RWPE-ERG cells (Figure 2e). To determine if these genes are direct targets of ERG, we performed ChIP, which demonstrated that ERG binds to the proximal promoter of both PLAU and MMP3 (Figure 2f). No enrichment of ERG binding was observed in RWPE-GUS cells or LNCaP (ETV1 rearrangement-positive [22]) for MMP3 or PLAU (Figure W4).
We next assessed the role of both MMPs and the plasminogen activator pathways in the invasive phenotype of RWPE-ERG cells using small molecule MMP inhibitors, amiloride (a specific PLAU inhibitor [38]), ectopic PAI-1 (which inhibits plasminogen activators [39]) and siRNA knockdown of PLAU. As shown in Figure 2g, whereas MMP inhibitors did not significantly inhibit invasion, both amiloride and PAI-1 significantly inhibited the invasiveness of RWPE-ERG cells. Similarly, siRNA knockdown of PLAU significantly inhibited the invasion of RWPE-ERG cells, whereas siRNA knockdown of the tissue plasminogen activator (PLAT) had no effect on RWPE-ERG invasion (Figure 2h). Similar effects on invasion were seen with independent siRNA duplexes directed against PLAU. Cytosine arabinoside (ARA-C), which has recently been identified as an inhibitor of the EWS-FLI fusion found in Ewing's sarcoma [27], also showed no effect on RWPE-ERG invasion (Figure 2g). Together, this work demonstrates that ERG directly induces PLAU expression in RWPE cells and that inhibition of PLAU blocks ERG-mediated invasion.
Knockdown of ERG in VCaP Cells Inhibits Invasion
Together, our in vivo and in vitro studies show that the most common TMPRSS2-ERG fusion product is unable to transform benign prostatic epithelial cell lines or induce the development of frank adenocarcinoma in the mouse prostate. However, our previous work, including expression profiling on laser-captured microdissected cell populations and a fluorescence in situ hybridization (FISH)-based study on prostate cancer progression, suggest that TMPRSS2-ERG gene fusions occur in the context of preexisting genetic lesions, often during the PIN to carcinoma transition [7,33].
To investigate the role of TMPRSS2-ERG in this context of preexisting genetic lesions, we used siRNA to knockdown ERG in VCaP (VCaP-siERG) cells that are known to harbor the TMPRSS2-ERG gene fusion [2]. Immunoblot analysis confirmed that siRNA directed against ERG reduced expression compared to nontargeting control siRNA (Figure 3a). Quantitative PCR also demonstrated a 63% decrease in ERG transcript expression in VCaP-siERG (Figure W5a, P = .009). ERG knockdown also significantly inhibited the invasion of VCaP cells (Figure 3b) without affecting proliferation (Figure W5b), similar to ERG over-expression in RWPE cells. Similar results were seen using a second siRNA targeting an independent sequence in ERG (data not shown).
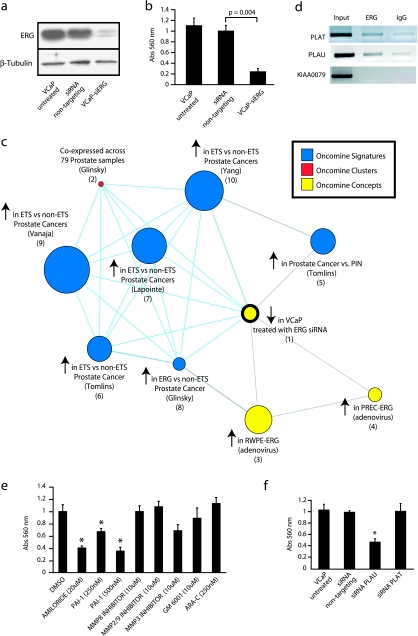
Figure 3.
Knockdown of ERG in VCaP cells attenuates a transcriptional program over-expressed in TMPRSS2-ETS-positive prostate cancers. (a) SiRNA knockdown of ERG in the TMPRSS2-ERG-positive prostate cancer cell line VCaP. VCaP cells were treated with transfection reagent alone (untreated), or transfected with nontargeting or ERG siRNA (VCaP-siERG) as indicated. ERG knockdown was confirmed by immunoblot analysis. (b) VCaP cells as indicated were assayed for invasion through a modified basement membrane. (c) VCaP-siERG and VCaP cells treated with nontargeting siRNA were profiled and a molecular concept map of the under-expressed in VCaP-siERG signature (ringed yellow node) was generated. Each edge represents a significant enrichment (P < .001). Blue edges indicate enrichments with in vivo ETS-positive versus negative prostate cancer signatures. (d) Chromatin immunoprecipitation identifies PLAT and PLAU as direct targets of ERG in VCaP cells, by enrichment of ERG binding to the proximal promoters of PLAT and PLAU compared to IgG control. The promoter of KIAA0089 was used as a negative control. (e) VCaP cells were treated with the indicated inhibitors (as in Figure 2g) and assessed for invasion. (f) VCaP cells were treated with transfection reagent alone (untreated), or transfected with nontargeting, PLAU or PLAT siRNA as indicated and assayed for invasion. For all invasion assays, mean (n = 3) ± SEM are shown; *P < .05.
