Abstract
Background
High-throughput tools for pan-genomic study, especially the DNA microarray platform, have sparked a remarkable increase in data production and enabled a shift in the scale at which biological investigation is possible. The use of microarrays to examine evolutionary relationships and processes, however, is predominantly restricted to model or near-model organisms.
Methodology/Principal Findings
This study explores the utility of Diversity Arrays Technology (DArT) in evolutionary studies of non-model organisms. DArT is a hybridization-based genotyping method that uses microarray technology to identify and type DNA polymorphism. Theoretically applicable to any organism (even one for which no prior genetic data are available), DArT has not yet been explored in exclusively wild sample sets, nor extensively examined in a phylogenetic framework. DArT recovered 1349 markers of largely low copy-number loci in two lineages of seed-free land plants: the diploid fern Asplenium viride and the haploid moss Garovaglia elegans. Direct sequencing of 148 of these DArT markers identified 30 putative loci including four routinely sequenced for evolutionary studies in plants. Phylogenetic analyses of DArT genotypes reveal phylogeographic and substrate specificity patterns in A. viride, a lack of phylogeographic pattern in Australian G. elegans, and additive variation in hybrid or mixed samples.
Conclusions/Significance
These results enable methodological recommendations including procedures for detecting and analysing DArT markers tailored specifically to evolutionary investigations and practical factors informing the decision to use DArT, and raise evolutionary hypotheses concerning substrate specificity and biogeographic patterns. Thus DArT is a demonstrably valuable addition to the set of existing molecular approaches used to infer biological phenomena such as adaptive radiations, population dynamics, hybridization, introgression, ecological differentiation and phylogeography.
Introduction
The development of innovative methods for detecting genetic variation has progressively enhanced the study of evolutionary relationships and processes [1]. High-throughput genomic tools such as the DNA microarray platform have sparked a remarkable increase in data production, leading to new evolutionary insights [2]. Their application is however nearly exclusively restricted to model and near-model organisms [2], [3]. In contrast, the detection of genetic variation in non-model plants and animal—the majority of life on earth—is largely restricted to direct sequencing of previously-identified variable loci or arbitrarily amplified dominant (AAD) markers, especially RAPDs, ISSRs and AFLPs [4].
Diversity Arrays Technology (DArT) was developed to overcome these and other limitations that prevent the application of microarray technology to non-model organisms. DArT is a hybridization-based genotyping technology which is currently implemented on the microarray platform to rapidly and simultaneously identify and type DNA polymorphism [5]. DArT detects primarily dominant markers, mostly resulting from single nucleotide polymorphisms at restriction sites [6] at hundreds to thousands of arbitrary genomic loci [6], [7]. First used to infer the genetic diversity in cultivated varieties of rice (Oryza sativa L.) [8], DArT was subsequently applied to barley (Hordeum vulgare L.) [7], grand eucalyptus (Eucalyptus grandis Hill ex Maiden) [9], cassava (Manihot esculenta Crantz) [10], wheat (Triticum aestivum L.) [11] and validated in the model organism thale cress (Arabidopsis thaliana L. Heynh) [6].
The application of DArT has so far been restricted to detection and mapping of DNA polymorphism in cultivated varieties of agriculturally important angiosperms. While some wild crop relatives have been included in previous DArT studies [10], [12], [13], the potential of DArT has not yet been explored in exclusively wild sample sets, nor extensively examined in a phylogenetic framework. Furthermore, because all existing DArT studies have been applied to flowering plants, its applicability to the rest of the plant kingdom and other organisms requires validation.
To explore whether DArT data can contribute to characterising evolutionary patterns and processes, we selected two lineages of seed-free land plants that provide two distinct opportunities to infer evolutionary hypotheses and detect hybridization by employing DArT: 1) the evolution of substrate specificity in the diploid fern Asplenium viride L. and 2) the phylogeography of the haploid moss Garovaglia elegans Dozy & Molk. Hampe ex Bosch & Sande Lac. These organisms were selected based on accessibility to well-preserved plant material and prior knowledge obtained by studying these organisms using conventional molecular evidence (allozymes and/or DNA sequencing). Furthermore, evolutionary interpretations have been hampered in both organisms by low DNA sequence diversity of a small number of selected loci.
