Summary
Two MOB1 genes, MOB1-A and MOB1-B, were identified in Trypanosoma brucei. MOB1-A of T. brucei was shown to form a complex with TbPK50, a functional homologue of the Schizosaccharomyces pombe protein kinase Orb6, and immune precipitated MOB1-A exhibited histone H1 protein kinase activity. MOB1-A and TbPK50 were also shown to bind p12cks1, a cyclin-dependent kinase accessory protein. Immune fluorescence of epitope-tagged MOB1-A and MOB1-B in bloodstream form trypanosomes showed they had a punctate distribution all through the cell cytoplasm and were excluded from the nucleus throughout the cell cycle. Using RNA interference, MOB1 was shown to be essential in both bloodstream and procyclic life cycle stages. In the bloodstream form, RNAi of MOB1 resulted, after 8 hours, in a significant increase in post-mitotic cells, the majority of which had a visible cleavage furrow. This was followed by the appearance of cells with abnormal complements of nuclei and kinetoplasts, often with the number of nuclei exceeding the number of kinetoplasts. Thus, downregulation of MOB1 in the bloodstream form results in a delay in cytokinesis, and leads to a de-regulation of the cell cycle, possibly through an inhibitory effect on kinetoplast replication. In contrast, downregulation of MOB1 in the procyclic form appears to impede the accuracy of cytokinesis, by allowing mis-positioning of the cleavage furrow and inappropriate cytokinesis. Unlike its counterpart in budding yeast, T. brucei MOB1 does not appear to be required for mitotic exit.
Introduction
The cell division cycle of Trypanosoma brucei, an African trypanosome, displays several unusual features compared to other eukaryotes (McKean, 2003). Discrete cell cycle phases exist for the nucleus and the kinetoplast (an organelle containing the DNA of the single mitochondrion) (Woodward et al., 1990), thus making it possible to determine the cell cycle phase of individual cells (Sherwin et al., 1989). The first detectable events in the procyclic form (insect stage) trypanosome cell cycle are the elongation and maturation of the pro-basal body, and the nucleation of a new flagellum during G1. Kinetoplast S phase commences just prior to nuclear S phase, and is substantially shorter. A tripartite attachment complex connects each kinetoplast to a basal body, ensuring that when the basal bodies separate early in G2, the kinetoplasts also segregate (Ogbadoyi et al., 2003). Mitosis then ensues, but without chromatin condensation (Galanti et al., 1998) or nuclear envelope break down (Ogbadoyi et al., 2000). A lateral stacking model has been proposed to account for chromosome segregation (Gull et al., 1998). In the procyclic form, one nucleus migrates to position itself between the two kinetoplasts in order to attain the required symmetry for cell division, although this is not seen in the bloodstream (mammalian infective) form (Hammarton et al., 2003b). Cytokinesis follows mitosis, with cleavage furrow ingression occurring unidirectionally from the anterior to posterior end along the longitudinal axis of the cell. Entry to cytokinesis may be more dependent on kinetoplast division and segregation than on mitosis (Ploubidou et al., 1999), but the flagellum is also important for this event (Moreira-Leite et al., 2001; LaCount et al., 2000; LaCount et al., 2002; Kohl et al., 2003). The flagellum is attached for most of its length to the cell body by the Flagellum Attachment Zone, which is thought to provide the necessary structural information to ensure correct positioning of the cleavage furrow (Robinson et al., 1995; Kohl et al., 2003).
Trypanosomes appear to lack some key eukaryotic cell cycle checkpoints, most notably the mitosis to cytokinesis checkpoint in the procyclic form (Ploubidou et al., 1999; Hammarton et al., 2003a; Hammarton et al., 2003b; Li et al., 2003). In this life cycle stage, inhibition of mitosis (e.g. through RNAi of the mitotic cyclin, CYC6 (Hammarton et al., 2003a; Li et al., 2003), or through incubation with the anti-microtubule agent, rhizoxin (Ploubidou et al., 1999)) does not inhibit cytokinesis, with the end result that a cell with 1 nucleus (with replicated DNA) and 2 kinetoplasts (1N*2K cell, where the asterisk represents replicated DNA) divides to give a 1N*1K daughter and a zoid (0N1K). In the bloodstream form, inhibition of mitosis does prevent cytokinesis, but not subsequent rounds of nuclear or kinetoplast S phase.
Cell division in T. brucei is regulated, as in other eukaryotes, by the action of cyclin-dependent kinases (Mottram et al., 1995; Hammarton et al., 2003b; Klingbeil et al., 2002; Tu et al., 2004). Here, we extend our knowledge of cell cycle regulators in T. brucei by investigating the role of MOB1, a member of the Mob protein family conserved in plants, animals and fungi. Mob proteins can be subdivided into Mob1 and Mob2 type proteins based on primary amino acid sequence (Stavridi et al., 2003). Mob proteins bind to and activate a protein kinase partner of the AGC group (NDR subfamily). Both cyclins and Mob proteins exhibit a tertiary structure that is rich in alpha-helices, but adopt distinct folds (Stavridi et al., 2003; Ponchon et al., 2004). Mob1 proteins possess an evolutionarily-conserved surface exhibiting a strong negative electrostatic potential which, in the case of human Mob1A and Schizosaccharomyces pombe Mob1, have been shown to interact with basic regions conserved at the N-termini of their partner kinases, NDR and Sid2 respectively (Ponchon et al., 2004; Stavridi et al., 2003; Bichsel et al., 2004; Hou et al., 2004). Binding of a Mob protein to its partner protein kinase is thought to induce a conformational change that ultimately stimulates its kinase activity (Komarnitsky et al., 1998; Mah et al., 2001; Hou et al., 2000; Bichsel et al., 2004; Devroe et al., 2004; Hou et al., 2004).
Mob1 is required for the initiation of cytokinesis (Luca et al., 2001; Salimova et al., 2000; Hou et al., 2000). In S. pombe, Mob1/Sid2 localise to the spindle pole bodies (SPBs) at mitosis but show a transient localisation to the cell division site during medial ring constriction and septation, suggesting that Mob1/Sid2 transduce the signal for initiation of cell division from the SPBs to the cell division site (Sparks et al., 1999; Hou et al., 2000; Salimova et al., 2000). In Saccharomyces cerevisiae, MOB1 with its partner kinase, DBF2, are localised to the SPBs for most of the cell cycle, but during late mitosis, localise to the bud neck (Frenz et al., 2000; Luca et al., 2001). S. cerevisiae MOB1 is also required for maintenance of ploidy (Luca et al., 1998) and for mitotic exit as part of the mitotic exit network (MEN) signalling cascade that results in the inactivation of mitotic cyclin-depdendent kinases (CDKs) (Luca et al., 1998; Luca et al., 2001). The yeast Mob2/MOB2 proteins are required for polarised cell growth and interact with the DBF2-related kinases Orb6 (Schizosaccharomyces pombe) and CBK1 (Saccharomyces cerevisiae) (Colman-Lerner et al., 2001; Weiss et al., 2002; Hou et al., 2003; Schneper et al., 2004). Schizosaccharomyces pombe Mob2 also regulates the onset of mitosis (Hou et al., 2003).
We have identified two MOB1 genes in T. brucei and have investigated their role in trypanosomes using RNA interference (RNAi). We show that MOB1 is required for cytokinesis, but not mitotic entry or exit in bloodstream and procyclic life cycle stages. We demonstrate that in the bloodstream form, epitope-tagged MOB1-A and MOB1-B have a punctate cytoplasmic localisation, and are excluded from the nucleus throughout the cell cycle. Additionally, we show that in procyclic trypanosomes, MOB1-A interacts with the NDR-family protein kinase TbPK50, which is a functional homologue of S. pombe Orb6.