To determine the transcriptional profile mediated by TMPRSS2-ERG in VCaP, we profiled VCaP-siERG cells. We identified 265 and 291 features over- and under-expressed (as described in the Materials and Methods section), respectively, in VCaP-siERG compared to VCaP treated with nontargeting siRNA, and uploaded these signatures into the OCM. The two most significantly enriched concepts in our under-expressed in VCaP-siERG signature were two signatures of genes over-expressed in ETS-positive versus -negative prostate cancers (GSE8218, OR = 5.73, P = 2.5 × 10−19 and Vanaja et al. [40], OR = 3.49, P = 3.9 × 10−11) (Figure 3c). All other over-expressed in ETS-positive versus -negative prostate cancer signatures [33,41,42] in the Oncomine database were also enriched in our under-expressed in VCaP-siERG signature, supporting VCaP as a highly relevant model of TMPRSS2-ERG-positive prostate cancers. Our under-expressed in VCaP-siERG signature also shared enrichment with our previous signature of genes over-expressed in laser-captured prostate cancer versus PIN (OR = 3.79, P = 4.5 × 10−6). In that study, we observed that PIN and prostate cancer had very similar expression signatures and hypothesized that TMPRSS2-ERG fusions occurred during the PIN to prostate cancer transition and dysregulated a limited number of transcripts, likely involved in invasion [33].
The Role of the Plasminogen Activator Pathway in VCaP Cells
Our under-expressed in VCaP-siERG signature also shared significant enrichment with our over-expressed in transient PrEC-ERG and transient RWPE-ERG signatures (Figure 3c; OR = 6.89 and 3.21, P = 1.4 × 10−5 and 7 × 10−5, respectively), suggesting common transcriptional programs controlled by ERG across cell types and genetic context. Interestingly, although PLAU was not significantly dysregulated in VCaP-siERG cells, the most strongly down regulated feature in VCaP-siERG cells was tissue plasminogen activator (PLAT). Similar to PLAU, which we showed to be strongly over-expressed and a direct target of ERG in RWPE cells, we confirmed that PLAT was strongly downregulated (Figure W5c). Quantitative PCR analysis showed that VCaP-siERG cells expressed very low levels of PLAU at baseline (Figure W6), however ChIP identified both PLAU and PLAT as direct targets of ERG in VCaP-siERG cells (Figure 3d). Whereas ectopic PAI-1, amiloride (which inhibits PLAU but not PLAT [38]) (Figure 3e), and siRNA knockdown of PLAU inhibited the invasion of VCaP cells, siRNA knockdown of PLAT had no effect on VCaP invasion (Figure 3f). Additionally, inhibitors of MMPs and ARA-C had no significant effect on VCaP invasion (Figure 3e), similar to RWPE-ERG. Together, these results support plasminogen activators as direct targets of ERG across multiple TMPRSS2-ERG model systems and demonstrate that inhibition of PLAU blocks ERG-induced invasion across TMPRSS2-ERG cell line models.
Transcriptional Signatures of ERG in VCaP Cells
Our under-expressed in VCaP-siERG signature also shared significant enrichment with a cluster of 18 genes coexpressed across 72 prostate cancer tissue samples [41], with eight genes shared (OR = 56.65, P = 7.2 × 10−10). Because this cluster contains ERG (Figure W5d), this result supports ERG knockdown in VCaP-modulating genes regulated by ERG in TMPRSS2-ERG-positive tumors. To identify such genes, we examined genes coexpressed with ERG across multiple prostate cancer profiling studies in the Oncomine database. We identified four genes, CACNA1D, KCNS3, LAMC2, and PLA1A, that were downregulated in VCaP-siERG cells and also showed greater than 0.5 correlation with ERG across multiple prostate cancer profiling studies (Figure W7). CACNA1D was significantly down regulated in three of four arrays, with the fourth array showing 0.54-fold expression in VCaP-siERG (P = .06). In addition, we also identified decreased expression of ARGHDIB in VCaP-siERG cells and over-expression in all ETS-positive versus -negative expression signatures (Figure W5d). By qPCR, we confirmed the decreased expression of these genes in VCaP-siERG cells (Figure W5e) and ChIP identified LAMC2, KCNS3, and PLA1A as direct targets of ERG (Figure W5f). By qPCR, we also confirmed the coexpression of ERG and PLA1A (R = 0.72, P = 6.1 × 10−8) in an independent set of prostate tissues (Figure W5g). Thus, our work provides direct ERG target genes over-expressed in TMPRSS2-ERG-positive prostate cancers for further functional study.