Materials and Methods
Preparation and evaluation of DArT Discovery Libraries
Independent DArT discovery arrays were constructed for two 16-specimen study groups: (1) 14 specimens of the diploid fern Asplenium viride, one specimen of diploid A. trichomanes L. to polarize genetic structuring within A. viride and one specimen of their naturally derived allotetraploid hybrid, A. adulterinum J. Milde [14], and (2) 15 specimens of the haploid moss Garovaglia elegans, collected from various locations in Australia and New Guinea, and one specimen of G. powellii Mitt. for polarization (Tables 1 and 2). At least 1 µg of total genomic DNA was extracted from silica gel desiccated (Asplenium) or air dried (Garovaglia) leaf material using a modification of the standard CTAB procedure [15] as specified in Trewick et al. [16] except that extractions were incubated in 500 µL CTAB buffer, 50 µl sarkosyl and 10 µl proteinase-K and purified by phenol-chloroform extraction. DNA quantity, quality and competence for restriction endonuclease digestion were confirmed by visualizing 2 µL of total genomic DNA alongside 2 µL of MseI restriction digested total genomic DNA by agarose gel electrophoresis and ethidium bromide staining. Genomic representations were produced from a mixture of either Asplenium or Garovaglia DNA samples according to Wenzl et al. [7]. Libraries were prepared as described [7], [8] either directly from PCR amplification products or from normalized amplification products subtracted using a modified version of suppression subtractive hybridization (SSH) [17] as described below. Polymorphic markers detected on the Asplenium and Garovaglia discovery arrays were re-arrayed onto “genotyping arrays” to enable genotyping of the individual samples. The in vivo genomic copy number of each DArT marker was approximated by hybridizing un-amplified metagenomic DNA to each genotyping array.
Table 1. Asplenium samples used in this study with voucher information, collecting locality, substrate specificity, ploidy level and GenBank accession numbers.
| sample | voucher info. | locality | substrate | ploidy | trnL-F | rps4-trnS | pgiC |
| Asplenium viride L. | |||||||
| Avi69 | 69 (BMa) | France | limestone | di- | EF645609 | EF645625 | EF645641 |
| Avi70 | 70C (BMa) | Norway | limestone | di- | EF645602 | EF645618 | EF645634 |
| Avi169 | not vouchered | Norway | serpentine | di- | EF645604 | EF645620 | EF645636 |
| Avi245 | 245(6) (BMa) | Morocco | limestone | di- | EF645608 | EF645624 | EF645640 |
| Avi255 | 255C (BMa) | Canada | limestone | di- | EF645598 | EF645614 | EF645630 |
| Avi268 | 268 (BMa) | Austria | limestone | di- | EF645600 | EF645616 | EF645632 |
| Avi272 | 272C (BMa) | Austria | serpentine | di- | EF645603 | EF645619 | EF645635 |
| Avi281 | 281 (BMa) | Croatia | limestone | di- | EF645601 | EF645617 | EF645633 |
| Avi284a | 284(1) (BMa) | Austria | magnesit | di- | EF645606 | EF645622 | EF645638 |
| Avi284b | 284A(2) (BMa) | Austria | magnesit | di- | EF645607 | EF645623 | EF645639 |
| Avi288b | 288 (BMa) | Switzerland | serp.-lime. cglm. | di- | EF645610 | EF645626 | EF645642 |
| Avi289b | 289 (BMa) | Switzerland | serp.-lime. cglm. | di- | EF645611 | EF645627 | EF645643 |
| Avi291 | 291A (BMa) | UK | serpentine | di- | EF645605 | EF645621 | EF645637 |
| Avi293 | 293A (BMa) | Canada | limestone | di- | EF645599 | EF645615 | EF645631 |
| Asplenium trichomanes L. | |||||||
| Atr120 | 120 (BMa) | Canada | unknown | di- | EF645613 | EF645629 | EF645644 |
| Asplenium adulterinum J. Milde | |||||||
| Aad79 | 79B (BMa) | Austria | serpentine | tetra- | EF645612 | EF645628 | N/Ac |
Unmounted specimens in the Molecular Lab Herbarium.
Avi288 and Avi289 were included in the Asplenium metagenome for the discovery, typing and quantitative evaluation of DArT markers, but were excluded from subsequent analyses due to substrate ambiguity.
As expected for an allopolyploid hybrid, the nuclear locus pgiC exhibits heterozygosity at the DNA sequence level and was thus not submitted to GenBank.
Table 2. Garovaglia samples used in this study with voucher information, collecting locality, substrate specificity, ploidy level and GenBank accession numbers.