Results
Cloning Trypanosoma brucei MOB1
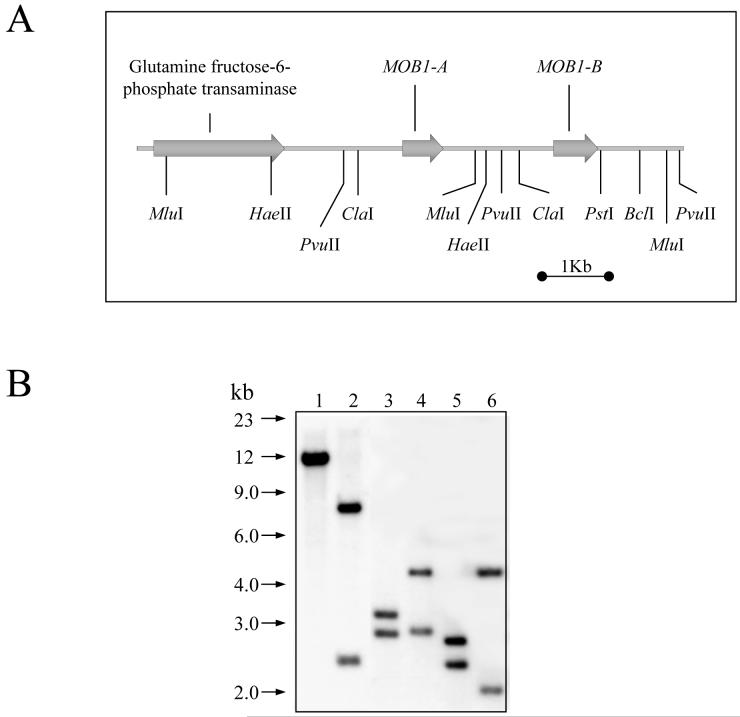
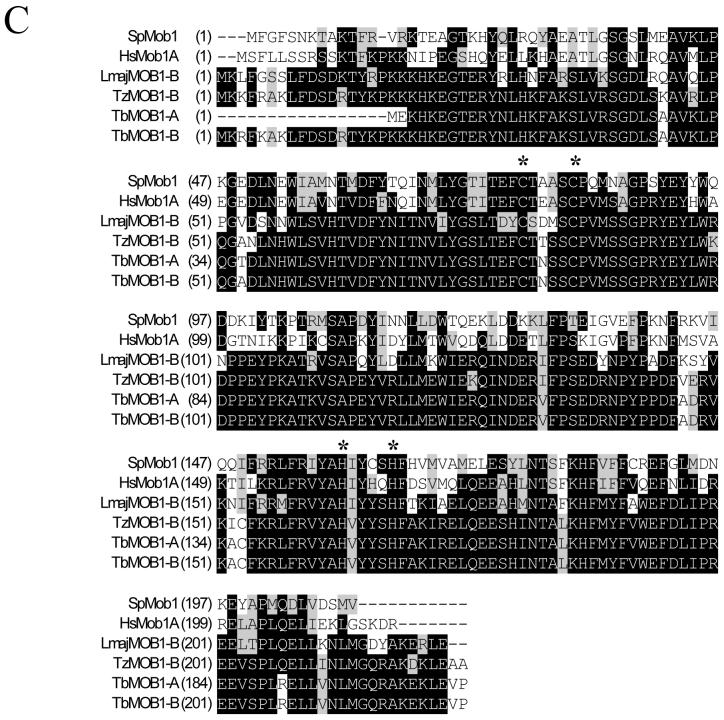
A clone containing a partial MOB1 sequence was isolated during a PCR screen for differentially expressed genes following concanavalin A treatment of procyclic T. brucei (Welburn et al., 1999). Southern blotting of T. brucei DNA using a MOB1 probe (Figure 1B), in conjunction with an analysis of clones isolated from a genomic library and sequence data subsequently published by the T. brucei genome sequencing projects (www.genedb.org), indicated that there are two MOB1 genes (MOB1-A and MOB1-B) in tandem (Figure 1A). The MOB1-A and MOB1-B proteins encoded by these genes (predicted molecular masses of 24.7 and 26.7 kDa respectively) exhibit significant sequence similarity to MOB1 proteins from other eukaryotes (Figure 1C), and are identical except at the N-terminus, where MOB1-B has an 18 amino acid extension relative to MOB1-A. MOB1-B is highly conserved amongst the kinetoplastids, whereas no MOB1-A homologues could be identified in either the Leishmania major or Trypanosoma cruzi genomes. Further, no trypanosome MOB2 homologues could be identified. T. brucei MOB1-B shares 75.6% identity with L. major MOB1-B and 92.4% identity with T. cruzi MOB1-B. All of the kinetoplastid MOB1 proteins contain the conserved cysteine and histidine residues (Figure 1C) that in human Mob1A have been shown to chelate the zinc atom that stabilises the four helix bundle (Stavridi et al., 2003). Additionally, analysis of the predicted secondary and tertiary structure of the T. brucei MOB1 proteins using the 3D-PSSM protein fold recognition (threading) server at http://www.sbg.bio.ic.ac.uk/~3dpssm/ (Kelley et al., 2000) revealed that they are likely to adopt a similar fold to human Mob1A (data not shown).
Figure 1.
Analysis of the MOB1 gene and protein sequences. A: The genomic organisation of the MOB1 genes on chromosome VII. Grey arrow: open reading frame. The positions of cleavage sites for restriction endonucleases used in (B) are given. B: Southern blot of genomic DNA, probed with open reading frame of MOB1. The DNA in lanes 1-6 was digested with Bcl I, Cla I, Hae II, Mlu I, Pvu II, Mlu I + Pst I respectively. C: Alignment of MOB1 protein sequences. The protein sequences of MOB1 homologues from T. brucei (TbMOB1-A and TbMOB1-B), T. cruzi (TzMOB1-B), L. major (LmajMOB1-B), Homo sapiens (HsMob1A) and S. pombe (SpMob1) are aligned. Identical residues are shaded black while conserved residues are shaded grey. Asterisks indicate the positions of the cysteine and histidine residues that tetrahedrally coordinate the zinc atom. Numbers refer to the amino acid residue.
Biochemical studies of MOB1
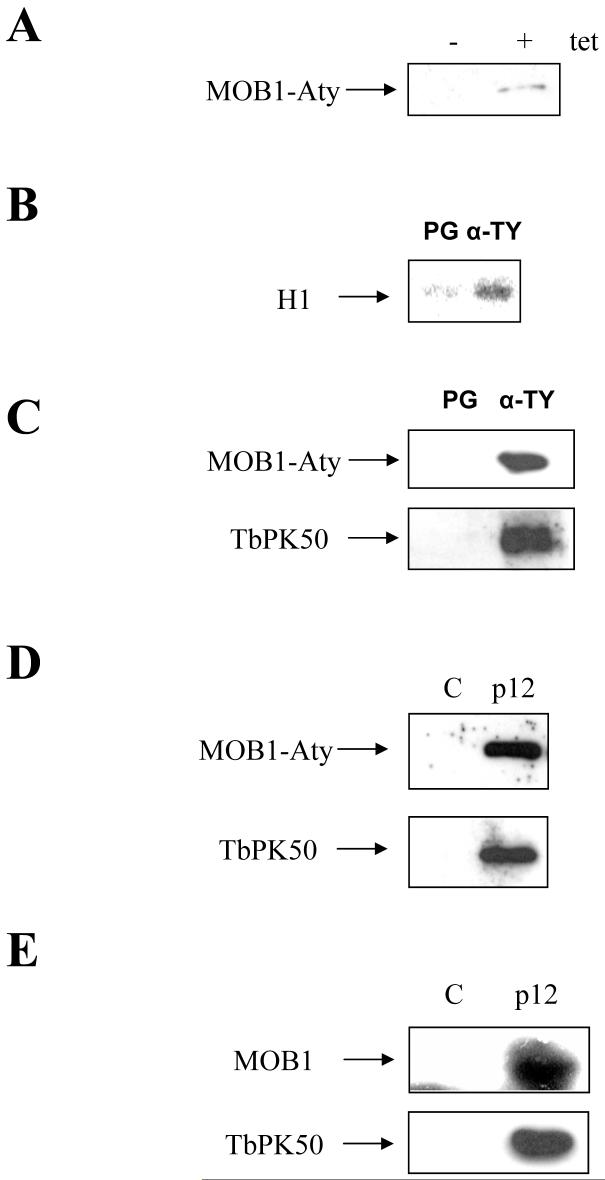
To facilitate biochemical studies on MOB1, a construct allowing tetracycline-inducible expression of TY-1-tagged MOB1-A (MOB1-Aty) was generated and transfected into the procyclic cell line EATRO 795 pHD449. Tetracycline-induced expression of MOB1-Aty was confirmed by Western blotting of cell lysates with the anti-TY antibody (Figure 2A). To determine whether MOB1 interacts with an active protein kinase, MOB1-Aty was precipitated from procyclic cell lysates with anti-TY antibody-conjugated protein G beads and assessed for protein kinase activity. An increased histone H1 kinase activity relative to unconjugated protein G beads was detected (Figure 2B). Yeast and mammalian MOB1 proteins interact with protein kinases of the AGC group, NDR family. Only two NDR-like protein kinases are evident in the T. brucei genome. One, TbPK50, was previously shown to complement a fission yeast Orb6 mutant (García-Salcedo et al., 2002). The other, PK4 (accession number AJ245823), was demonstrated to be essential in bloodstream form T. brucei, but otherwise has not been studied in detail (C. Schulte zu Sodingen and M. Boshart, pers. comm.). Western blotting showed both TbPK50 and MOB1-Aty bound to anti-TY antibody conjugated beads, but not to unconjugated protein G beads following incubation of the beads with MOB1-Aty-expressing cell lysates (Figure 2C), indicating that MOB1-Aty can interact in a complex with TbPK50 in vivo. Next, to further examine the properties of the MOB1-Aty-associated protein kinase complex, it was investigated whether the complex could bind to leishmanial p12cks1 beads as is the case with cyclin-CDK complexes of T. brucei (Van Hellemond et al., 2000; Hammarton et al., 2003a). p12cks1 beads are a commonly used in vitro tool for analysing trypanosome cyclin-CDK complexes and it was therefore desirable to know whether the MOB1-A:PK50 complex could also bind. Western blotting revealed that both MOB1-Aty and TbPK50 bound to p12cks1 but not to control beads (Figure 2D) in cells expressing MOB1-Aty. To confirm that these findings were not an artefact of MOB1-Aty expression, the experiment was repeated with wild type EATRO 795 cell lysates. Western blotting with affinity-purified anti-MOB1 antiserum and with anti-TbPK50 antiserum revealed that both native MOB1 and TbPK50 from wild type lysates bound p12cks1 beads (Figure 2E).