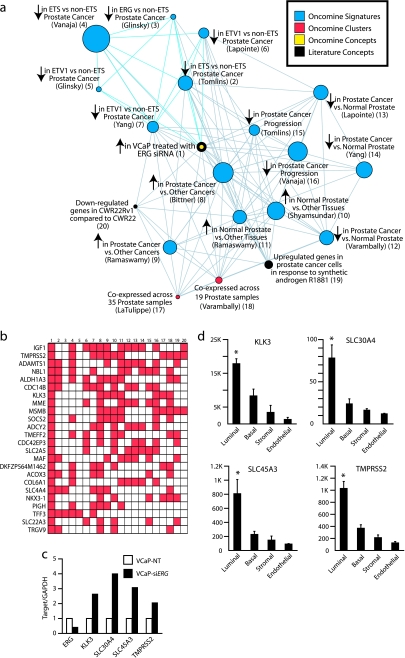
We next examined our over-expressed in VCaP-siERG signature using the OCM. Consistent with the results described above, all under-expressed in ETS-positive versus -negative prostate cancer signatures in the Oncomine database (GSE8218 and [33,40–42]) were enriched in our over-expressed in VCaP-siERG signature (OR = 6.41−2.71, P = 5.2 × 10−15 to 7.0 × 10−5). Intriguingly, OCM analysis revealed that the most significantly enriched concept in our over-expressed in VCaP-siERG signature was a signature of genes over-expressed in prostate cancers compared to 28 other cancer types (GSE2109) (OR = 4.46, P = 5.8 × 10−18) (Figure 4a). Several other concepts representing genes over-expressed in prostate cancer compared to other cancers, normal prostate tissue compared to other normal tissues and normal prostate compared to prostate cancer were also strongly enriched in our signature. Examining the genes common to these concepts and VCaP-siERG, we identified numerous archetypal prostate epithelial cell transcripts, including KLK3 (PSA), MSMB, NKX3-1, TMPRSS2, TRGV9 (also known as TARP) [43], SLC30A4 (also known as ZnT4) [44], and SLC45A3 [22] (Figure 4b and Figure W8). We confirmed the over-expression of this transcriptional program by qPCR (Figure 4c), and confirmed that these genes are normally expressed specifically in luminal epithelial prostate cells using an independent data set containing expression profiling data from magnetically sorted prostate luminal epithelial, basal epithelial, stromal fibromuscular, and endothelial cells (Figure 4d and Figure W8). Because ERG knockdown in VCaP results in the increased expression of genes associated with differentiated luminal prostate epithelial cells, we hypothesize that TMPRSS2-ERG fusion may function to keep prostate cancer cells in a dedifferentiated state. Future experiments will be needed to address this hypothesis.
Figure 4.
ERG knockdown in VCaP cells derepresses a transcriptional program associated with normal prostatic epithelial cell differentiation. (a) VCaP-siERG and VCaP cells treated with nontargeting siRNA were profiled and a molecular concept map of the over-expressed in VCaP-siERG signature (ringed yellow node) was generated. Each edge represents a significant enrichment (P < .001). Blue edges indicate enrichments with in vivo ETS-positive versus -negative prostate cancer signatures. (b) Overlay map identifying genes present (red cells), including KLK3 (PSA), across multiple concepts in the over-expressed in VCaP-siERG enrichment network (indicated by number). (c) qPCR confirmation of increased expression in VCaP-siERG cells (black) compared to VCaP-NT cells (white) of transcripts strongly expressed in prostatic epithelial cells. (d) Analysis of prostate cell type specificity using a microarray data set profiling magnetically sorted prostate cell populations. Mean RMA normalized fluorescent intensities (n = 5 ± SEM) are shown. *P < .05, for all pairwise t tests involving luminal cells.