| sample | voucher info | locality | trnG |
| Garovaglia elegans (Dozy & Molk.) Bosch & Sande Lac. subsp. dietrichiae (Müll. Hal.) During | |||
| Gel5465 | Newton 5465 (BM1) | Australia | DQ194243 |
| Gel6446 | Newton 6446 (BM1) | Australia | EF551190 |
| Gel6434a | Newton 6434 (BM1) | Australia | not sequenced |
| Gel6485 | Newton 6485 (BM1) | Australia | not sequenced |
| Gel6504 | Newton 6504 (BM1) | Australia | EF551192 |
| Gel6516 | Newton 6516 (BM1) | Australia | EF551196 |
| Gel6520 | Newton 6520 (BM1) | Australia | EF551193 |
| Gel6524 | Newton 6524 (BM1) | Australia | EF551198 |
| Gel6532 | Newton 6532 (BM1) | Australia | EF551191 |
| Gel6547 | Newton 6547 (BM1) | Australia | EF551197 |
| Gel6550 | Newton 6550 (BM1) | Australia | EF551194 |
| Gel6560 | Newton 6560 (BM1) | Australia | EF551195 |
| Garovaglia elegans subsp. elegans (Müll. Hal.) During | |||
| Gel42.588 | Sloover 42.588 (NY) | Papua New Guinea | EF551189 |
| Garovaglia elegans fo. latifolia (E.B. Bartram) During | |||
| Gel40397 | Streimann 40397 (NY) | Papua New Guinea | EF551188 |
| Gel40482 | Streimann 40482 (NY) | Papua New Guinea | EF551187 |
| Garovaglia powellii ssp. muelleri (Hampe) During | |||
| Gpo6496 | Newton 6496 (BM1) | Australia | DQ194245 |
Gel6434 was discovered to be derived from a mixed-taxon cushion containing individuals of both G. elegans ssp. dietrichiae and G. powelli.
Genotyping and analysis of DArT images
The genotypes of all individual Asplenium and Garovaglia samples were scored as absence/presence for 444 and 905 markers, respectively, and formatted as a binary data matrix (Table S1). The reproducibility of the two DArT genotyping arrays was examined by independent assay using the same DNA. The genotyping of genomic representations of individual samples was performed substantially as described [7], with the exception that the polylinker fragment (reference in DArT assay) was labelled with FAM instead of Cy5 dye. Arrays were scanned with 10 µm resolution at 543 nm (Cy3) and 488 nM (FAM) on a LS300 confocal laser scanner (Tecan, Grödig, Austria) as described in Akbari et al. [11]. Array images were analyzed with DArTsoft 7.4 (Diversity Arrays Technology P/L, Canberra, Australia). The program automatically recognized array features using a seeded-region-growth algorithm and reported, for each fluorescent channel, the average and standard deviation (SD) of pixel intensities within and around each array feature, the fraction of saturated pixels within each feature and the number of pixels of each feature, amongst other parameters (C. Cayla, personal communication). Clones with variable relative hybridization intensity across slides were subjected to fuzzy k-means clustering to convert relative hybridization intensities into binary scores (presence vs. absence). The quality of each marker was then determined using several parameters including 1) p-value, the variance of the relative target hybridization intensity between allelic states as a percentage of the total variance, 2) call rate, the percentage of DNA samples with binary (1 or 0) allele calls and 3) reproducibility, the fraction of concordant calls for replicate assays (C. Cayla, personal communication). Samples with cluster membership probability calculated by DArTsoft's clustering algorithm below the threshold of 0.8 were not classified (“X”). The frequencies of Asplenium and Garovaglia DArT markers with at least one X score across the sample set were 178 and 1 respectively. This discrepancy may be explained by differences in ploidy between Asplenium (diploid sporophyte) and Garovaglia (haploid gametophyte) because scoring efficiency would be expected to diminish as a result of intermediate signal intensities corresponding to copy number variation in polyploid samples. Indeed, the majority (56%) of X scores in the Asplenium data set were detected in the genotype of the known allotetraploid F1 hybrid sample of A. adulterinum.
Suppression subtractive hybridization (SSH)
SSH was performed using a modification of the protocol given by Diatchenko et al. [17]. Asplenium viride sample Avi284b and G. elegans ssp. dietrichiae sample Gel6504 were selected arbitrarily as subtraction drivers for the Asplenium and Garovaglia experiments respectively. The following different mixtures (“testers”) of Asplenium and Garovaglia samples were used: Asplenium tester 1 (all A. viride amplification products excluding the driver Avi284b), Asplenium tester 2 (all Asplenium amplification products excluding the driver Avi284b) and Garovaglia tester (all Garovaglia amplification products excluding the driver Gel6504). All Asplenium and Garovaglia tester and driver digestion products were phenol chloroform extracted, isopropanol precipitated and resuspended in deionized water. Digested testers were ligated to adapters with either Core 1 or Core 2, corresponding to the relevant digesting enzymes, in separate reactions (Table 3). Subtraction was carried out in either one or two stages. 600 ng driver was mixed with 20 ng tester-Core 1 and 20 ng tester-Core 2, isopropanol precipitated and resuspended in Subtraction Hybridization Buffer (50 mM HEPES pH 8.3, 0.5 M NaCl, 0.02 M EDTA pH 8.0, 10% PEG 8000 w/v). The dissolved pellet was denatured and hybridized for 5 hr at 68°C (for two-stage subtraction, 300ng freshly denatured driver was added and hybridized for a further 5 hr at 68°C). Subtraction products were used as a template in an amplification reaction using Core 1 and 2 primers (Core 1 5′-GAGTAGTGCCAGAACGGTC-3′, Core 2 5′-TCGTAGACTGCGTATCCG-3′).