Figure 2.
Biochemical analysis of MOB1-A in procyclic cells. A: Western blot of cell extracts from procyclic cells expressing MOB1-Aty, +/- induction with tetracycline, probed with anti-TY antibody. B: Autoradiograph of histone H1 kinase assays of complexes bound to protein G (PG) and anti-TY (α-TY) antibody-conjungated beads following incubation of beads with MOB1-Aty-expressing cell lysates. C: Western blots of protein G and anti-TY antibody-conjugated beads following incubation with MOB1-Aty-expressing cell lysates, probed with anti-TY (upper panel) and anti-TbPK50 (lower panel) antibodies. D: Western blots of control and p12cks1 beads incubated with MOB1-Aty cell lysates (+ tetracycline), probed with anti-TY (upper panel) and anti-TbPK50 (lower panel) antibodies. E: Western blots of control and p12cks1 beads incubated with EATRO 795 wild type cell lysates, probed with anti-MOB1 (upper panel) and anti-TbPK50 (lower panel) antibodies. tet: tetracycline.
Localisation of MOB1
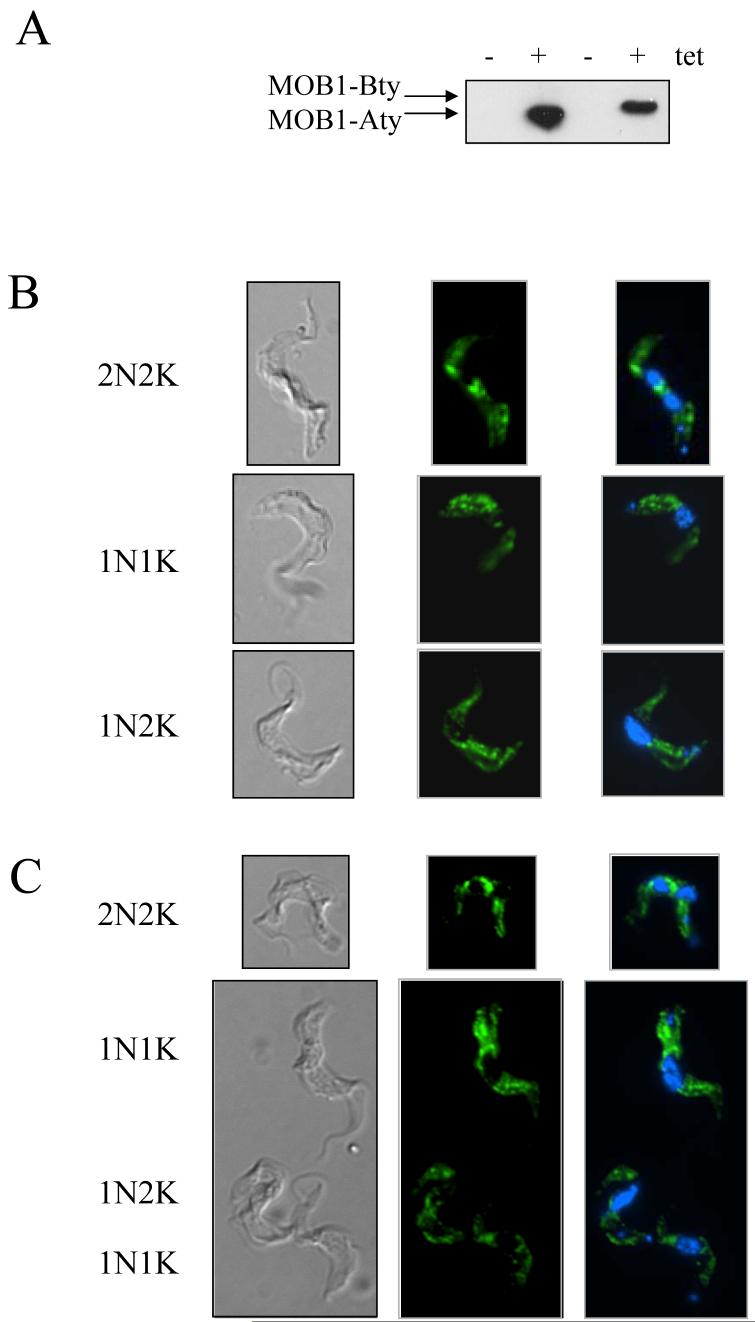
Bloodstream form cell lines expressing MOB1-Aty and MOB1-Bty were generated and the localisation of the two MOB1 proteins in the trypanosome investigated. Tetracycline-induced expression of MOB1-Aty and MOB1-Bty was confirmed by Western blotting (Figure 3A) and the cells were examined by immune fluorescence using the anti-TY antibody (Figure 3B and 3C). Both MOB1-Aty and MOB1-Bty showed similar punctate distributions throughout the cytoplasm, but were excluded from the nucleus at all cell cycle stages, as shown by monitoring the number of nuclei and kinetoplasts present per cell by co-staining with DAPI. Specific staining of particular organelles or structures such as the basal bodies or the cytokinesis furrow was not observed. Control slides processed with secondary antibody alone showed negligible fluorescence (data not shown).
Figure 3.
Immune fluorescence of MOB1 in bloodstream form cells. A: Western blot of cell extracts from bloodstream form cells expressing MOB1-Aty (left two lanes) and MOB1-Bty (right two lanes), +/- induction with tetracycline, probed with anti-TY antibody. B: Images of cells induced to express MOB1-Aty, co-stained with anti-TY antibody (green) and DAPI (blue). Examples of cells from different cell cycle stages are shown. From left to right: DIC image, anti-TY, anti-TY and DAPI overlay. C: Images of cells induced to express MOB1-Bty. Order as for (B). tet: tetracycline; N: nucleus; K: kinetoplast.
MOB1 RNAi in bloodstream form trypanosomes
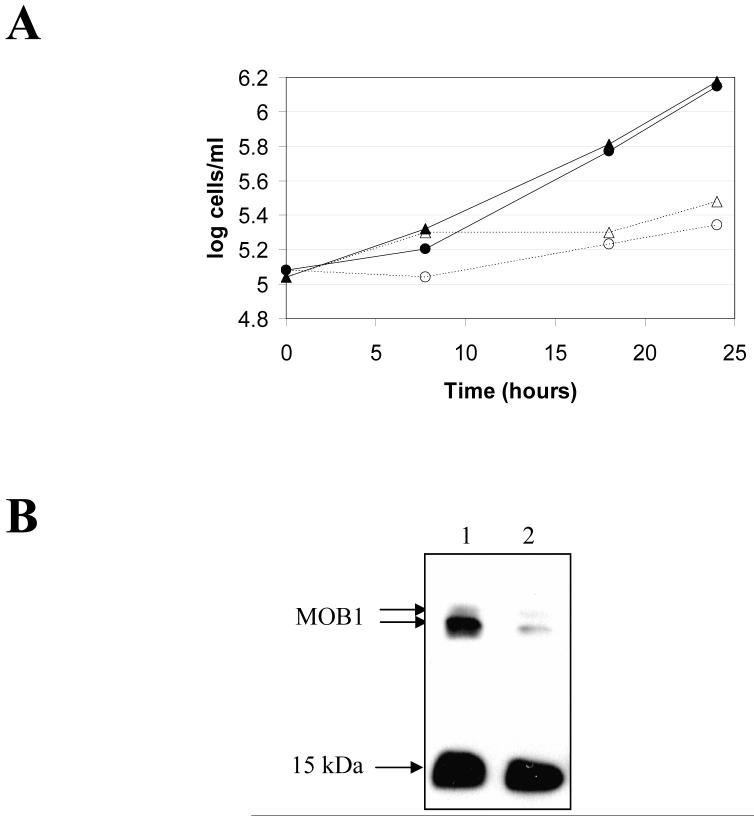
An RNAi approach was taken to investigate the role of MOB1 in vivo. A fragment of MOB1 was cloned into vector p2T7ti and transfected into the T. brucei bloodstream from 427 pLew13 pLew90-6 cell line. Two independent clones were selected, and growth curves generated in the absence or presence of tetracycline, which induces expression of the MOB1 RNAi (Figure 4A). Since the fragment used for RNAi is present in both MOB1-A and MOB1-B, induction of the RNAi would be expected to knockdown expression of both genes. Induction of MOB1 RNAi resulted in a growth arrest in bloodstream form cells 18-24 hours post-induction. Both clones had comparable phenotypes, so data for one clone only is presented. Western blotting of RNAi induced cell lysates with the anti-MOB1 antibody showed a significant decrease in MOB1 protein levels relative to uninduced cell lysates (Figure 4B). Equal loading was confirmed by virtue of the anti-MOB1 antibody exhibiting cross-reactivity to an unknown 15 kDa protein.