Discussion
Our in vitro and in vivo studies on the TMPRSS2-ERG fusion described here support the functional similarity between ETS gene fusions, consistent with our initial observation of mutually exclusive ERG or ETV1 over-expression in prostate cancers [2]. This includes the similar phenotypic and transcriptional programs induced by ERG, ETV1, and ETV5 over-expression in benign prostate cells, the similar phenotype of transgenic mice expressing ERG or ETV1 in the prostate [22], and the enrichment of genes over-expressed in ERG or ETV1-positive versus ETS-negative prostate cancers in our VCaP-siERG signature (see Figure 3c).
Importantly, our in vivo and in vitro studies show that the most common TMPRSS2-ERG fusion product is unable to transform benign prostatic epithelial cell lines or induce the development of frank adenocarcinoma in the mouse prostate. However, both of these results are consistent with the occurrence of TMPRSS2-ERG fusions in the context of preexisting genetic lesions during the course of human prostate cancer development.
Similar to the expression of the TMPRSS2-ETV1 fusion product in the mouse prostate [22], expression of the TMPRSS2-ERG fusion product in the mouse prostate resulted in the development of PIN, without the development of frank adenocarcinoma. As described below, in human prostate cancer development, TMPRSS2-ERG fusions occur in the context of earlier lesions, such as loss of single NKX3-1 and/or PTEN alleles [45]. Importantly, mouse models of such early lesions, such as NKX3-1+/− and PTEN+/− mice [46–48], also only develop mPIN without frank adenocarcinoma. Together, these results are consistent with the development of invasive prostate cancer requiring multiple genetic lesions. Importantly, these results also suggest that crosses between ARR2Pb-ERG mice and transgenic mice modeling earlier lesions should produce highly relevant oncogene/tumor suppressor models mimicking early events in human prostate cancer development.
In this study, over-expression of ERG in benign prostate cells markedly increased invasion but did not result in transformation, similar to experiments with ETV1 and ETV5 [3,33]. These results support our previous hypothesis that ETS gene rearrangements mediate invasion in human prostate cancer development. For example, using expression profiling on laser captured microdissected cell populations, we demonstrated that whereas benign prostatic epithelial cells and epithelial cells in PIN lesions have distinct expression profiles, PIN and cancerous epithelium share remarkably similar expression profiles [33]. This suggests that PIN and cancerous cells share many genetic lesions, with a limited number of genetic events likely mediating the PIN to prostate cancer transition (defined histologically by the presence of invasion). Importantly in our profiled samples, TMPRSS2-ERG fusions only occurred in prostate cancer and not in PIN lesions (as evidenced by ERG outlier expression), suggesting that it might be the key lesion driving the invasive transition.
Further supporting a role for TMPRSS2-ERG in invasion, we previously demonstrated in a FISH-based study that TMPRSS2-ERG fusion was not identified in benign prostate cells or proliferative inflammatory atrophy, which may be an early precursor of PIN/prostate cancer. However, TMPRSS2-ERG fusion could be detected in 19% of PIN lesions, but these foci were intermingling with cancerous glands that were similarly TMPRSS2-ERG-positive [7]. TMPRSS2-ERG fusion was not identified in PIN lesions distant to prostate cancer, even if the cancerous lesion from the same individual demonstrated the TMPRSS2-ERG fusion. Together, this FISH-based study suggested that TMPRSS2-ERG fusions may directly mediate the development of prostate cancer from PIN lesions.
Thus, to study TMPRSS2-ERG function in a more realistic cellular context, we investigated the effects of ERG knockdown in the TMPRSS2-ERG-positive VCaP cell line. These experiments confirmed VCaP as a highly relevant prostate cancer cell line model, as siRNA knockdown of ERG inhibited invasion and modulated transcriptional programs activated in TMPRSS2-ERG-positive tumors. Additionally, ERG knockdown also modulated the transcriptional program that differentiated our laser captured PIN and prostate cancer cell populations, consistent with TMPRSS2-ERG driving this important transition. Importantly, these programs were not modulated by ERG over-expression in RWPE cells, further supporting VCaP as a more realistic model of TMPRSS2-ERG prostate cancer. Interestingly, in both RWPE-ERG and VCaP cells, we demonstrate that the plasminogen activator pathway is crucial to ERG-mediated invasion, similar to ETV5-mediated invasion [3]. Thus, this pathway warrants further investigation as a therapeutic target for TMPRSS2-ERG-positive prostate cancer.
Supplementary Material
Acknowledgments
We thank S. Dhanasekaran, J. Siddiqui, M. LeBlanc, and N. Singla for technical assistance; K. Pienta for the VCaP cell line; and R. Craig and L. Stoolman for the FACS analysis. We thank the University of Michigan Transgenic Animal Model Core for ERG mouse generation and the University of Michigan Vector Core for ERG virus generation.
Abbreviations
- MMP
matrix metalloproteinase
- mPIN
mouse prostatic intraepithelial neoplasia
- OR
odds ratio
- PLAT
tissue plasminogen activator
- PLAU
urokinase plasminogen activator
- PrEC
primary benign prostate epithelial cell
- qPCR
quantitative polymerase chain reaction
Footnotes
Supported in part by the Department of Defense (PC040517 to R. M., W81XWH-06-1-0224 to A. M. C. and S. V., and PC060266 to J. Y.), the National Institutes of Health (Prostate Cancer Specialized Program of Research Excellence P50CA69568 to A. M. C. and R. B. S. and R01 CA102872 to A. M. C.), the Early Detection Research Network (UO1 CA111275-01 to A. M. C.), and the Prostate Cancer Foundation (to A. M. C.). A. M. C. is supported by a Clinical Translational Research Award from the Burroughs Welcome Foundation. S. A. T. is supported by a Rackham Predoctoral Fellowship. S. A. T. is a Fellow of the Medical Scientist Training Program.
This article refers to supplementary materials, which are designated by Tables W1, W2, and W3 and Figures W1, W2, W3, W4, W5, W6, W7, and W8 and are available online at www.neoplasia.com.
Disclosure: The University of Michigan has filed a patent on ETS gene rearrangements and SPINK1 over-expression in prostate cancer, on which S. A. T., R. M., and A. M. C. are co-inventors, and the diagnostic field of use has been licensed to Gen-Probe Incorporated. Gen-Probe also has a sponsored research agreement with A. M. C., however Gen-Probe has not played a role in the design and conduct of the study, in the collection, analysis, or interpretation of the data, and in the preparation, review, or approval of the manuscript.
References
- 1.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 3.Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, Cao X, Singla N, Montie JE, Varambally S, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 4.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 5.Lapointe J, Kim YH, Miller MA, Li C, Kaygusuz G, van de Rijn M, Huntsman DG, Brooks JD, Pollack JR. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol. 2007;20:467–473. doi: 10.1038/modpathol.3800759. [DOI] [PubMed] [Google Scholar]
- 6.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 7.Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 8.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717–719. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 10.Cerveira N, Ribeiro FR, Peixoto A, Costa V, Henrique R, Jeronimo C, Teixeira MR. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, Eeles R, Martin FL, Phillips DH, Crundwell M, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2006;26:2667–2673. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 12.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66:10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 13.Iljin K, Wolf M, Edgren H, Gupta S, Kilpinen S, Skotheim RI, Peltola M, Smit F, Verhaegh G, Schalken J, et al. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66:10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- 14.Yoshimoto M, Joshua AM, Chilton-Macneill S, Bayani J, Selvarajah S, Evans AJ, Zielenska M, Squire JA. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia. 2006;8:465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehra R, Tomlins SA, Shen R, Nadeem O, Wang L, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 16.Nam RK, Sugar L, Wang Z, Yang W, Kitching R, Klotz LH, Venkateswaran V, Narod SA, Seth A. Expression of TMPRSS2 ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther. 2007;6:40–45. doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- 17.Rajput AB, Miller MA, De Luca A, Boyd N, Leung S, Hurtado-Coll A, Fazli L, Jones EC, Palmer JB, Gleave ME, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winnes M, Lissbrant E, Damber JE, Stenman G. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncol Rep. 2007;17:1033–1036. [PubMed] [Google Scholar]
- 19.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 20.Mertz KD, Setlur SR, Dhanasekaran SM, Demichelis F, Perner S, Tomlins S, Tchinda J, Laxman B, Vessella RL, Beroukhim R, et al. Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model. Neoplasia. 2007;9:200–206. doi: 10.1593/neo.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendriksen PJ, Dits NF, Kokame K, Veldhoven A, van Weerden WM, Bangma CH, Trapman J, Jenster G. Evolution of the androgen receptor pathway during progression of prostate cancer. Cancer Res. 2006;66:5012–5020. doi: 10.1158/0008-5472.CAN-05-3082. [DOI] [PubMed] [Google Scholar]