Figure 4.
MOB1 RNAi in bloodstream form cells. A: Growth curves. Filled triangles: clone 1 − tetracycline; open triangles; clone 1 + tetracycline; filled circles: clone 2 − tetracycline; open circles: clone 2 + tetracycline. B: Western blot of cell extracts of clone 1 probed with anti-MOB1 antibody following induction of RNAi for 24 hours. Lane 1: uninduced; lane 2: induced.
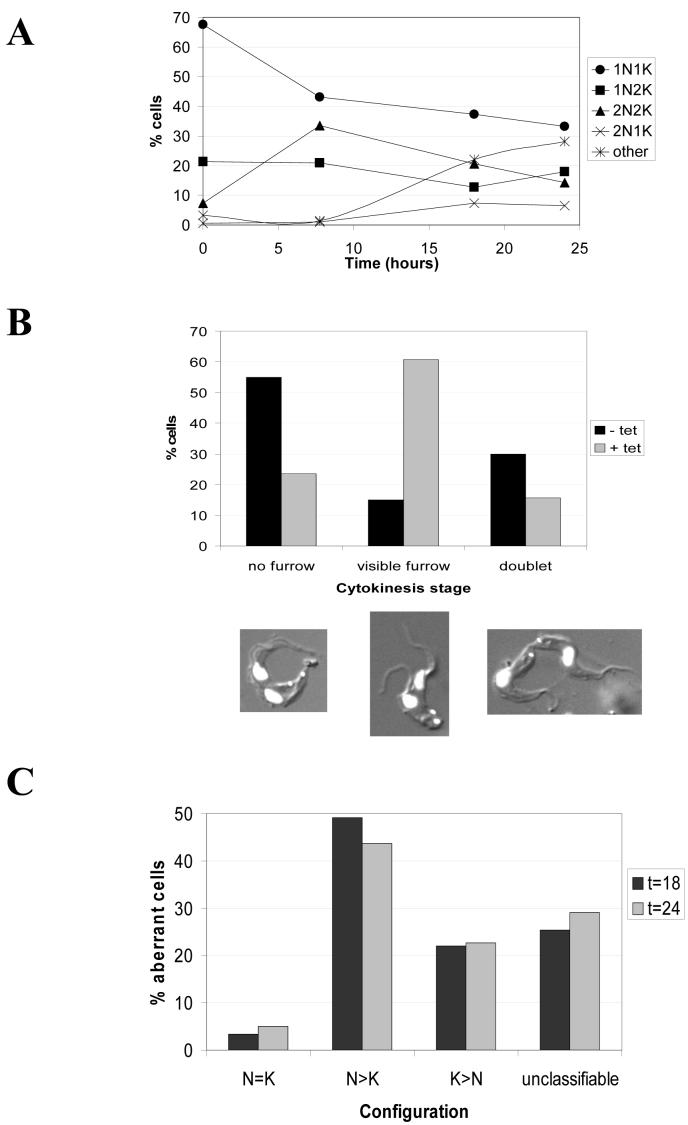
To examine the phenotype of MOB1 RNAi cells, nucleus (N) and kinetoplast (K) configurations of cells were monitored over time (Figure 5). Initially, a significant increase in post-mitotic cells (2N2K configuration) was observed, with a corresponding decrease in cells exhibiting other normal configurations (1N1K and 1N2K) such that by 8 hours post-induction, 2N2K cells comprised approximately a third of the population (Figure 5A). Cells exhibiting aberrant N-K configurations were not observed at 8 hours post-induction, but thereafter an increase in aberrant cells occurred such that they comprised around 35% of the population by 24 hours post-induction.
Figure 5.
Analysis of nucleus and kinetoplast configurations of MOB1 bloodstream form RNAi cells, clone 1. A: N-K configurations over time following induction of RNAi as revealed by DAPI staining. B: Classification of post-mitotic cells for stage of cytokinesis. C: Analysis of abnormal N-K configurations in induced populations at 18 and 24 hours post-induction. tet: tetracycline.
Cells with a 2N2K configuration from uninduced (n=100) and induced (n=200; T=8 hours) populations were scored according to whether they had not yet commenced cytokinesis (no cleavage furrow visible), were midway through cytokinesis (cleavage furrow visible), or were in the latter stages of cytokinesis (doublets: cleavage furrow no longer present, but cells still joined at their posterior ends (see examples of cells in Figure 5B). In uninduced MOB1 RNAi cells, the majority of 2N2K cells (approximately 85%) had either not commenced cytokinesis or were in the latter stages of cytokinesis, with only about 15% cells having a visible cleavage furrow. In contrast, following induction of MOB1 RNAi, just over 60% of the accumulating 2N2K cells had a visible cleavage furrow, indicating that there was a delay in furrow ingression in these cells. A complete block in cytokinesis was not observed, as significant numbers of 1N1K and 1N2K cells were present still in the population 24 hours post-induction.
Within the subset of cells displaying an abnormal N-K configuration at 18 and 24 hours post-induction, many variations of N-K configuration were observed. The most abundant abnormal configuration was 2N1K, comprising 8% of the total cell population at 18 and 24 hours post-induction (Figure 5A). No other configurations were consistently observed in large numbers, and to aid interpretation of these results, abnormal cells were grouped according to the relative numbers of N and K in each cell (Figure 5C). At both 18 and 24 hours post-induction, just under 30% of the abnormal cells had kinetoplast numbers exceeding or equalling the number of nuclei, which would be expected if cells were merely inhibited in cell division, but DNA replication and organelle segregation proceeded unimpeded. Instead, 40-50% of the abnormal cells had more nuclei than kinetoplasts, indicating some degree of inhibition of kinetoplast replication following the initial delay in cytokinesis. It was not possible to classify the N-K configuration of some cells due to the severity of abnormality.
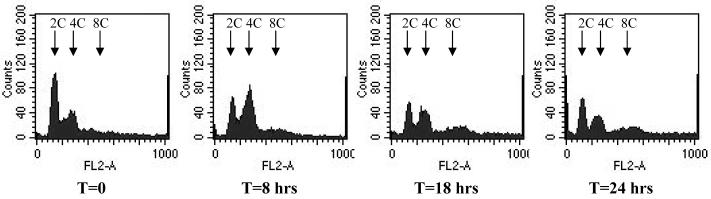
The ploidy of MOB1 RNAi bloodstream form cells was investigated using flow cytometry (Figure 6). At 8 hours post-induction, a significant increase in cells with 4C DNA content (where C represents haploid DNA content) was detected, matching the increase in post-mitotic cells observed by microscopy. The 4C peak decreased at later time points, and a peak representing cells with 8C DNA content emerged, consistent with the aberrant N-K distributions seen by microscopy, and indicating that re-replication of DNA had occurred in a subset of cells within the population.
Figure 6.
Flow cytometry of bloodstream form MOB1 RNAi cells. Flow cytometry profiles are given for cells over time following induction of the RNAi. The DNA content of each peak is indicated by the arrows.