- 22.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Thomas TZ, Kasper S, Matusik RJ. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology. 2000;141:4698–4710. doi: 10.1210/endo.141.12.7837. [DOI] [PubMed] [Google Scholar]
- 24.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 25.Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, Humphrey PA, Sundberg JP, Rozengurt N, Barrios R, et al. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor Meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 2004;64:2270–2305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
- 26.Korenchuk S, Lehr JE, MClean L, Lee YG, Whitney S, Vessella R, Lin DL, Pienta KJ. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15:163–168. [PubMed] [Google Scholar]
- 27.Stegmaier K, Wong JS, Ross KN, Chow KT, Peck D, Wright RD, Lessnick SL, Kung AL, Golub TR. Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Med. 2007;4:e122. doi: 10.1371/journal.pmed.0040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, Barrette TR, Ghosh D, Varambally S, Chinnaiyan AM. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–454. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shyamsundar R, Kim YH, Higgins JP, Montgomery K, Jorden M, Sethuraman A, van de Rijn M, Botstein D, Brown PO, Pollack JR. A DNA microarray survey of gene expression in normal human tissues. Genome Biol. 2005;6:R22. doi: 10.1186/gb-2005-6-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oudes AJ, Campbell DS, Sorensen CM, Walashek LS, True LD, Liu AY. Transcriptomes of human prostate cells. BMC Genomics. 2006;7:92. doi: 10.1186/1471-2164-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- 33.Tomlins SA, Mehra R, Rhodes DR, Cao X, ang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 34.Dechow TN, Pedranzini L, Leitch A, Leslie K, Gerald WL, Linkov I, Bromberg JF. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C. Proc Natl Acad Sci USA. 2004;101:10602–10607. doi: 10.1073/pnas.0404100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laufs S, Schumacher J, Allgayer H. Urokinase-receptor (u-PAR): an essential player in multiple games of cancer: a review on its role in tumor progression, invasion, metastasis, proliferation/dormancy, clinical outcome and minimal residual disease. Cell Cycle. 2006;5:1760–1771. doi: 10.4161/cc.5.16.2994. [DOI] [PubMed] [Google Scholar]
- 36.Fingleton B. Matrix metalloproteinases: roles in cancer and metastasis. Front Biosci. 2006;11:479–491. doi: 10.2741/1811. [DOI] [PubMed] [Google Scholar]
- 37.de Launoit Y, Baert JL, Chotteau-Lelievre A, Monte D, Coutte L, Mauen S, Firlej V, Degerny C, Verreman K. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim Biophys Acta. 2006;1766:79–87. doi: 10.1016/j.bbcan.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Vassalli JD, Belin D. Amiloride selectively inhibits the urokinasetype plasminogen activator. FEBS Lett. 1987;214:187–191. doi: 10.1016/0014-5793(87)80039-x. [DOI] [PubMed] [Google Scholar]
- 39.Gils A, Declerck PJ. Plasminogen activator inhibitor-1. Curr Med Chem. 2004;11:2323–2334. doi: 10.2174/0929867043364595. [DOI] [PubMed] [Google Scholar]
- 40.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- 41.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfgang CD, Essand M, Vincent JJ, Lee B, Pastan I. TARP: a nuclear protein expressed in prostate and breast cancer cells derived from an alternate reading frame of the T cell receptor gamma chain locus. Proc Natl Acad Sci USA. 2000;97:9437–9442. doi: 10.1073/pnas.160270597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henshall SM, Afar DE, Rasiah KK, Horvath LG, Gish K, Caras I, Ramakrishnan V, Wong M, Jeffry U, Kench JG, et al. Expression of the zinc transporter ZnT4 is decreased in the progression from early prostate disease to invasive prostate cancer. Oncogene. 2003;22:6005–6012. doi: 10.1038/sj.onc.1206797. [DOI] [PubMed] [Google Scholar]
- 45.Tomlins SA, Rubin MA, Chinnaiyan AM. Integrative biology of prostate cancer progression. Annu Rev Pathol. 2006;1:243–271. doi: 10.1146/annurev.pathol.1.110304.100047. [DOI] [PubMed] [Google Scholar]
- 46.Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM, Abate-Shen C. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2002;99:2884–2889. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdulkadir SA, Magee JA, Peters TJ, Kaleem Z, Naughton CK, Humphrey PA, Milbrandt J. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol Cell Biol. 2002;22:1495–1503. doi: 10.1128/mcb.22.5.1495-1503.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.