MOB1 RNAi in procyclic form trypanosomes
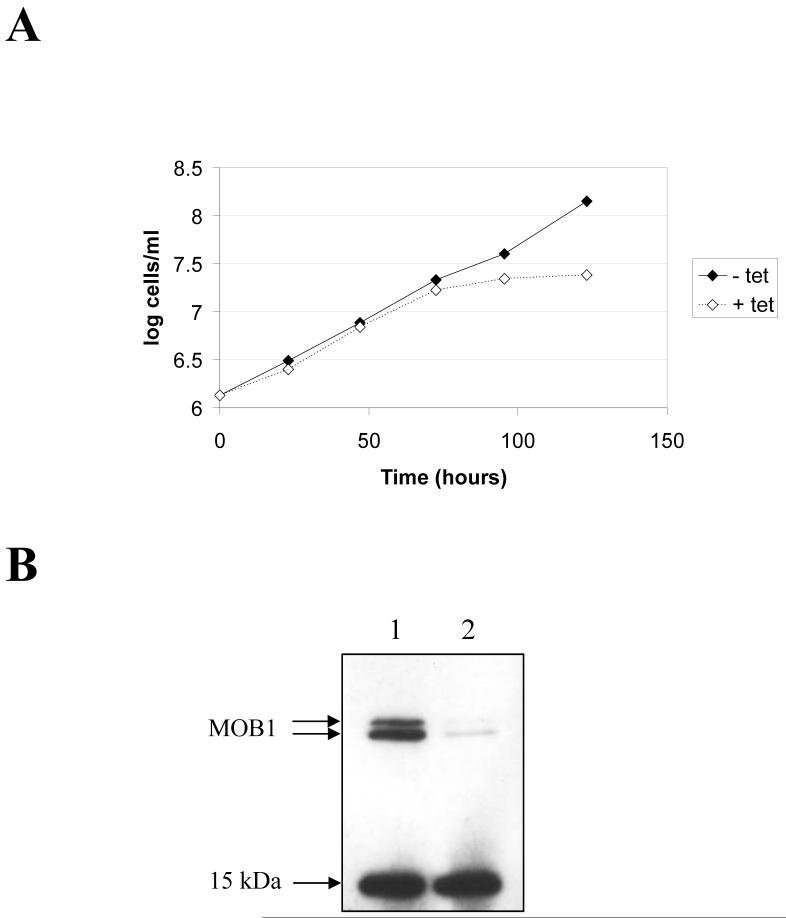
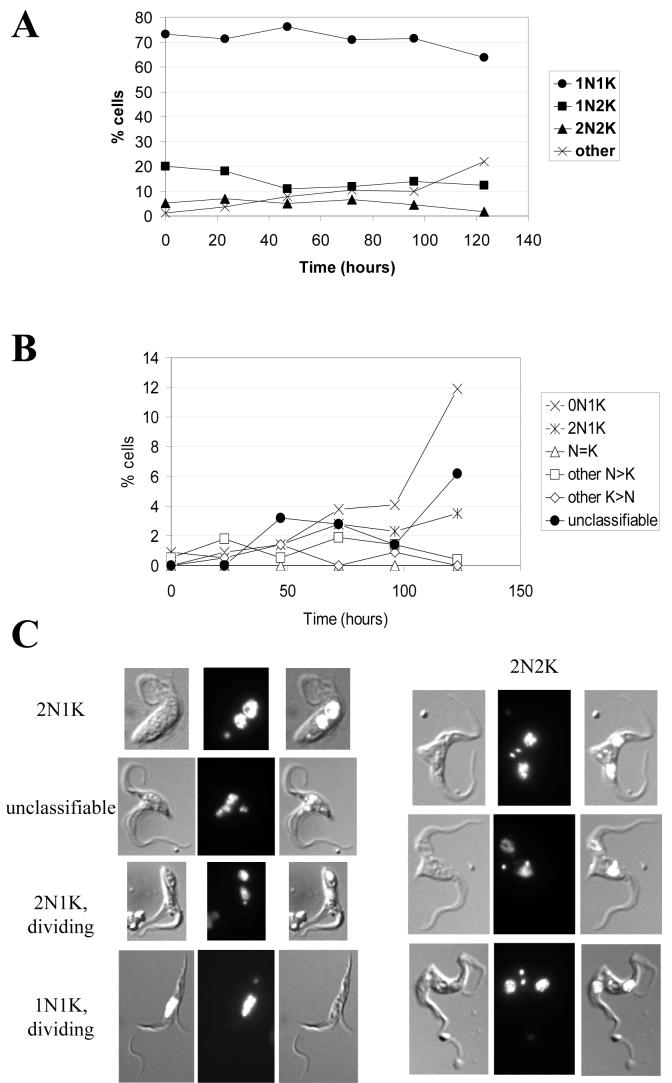
In the procyclic form, induction of MOB1 RNAi led to a growth arrest 72-96 hours post-induction (Figure 7A), which was accompanied by a significant decrease in MOB1 protein levels (Figure 7B). No increase in post-mitotic (2N2K) cells was observed (Figure 8), and overall, the changes in N-K configurations were more subtle than those observed in the bloodstream form. Nevertheless, by 120 hours post-induction, over 20% cells exhibited an abnormal configuration (Figure 8A). The two most dominant abnormal configurations were zoids (0N1K) and 2N1K cells (Figure 8B), which comprised up to 12% and 5% of the total cell population respectively. The configurations of the majority of the remaining abnormal cells were unclassifiable. Some 2N2K cells (up to 1% of the total cell population, but up to 20% of the 2N2K population) were observed to have commenced cytokinesis, since the anterior ends of mother and daughter were separated, but appeared to no longer display a cytokinesis furrow (Figure 8C), suggesting a stalling or even a block in the latter stages of cytokinesis occurred in a subset of post-mitotic cells. In some 2N2K cells it appeared that the furrow was mis-positioned at the last stages of cytokinesis to result in a 2N2K cell dividing to give a 1N2K and 1N0K daughters (Figure 8C, bottom 2N2K cell). Additionally, examples of cells commencing inappropriate cytokinesis could be found, such as the 2N1K and 1N1K cells undergoing cytokinesis shown in Figure 8C. Flow cytometry of MOB1 RNAi procyclic cells revealed no increase in ploidy, but a peak of low fluorescence, representing the zoids, was visible in induced populations (Figure 9).
Figure 7.
MOB1 RNAi in procyclic cells. A: Growth curves. Filled diamonds: clone 1 − tetracycline; open diamonds; clone 1 + tetracycline. B: Western blot of cell extracts of clone 1 probed with anti-MOB1 antibody following induction of RNAi for 24 hours. Lane 1: uninduced; lane 2: induced.
Figure 8.
Analysis of nucleus and kinetoplast configurations of MOB1 procyclic RNAi cells, clone 1. A: N-K configurations over time following induction of RNAi as revealed by DAPI staining. B: Abnormal N-K configurations over time following induction of RNAi as revealed by DAPI staining. C: Images of abnormal RNAi cells (DIC image, DAPI image, merge).
Figure 9.
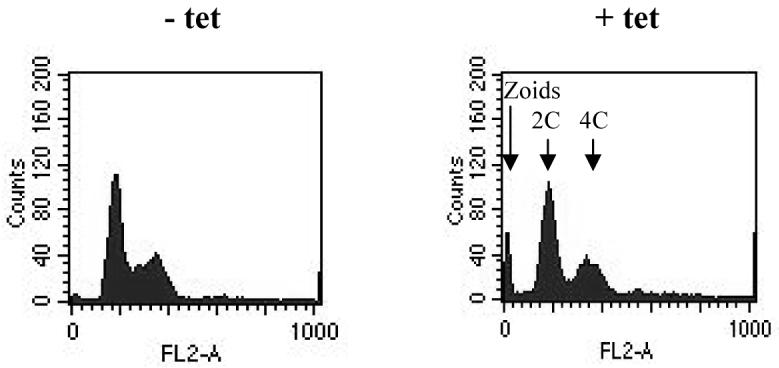
Flow cytometry of procyclic MOB1 RNAi cells. Flow cytometry profiles are given for cells at 123 hours following induction of RNAi. Left: − tetracycline (tet); right: + tetracycline. The DNA content of each peak is shown by the arrows.
Discussion
We have demonstrated that MOB1 is essential for accurate and timely cytokinesis in T. brucei. In the bloodstream form, induction of MOB1 RNAi resulted after 8 hours in a build-up of post-mitotic cells, many of which were in the process of cleavage furrow ingression, indicating a delay in cytokinesis. At later time points a general de-regulation of cell cycle progression occurred, apparently due to impairment of kinetoplast replication and/or segregation (demonstrated by the majority of aberrant cells containing more nuclei than kinetoplasts), in combination with the delay in cytokinesis leading to re-initiation of DNA synthesis in the same cell cycle. In procyclic trypanosomes, RNAi of MOB1 resulted in the accumulation of significant numbers of zoids as well as other abnormal cells, indicating a defect in cytokinesis. Microscopically, examples of cells undergoing inappropriate cytokinesis as well as cells where the cytokinesis furrow appeared to be mis-positioned, likely leading to the production of aberrant daughter cells were observed. Further, the partially separated morphology of some dividing 2N2K cells coupled with a lack of visible further furrow ingression (Fig. 8C), which was not seen in uninduced populations, leads us to speculate that in some cells at least, there was a defect in completion of furrow ingression, resulting in cytokinesis being delayed in its latter stages to some degree. However, unlike with the bloodstream form, no increase in ploidy was observed, arguing that any delay in cytokinesis was slight, or occurred after a point where re-initiation of DNA synthesis in the same cell cycle was possible. This provides further evidence that cytokinesis is far more easily impeded in the bloodstream form than the procyclic form (Hammarton et al., 2003b).
Does MOB1 form part of a cytokinesis checkpoint in trypanosomes? We have provided evidence that cytokinesis is delayed, but not prevented, following MOB1 knockdown. If MOB1 does contribute to the control of a cytokinesis checkpoint, it appears that following activation of the checkpoint, it is eventually over-ridden, although in the procyclic form at least, the accuracy of cytokinesis may then be sacrificed. However, it does seem that T. brucei MOB1 is not involved in exit from mitosis, and the exclusion of both MOB1 proteins from the nucleus throughout the cell cycle is consistent with this hypothesis. In this respect T. brucei MOB1 appears to have functions more in common to Mob1 from Schizosaccharomyces pombe than Saccharomyces cerevisiae. Absence of Mob1 in fission yeast prevents septum formation, but the rest of the yeast cell cycle continues unimpeded to generate multi-nucleated cells. The more complex phenotype seen in trypanosomes following knockdown of MOB1 emphasises the intricacies of coordinating the cell cycles of both the nucleus and kinetoplast with the spatial and structural requirements of cytokinesis in T. brucei. Indeed, the possibility that MOB1 is also involved in the control of kinetoplast replication and/or segregation, at least in the bloodstream form, is intriguing. This phenotype is unusual as a block in cytokinesis caused by RNAi of CYC6 (Hammarton et al., 2003a) or GPI8 (Lillico et al., 2003) in the bloodstream form, or by RNAi of FLA1 (LaCount et al., 2002) in bloodstream or procyclic cells resulted in the production of cells with multiple nuclei and multiple kinetoplasts. Structurally, in procyclic cells, it has been shown that segregation of the kinetoplasts requires microtubule-mediated separation of the basal bodies (Robinson et al., 1991; Robinson et al., 1995), and that the kinetoplasts are linked to the basal bodies through the tripartite attachment complex (Ogbadoyi et al., 2003). However, whereas many of the enzymes involved in the replication of kinetoplast DNA have been described (Klingbeil et al., 2002), little is known about the molecular control of kinetoplast segregation in T. brucei. Since mitosis and kinetoplast replication are separable events, it has been suggested that these processes are controlled by distinct regulatory molecules (Hammarton et al., 2003a) and MOB1 with its partner kinase would appear to be a good candidate for such a regulator. The immune fluorescence data presented here showed MOB1-Aty and MOB1-Bty to be distributed all through the cell body throughout the cell cycle. No specific localisation to the basal bodies or to the cytokinesis furrow was detected, suggesting that trypanosome MOB1 may not be recruited to the site of cell cleavage to initiate cytokinesis and bring about furrow ingression as it is in yeast (Salimova et al., 2000; Luca et al., 2001).
We have demonstrated that MOB1-Aty associates with the TbPK50 protein kinase (an Orb6 functional homologue) in procyclic T. brucei and that immune-precipitated MOB1-Aty from procyclic cell lysates has histone H1 kinase activity. It is likely that the kinase activity detected was that of the MOB1-Aty/TbPK50 protein kinase complex. However, previously, TbPK50 immune precipitated from bloodstream form lysates had autophosphorylation activity, but was unable to phosphorylate a range of kinase substrates including histones (García-Salcedo et al., 2002). It is possible that these immune precipitates (due to differences in assay conditions) contained only monomeric TbPK50 and that association with MOB1 is required for transphosphorylation activity, making TbPK50 a MOB-dependent kinase. Indeed, the Sid2p protein kinase of S. pombe requires disruption of self-association, Mob1 binding and phosophorylation to achieve its full activity (Hou et al., 2003). However, we cannot rule out the possibility that MOB1-Aty forms a complex with another protein kinase that displays histone H1 kinase activity, and indeed a second NDR-family kinase, TbPK4, exists in T. brucei.
In S. pombe, Orb6 complexed with Mob2 rather than Mob1 is required for switching from polarised to bipolar growth during G2 phase, and may mark the new cell end following cell division. In Saccharomyces cerevisiae, the CBK1/MOB2 complex, (the functional homologue of S. pombe Orb6/Mob2), is not only required for maintaining polarised growth during bud morphogenesis and mating projection formation (Weiss et al., 2002), but also to effect the transcription of genes required for cell separation following cytokinesis (Weiss et al., 2002), and may even form part of a cytokinesis checkpoint ensuring the cell wall is not degraded until membrane fission is complete (Colman-Lerner et al., 2001). Orb6 and CBK1 have been shown to interact via the yeast two-hybrid system with Mob1/MOB1 as well as Mob2/MOB2 although it is not clear whether the interaction with Mob1/MOB1 is physiological (Hou et al., 2004). Instead Mob1 is known to associate with the NDR subfamily protein kinases Sid2 and DBF2 in S. pombe and S. cerevisiae respectively. Analysis of the genome suggests that T. brucei has only two NDR subfamily protein kinases, both of which are more similar in sequence to Orb6/CBK1 than to Sid2/DBF2. As T. brucei appears also to lack a MOB2 homologue, the TbPK50/MOB1-A complex in T. brucei might fulfil some of the functions of yeast MOB1 and MOB2. Since TbPK50 was found to functionally complement an S. pombe Orb6 mutant, this demonstrates that it is able to carry out cell polarity regulatory functions, at least in yeast. TbPK50 is only expressed in proliferative life cycle stages in the parasite and displays a cytoplasmic localisation (García-Salcedo et al., 2002), consistent with the notion that it may be required for cytokinesis with MOB1. It is possible that, due to the very precise demands on the trypanosome of the cytokinesis process, coupling of a cytokinesis-regulating protein such as MOB1 to a cell polarity-controlling homologue such as TbPK50 was found to be highly advantageous at some point in evolution.
Finally, Mob1 proteins resemble cyclins in as far as they induce an activatory conformational switch in their kinase partner upon binding (Mah et al., 2001). Recent X-ray and NMR analyses of MOB1 from human and Xenopus respectively suggest that although MOB1 shares a high α-helical content with cyclins, they have distinct folds (Stavridi et al., 2003; Ponchon et al., 2004). In this study we have demonstrated that MOB1-A and TbPK50 bind to the CDK accessory protein p12cks1 in vitro. Previously, only CDK complexes have been demonstrated to bind p12cks1 in trypanosomes, and beads conjugated to p12cks1 and the related protein, p13suc1, have been used as an in vitro tool to distinguish between different CDK complexes (Hammarton et al., 2003a). Thus, these data indicate that p12cks1-binding kinase activity is not solely CDK-derived. However, the in vivo significance of these data is not yet clear. CKS1 is a highly conserved protein that acts as an adaptor in the ubiquitin-mediated degradation of cyclins by the anaphase promoting complex (APC) at the end of mitosis (Patra et al., 1998). CKS1 and other components of the APC are present in T. brucei, and CKS1 has been shown to bind several cyclin-dependent kinase complexes (Van Hellemond et al., 2000; Hammarton et al., 2003a). It is possible, therefore, that the activity of the MOB1/TbPK50 complex is regulated in part by the APC-dependent proteolysis of the MOB1 subunit, which would indicate a greater level of similarity in the mode of activation of MOB-dependent and cyclin-dependent protein kinase complexes than previously recognised.
Experimental procedures
Recombinant DNA techniques
Standard DNA techniques were performed as described by (Sambrook et al., 1989). Plasmid isolation from Escherichia coli, and DNA extraction from agarose gels used Qiaprep and Qiaex kits from Qiagen respectively. DNA sequencing was performed by the MBSU DNA sequencing service (University of Glasgow) using an ABI automatic sequencer.
Cloning MOB1
During a randomly amplified differentially expressed sequences PCR (RADES-PCR) screen for differentially expressed transcripts following treatment of procyclic T. brucei with concanavalin A, clone 1365 (2) was isolated and shown to display significant similarity to MOB1 from Dictyostelium discoideum (Welburn et al., 1999). The 693 bp insert of clone 1365 (2) was used as a probe to screen a T. brucei λgt11 library. A single positive plaque was isolated and the corresponding 4.3 kb insert excised by restriction digestion with Eco RI and subcloned into pUC18 generating plasmid pMOB1. Sequencing of the 4.3 kb insert revealed it to contain most of the MOB1 ORF and 800 bp of 3′ UTR, but sequencing of the 5′ end of the ORF repeatedly failed. 5′ RACE was used to confirm the start codon of the MOB1 ORF. Southern blotting revealed that there were most likely two copies of MOB1 located in close proximity, thus potentially explaining the sequencing problems with pMOB1. Further screening of a trypanosome ILTat 1.2 genomic library using the insert of clone 1365 (2) as probe isolated a clone whereby the two copies of MOB1 could be separated by digestion with Pvu II allowing them to be cloned separately into pGEM-T and sequenced. The sequences for the two genes, named MOB1-A and MOB1-B have been submitted to NCBI (accession numbers AY046919 and AY046920). Subsequent sequencing by the trypanosome genome project (www.genedb.org) has revealed that MOB1-A and MOB1-B exist on chromosome 7 as a tandem array downstream of an ORF encoding for a putative glucosamine-fructose-6-phosphate aminotransferase.
Antibodies and immunoblotting
The AB1 antibody specific for T. brucei MOB1 was generated by immunising a rabbit with the peptide, PEP1 (MEKHKEGTERYNLHK), corresponding to residues 1-15 of MOB1A. In MOB1-B, only amino acids 3-15 of PEP1 are conserved. Anti-MOB1 antibody was affinity purified against a PEP1 column prior to use. The mouse monoclonal antibody BB2 (Bastin et al., 1996) was used as the anti-TY antibody. Anti-TbPK50 antibody (García-Salcedo et al., 2002) was a kind gift from José García-Salcedo. Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane. Western blots were performed as described previously (Mottram et al., 1996) with a 1/20 dilution of anti-TY antibody or a 1/200 dilution of AB1 or a 1/2000 dilution of anti-TbPK50 antibody followed by a 1/20,000 dilution of the appropriate horseradish peroxidase-conjugated secondary antibody. The West-Pico and West-Dura chemiluminescence detection systems (Pierce) were used to visualise antigens.
Transfection of T. brucei
Culture and transfection of T. brucei was carried out essentially as described previously (Vassella et al., 2001). Where necessary, hygromycin was added at a concentration of 50 μgml-1 (procyclic cells) or 5 μgml-1 (bloodstream form cells), G418 at 10 μgml-1 (procyclic cells) or 2.5 μgml-1 (bloodstream form cells), zeocin at 10 μgml-1 (procyclic cells) and phleomycin at 2.5 μgml-1 (bloodstream form cells).
Generation of T. brucei MOB1ty-overexpressing cell lines
To generate TY-tagged MOB1-A, the primers pr5-29 (5′- CGAAGCTTATGGAAAAGCATAAGGAAGGAACTG-3′) and pr5-30 (incorporating sequence coding for the TY tag at the C terminus of MOB1-A) (5′- CGACGCGTTTAGTCAAGTGGATCCTGGTTAGTATGGACCTCCGGAACCTCCAACTTTTCCTTCG-3′) were used to PCR-amplify MOB1-Aty from EATRO 795 genomic DNA before cloning the fragment into pGEM-T Easy (Promega) generating pTyMobGEM and sequencing it to confirm correct integration of the TY tag. MOB1-Aty was excised from pTyMobGEM as a Hind III-Mlu I fragment and subcloned into Hind III-Mlu I –cut pHD675 (Biebinger et al., 1997) to generate pGL850. To generate TY-tagged MOB1-B, MOB1-B was PCR-amplified from EATRO 795 genomic DNA using the primers OL1533 (5′- GGAGAAGCTTATGAAGCGTTTTAAGGCAAAG-3′) and OL1534 (5′- CCACCTCGAGTTACGGAACCTCCAACTTTTC-3′) before being cloned into pCR2.1 TOPO (Invitrogen) generating pGL1093 and sequenced to confirm there were no PCR-introduced errors. A Hind III - Bss HII fragment of MOB1-B containing most of the ORF (just lacking the 3′ end) was used to replace the same fragment in pGL850 (containing the 5′ end of MOB1-A), thereby generating pGL1156 (MOB1-Bty in pHD675).
To generate trypanosomes expressing MOB1-Aty or MOB1-Bty, plasmids pGL850 and pGL1156 were linearised by digestion with Not I and transfected into the procyclic cell line EATRO 795 pHD449 (Van Hellemond et al., 2000) or the bloodstream cell line EATRO 2965.3 pHD449 (a kind gift from Dr Keith Matthews). Expression of tagged proteins was induced in mid-log phase cultures by growing cells in medium containing 50 ngml-1 (procyclic cells) or 1 μgml-1 (bloodstream form cells) tetracycline for 24-48 hours. Cells were harvested by centrifugation, washed in phosphate-buffered saline (PBS) or trypanosome dilution buffer (Hammarton et al., 2004) and either snap-frozen in a dry ice/ethanol bath (for immune precipitations and kinase assays) or resuspended in Laemmli buffer (for SDS-PAGE and Western blot analysis).
Immune precipitations and histone H1 kinase assays
To aid immune precipitation of MOB1ty kinase complexes from MOB1ty-expressing cell lines, the BB2 monoclonal antibody was crosslinked to protein G sepharose beads with the crosslinker disuccinimidyl suberate (Pierce), according to the manufacturer’s protocol. Trypanosome S100 lysates (prepared as described previously (Van Hellemond et al., 2000) were incubated with anti-TY or protein G control beads overnight at 4°C, before being extensively washed and assayed for histone H1 kinase activity (Van Hellemond et al., 2000) or resuspended in Laemmli buffer and analysed by Western blotting.
p12cks1 binding assays
Leishmanial p12cks1 protein was coupled to Amino-link beads at a concentration of 5 mgml-1 as described previously (Mottram et al., 1996). Amino-link beads coupled to Tris-HCl pH7.4 to block the reactive sites were used as a negative control. S100 lysates were incubated with 50 μl of control or p12cks1 beads overnight at 4°C. Beads were extensively washed with lysis buffer and resuspended in Laemmli buffer for analysis by immunoblotting.
Generation and induction of RNAi cell lines
For RNAi, the procyclic 427 pLew13 pLew29 and the bloodstream form 427 pLew13 pLew90-6 cell lines (Wirtz et al., 1999) were transfected with pGL646. Plasmid pGL646 was generated by PCR-amplifying a 592 bp fragment of MOB1 (common to both MOB1-A and MOB1-B) with oligonucleotides OL868 (5′- CGGATCCACAACCTCCACAAGTTTGCC-3′) and OL869 (5′- CAAGCTTTTCCTTCGCGCGCTGACC-3′) incorporating Bam HI and Hind III sites respectively, allowing cloning into these sites in vector p2T7ti (LaCount et al., 2000) in place of the GFP insert. Independent clones were generated by limited dilution cloning. To induce RNAi in procyclic forms, the cell line was grown to a density of 0.5-1 × 107 cells ml-1, diluted to give a density of 5 × 105 cells ml-1 and cultured in the presence or absence of 50 ngml-1 tetracycline. Cells were counted daily using a Neubauer improved haemocytometer, and cultures were diluted to 106 cells ml-1 when the density approached 107 cells ml-1. Bloodstream form cells were induced at a density of 105 cells ml-1 with 1 μgml-1 tetracycline, and were diluted back to 105 cells ml-1 whenever the cell density approached 106 cells ml-1.
DAPI staining and flow cytometry
DAPI staining and flow cytometry were carried out exactly as described previously (Hammarton et al., 2003a).
Immune fluorescence
Cells were spread on microscope slides, allowed to air dry and fixed overnight in methanol at −20°C. For immune fluorescence, slides were removed from methanol, allowed to air dry, and cells were then rehydrated in PBS for 5 minutes. Non-specific sites were blocked by incubating slides for 1 hr in PBS containing 5% skimmed milk (w/v). Slides were washed 3 times in PBS, incubated for 1 hr in anti-TY antibody, washed 3 times in PBS and then incubated for 1 hr in Alexa Fluor 488 goat anti-mouse IgG secondary antibody (Invitrogen) diluted 1/200 in PBS-5% skimmed milk (w/v). Following a further 3 washes in PBS, 50 μl DAPI (1 μgml-1) containing 0.5% 1,4-diazabicyclo[2.2.2]octane as anti-fading agent was added to each slide and spread by addition of a coverslip. Slides were examined on a Zeiss Axioplan microscope, and images taken with a Hamamatsu ORCA-ER digital camera. A series of images in the z plane were taken at 0.2 μm intervals and were deconvolved and processed using Openlab version 4.0.2beta software.
Acknowledgements
This work was funded by the Medical Research Council and the Wellcome Trust. We thank José García-Salcedo for providing us with the anti-TbPK50 antibody.
References
- Bastin P, Bagherzadeh A, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- Bichsel SJ, Tamaskovic R, Stegert MR, Hemmings BA. Mechanism of activation of nuclear Dbf2-related (NDR) kinase by the hMOB1 protein. J. Biol. Chem. 2004;279:35228–35235. doi: 10.1074/jbc.M404542200. [DOI] [PubMed] [Google Scholar]
- Biebinger S, Wirtz E, Lorenz P, Clayton CE. Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 1997;85:99–112. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- Devroe E, Erdjument-Bromage H, Tempst P, Silver PA. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J Biol. Chem. 2004;279:24444–24451. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- Frenz LM, Lee SE, Fesquet D, Johnston LH. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J. Cell Sci. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- Galanti N, Galindo M, Sabaj V, Espinoza I, Toro GC. Histone genes in trypanosomatids. Parasitol. Today. 1998;14:64–70. doi: 10.1016/s0169-4758(97)01162-9. [DOI] [PubMed] [Google Scholar]
- García-Salcedo JA, Nolan DP, Gijón P, Gómez-Rodriguez J, Pays E. A protein kinase specifically associated with proliferative forms of Trypanosoma brucei is functionally related to a yeast kinase involved in the co-ordination of cell shape and division. Mol. Microbiol. 2002;45:307–319. doi: 10.1046/j.1365-2958.2002.03019.x. [DOI] [PubMed] [Google Scholar]
- Gull K, Alsford S, Ersfeld K. Segregation of minichromosomes in trypanosomes: implications for mitotic mechanisms. Trends Microbiol. 1998;6:319–323. doi: 10.1016/s0966-842x(98)01314-6. [DOI] [PubMed] [Google Scholar]
- Hammarton TC, Clark J, Douglas F, Boshart M, Mottram JC. Stage-specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J. Biol. Chem. 2003a;278:22877–22886. doi: 10.1074/jbc.M300813200. [DOI] [PubMed] [Google Scholar]
- Hammarton TC, Engstler M, Mottram JC. The Trypanosoma brucei cyclin, CYC2, is required for cell cycle progression through G1 phase and maintenance of procyclic form cell morphology. J. Biol. Chem. 2004;279:24757–24764. doi: 10.1074/jbc.M401276200. [DOI] [PubMed] [Google Scholar]
- Hammarton TC, Mottram JC, Doerig CD. The cell cycle of parasitic protozoa: potential for chemotherapeutic exploitation. Prog. Cell Cycle Res. 2003b;5:91–101. [PubMed] [Google Scholar]
- Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- Hou MC, Wiley DJ, Verde F, McCollum D. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J Cell Sci. 2003;116:125–135. doi: 10.1242/jcs.00206. [DOI] [PubMed] [Google Scholar]
- Hou MC, Guertin DA, McCollum D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol. Cell. Biol. 2004;24:3262–3276. doi: 10.1128/MCB.24.8.3262-3276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Klingbeil MM, Motyka SA, Englund PT. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol. Cell. 2002;10:175–186. doi: 10.1016/s1097-2765(02)00571-3. [DOI] [PubMed] [Google Scholar]
- Kohl L, Robinson D, Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 2003;22:5336–5346. doi: 10.1093/emboj/cdg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky SI, Chiang YC, Luca FC, Chen JJ, Toyn JH, Winey M, Johnston LH, Denis CL. DBF2 protein kinase binds to and acts through the cell cycle- regulated MOB1 protein. Mol. Cell. Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount DJ, Barrett B, Donelson JE. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J Biol. Chem. 2002;277:17580–17588. doi: 10.1074/jbc.M200873200. [DOI] [PubMed] [Google Scholar]
- LaCount DJ, Bruse S, Hill KL, Donelson JE. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol. Biochem. Parasitol. 2000;111:67–76. doi: 10.1016/s0166-6851(00)00300-5. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang CC. A PHO80-like cyclin and a B-type cyclin control the cell cycle of the procyclic form of Trypanosoma brucei. J. Biol. Chem. 2003;278:20652–20658. doi: 10.1074/jbc.M301635200. [DOI] [PubMed] [Google Scholar]
- Lillico SG, Field MC, Blundell P, Coombs GH, Mottram JC. Essential roles for GPI-anchored proteins in African trypanosomes revealed using mutants deficient in GPI8. Mol. Biol. Cell. 2003;14:1182–1194. doi: 10.1091/mbc.E02-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol. Cell Biol. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca FC, Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah AS, Jang J, Deshaies RJ. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA. 2001;98:7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean PG. Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr. Opin. Microbiol. 2003;6:600–607. doi: 10.1016/j.mib.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Moreira-Leite FF, Sherwin T, Kohl L, Gull K. A trypanosome structure involved in transmitting cytoplasmic information during cell division. Science. 2001;294:610–612. doi: 10.1126/science.1063775. [DOI] [PubMed] [Google Scholar]
- Mottram JC, Grant KM. Leishmania mexicana p12cks1, a functional homologue of fission yeast p13suc1, associates with a stage-regulated histone H1 kinase. Biochem. J. 1996;316:833–839. doi: 10.1042/bj3160833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram JC, Smith G. A family of trypanosome cdc2-related protein kinases. Gene. 1995;162:147–152. doi: 10.1016/0378-1119(95)00350-f. [DOI] [PubMed] [Google Scholar]
- Ogbadoyi E, Ersfeld K, Robinson D, Sherwin T, Gull K. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma. 2000;108:501–513. doi: 10.1007/s004120050402. [DOI] [PubMed] [Google Scholar]
- Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell. 2003;14:1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra D, Dunphy WG. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase-promoting complex at mitosis. Genes Dev. 1998;12:2549–2559. doi: 10.1101/gad.12.16.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploubidou A, Robinson DR, Docherty RC, Ogbadoyi EO, Gull K. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci. 1999;112:4641–4650. doi: 10.1242/jcs.112.24.4641. [DOI] [PubMed] [Google Scholar]
- Ponchon L, Dumas C, Kajava AV, Fesquet D, Padilla A. NMR solution structure of Mob1, a mitotic exit network protein and its interaction with an NDR kinase peptide. J. Mol. Biol. 2004;337:167–182. doi: 10.1016/j.jmb.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell-cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J. Cell Biol. 1995;128:1163–1172. doi: 10.1083/jcb.128.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimova E, Sohrmann M, Fournier N, Simanis V. The S-pombe orthologue of the S-cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J. Cell Sci. 2000;113:1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. In: Molecular cloning: A laboratory manual. Nolan C, editor. Cold Spring Harbor, NY: Cold Spring Laboratory Press; 1989. [Google Scholar]
- Schneper L, Krauss A, Miyamoto R, Fang S, Broach JR. The Ras/protein kinase A pathway acts in parallel with the Mob2/Cbk1 pathway to effect cell cycle progression and proper bud site selection. Eukaryot. Cell. 2004;3:108–120. doi: 10.1128/EC.3.1.108-120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin T, Gull K. The cell-division cycle of Trypanosoma brucei brucei - timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. Lond. Ser. B. 1989;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146:777–790. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavridi ES, Harris KG, Huyen Y, Bothos J, Verwoerd PM, Stayrook SE, Pavletich NP, Jeffrey PD, Luca FC. Crystal structure of a human Mob1 protein: Toward understanding mob-regulated cell cycle pathways. Structure. 2003;11:1163–1170. doi: 10.1016/s0969-2126(03)00182-5. [DOI] [PubMed] [Google Scholar]
- Tu X, Wang CC. The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell-cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J. Biol. Chem. 2004;279:20519–20528. doi: 10.1074/jbc.M312862200. [DOI] [PubMed] [Google Scholar]
- Van Hellemond JJ, Neuville P, Schwartz RJ, Matthews KR, Mottram JC. Isolation of Trypanosoma brucei CYC2 and CYC3 cyclin genes by rescue of a yeast G1 cyclin mutant. Functional characterisation of CYC2. J. Biol. Chem. 2000;275:8315–8323. doi: 10.1074/jbc.275.12.8315. [DOI] [PubMed] [Google Scholar]
- Vassella E, Krämer R, Turner CMR, Wankell M, Modes C, Van den Bogaard M, Boshart M. Deletion of a novel protein kinase with PX and FYVE-related domains increases the rate of differentiation of Trypanosoma brucei. Mol. Microbiol. 2001;41:33–46. doi: 10.1046/j.1365-2958.2001.02471.x. [DOI] [PubMed] [Google Scholar]
- Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 2002;158:885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn SC, Lillico SG, Murphy NB. Programmed cell death in procyclic form Trypanosoma brucei rhodesiense --identification of differentially expressed genes during con A induced death. Mem. Inst. Oswaldo Cruz. 1999;94:229–234. doi: 10.1590/s0074-02761999000200020. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell-cycle of Trypanosoma brucei. J. Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]