Table 3. Subtraction adapters (5′ to 3′).
| DpnII | Core 1 | GATCGACCGTTCTGGCA annealed to CTGAGTAGTGCCAGAACGGTC |
| Core 2 | GATCCGGATACGCAGTCTA annealed to CCTCGTAGACTGCGTATCCG | |
| HpyCH4IV | Core 1 | GCGACCGTTCTGGCA annealed to CTGAGTAGTGCCAGAACGGTC |
| Core 2 | CGCGGATACGCAGTCTA annealed to CCTCGTAGACTGCGTATCCG | |
| MseI | Core 1 | TAGACCGTTCTGGCA annealed to CTGAGTAGTGCCAGAACGGTC |
| Core 2 | TACGGATACGCAGTCTA annealed to CCTCGTAGACTGCGTATCCG | |
| NlaIII | Core 1 | CTGAGTAGTGCCAGAACGGTCCATG annealed to GACCGTTCTGGCA |
| Core 2 | CCTCGTAGACTGCGTATCCGCATG annealed to CGGATACGCAGTCTA |
Quantitative evaluation of DArT data
Polymorphism information content (PIC) [18], P and Q, reproducibility and call rate (C. Cayla, personal communication) were examined for both Asplenium and Garovaglia datasets to assess the distribution and reliability of hybridization (Table S1). The distribution of Asplenium DArT polymorphisms was examined on a per species basis according the following four categories: A. trichomanes private, A. viride private, A. trichomanes/A. viride shared, or A. viride “X”s but no “1”s. The A. adulterinum genotype was excluded from this process, as it contained polymorphisms shared by and private to both A. viride and A. trichomanes. Also excluded were 84 subtraction-derived DArT markers in the Asplenium data set that displayed an identical scoring pattern across all samples (this pattern did not match any known geographic patterns or systematic relationships and, when these 84 markers were included in a comparison of the diversity patterns between subtracted and non-subtracted genomic representations, substantial differences in inferred relationships were observed). Samples Avi288 and Avi289 were also excluded due to substrate ambiguity. The distribution of Garovaglia DArT polymorphisms was similarly examined on a per species basis according the following four categories: G. powellii private, G. elegans ssp. dietrichiae (Australia) private, G. elegans ssp. elegans and G. elegans fo. latifolia (Papua New Guinea) shared, or unassignable by taxonomy or geography. The G. elegans sample Gel6434 was excluded from this process, as it was derived from a mixed-taxon cushion, containing individuals of both G. elegans ssp. dietrichiae and G. powellii.
Direct sequencing of chloroplast and nuclear loci
For Asplenium, total genomic DNA was extracted from silica desiccated leaf material as described above. The following portions of the chloroplast genome were amplified by the polymerase chain reaction: the combined trnL(CAA) gene and trnL-trnF(GAA) intergenic spacer (IGS) region (trnL-F), using primers FERN1 [16] and F [19] as specified in Trewick et al. [16] and the combined rps4 gene and rps4-trnS(GAA) IGS region (rps4-trnS) following the primers and conditions specified in Schneider et al. [20]. Exons 14 to 16 of the single-copy nuclear locus pgiC were amplified using primers 14F and16R according to Ishikawa et al. [21]. Bidirectional cycle sequencing was carried out on an ABI 3730 capillary DNA sequencer using ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer) and the same primers used for amplification. Sequence contigs were assembled and automatically aligned with subsequent manual corrections using SeqMan and MegAlign respectively (v. 6.00 Lasergene, DNAstar, Madison, WI, USA). For Garovaglia, total genomic DNA was extracted using a modification of the standard CTAB procedure [15] as specified in Pedersen and Newton [22]. The chloroplast encoded trnG(UCC) intron (trnG) was amplified, sequenced and aligned using primers and conditions as specified in Pedersen and Newton [22]. All sequences were deposited in GenBank (Tables 1 and 2).
Direct sequencing of selected DArT markers
Inserts and part of the polylinker region were amplified for a representative selection of 96 DArT marker clones from each study using M13 forward and reverse primers as reported in Jaccoud et al [8]. Amplification products were bidirectionally sequenced as described above using the same primers used for amplification. Successful sequence reads were obtained for 74 Asplenium and 74 Garovaglia DArT markers. Sequence contigs were assembled (vectors removed) and aligned as described above. All sequences were deposited in GenBank (Table S1).
Sequence similarity searches and redundancy estimates
Each of the 148 DArT marker sequences were queried against 1) all other sequenced DArT markers in the same study and 2) GenBank (www.ncbi.nlm.nih.gov) to identify putative sequence identities using the Basic Local Alignment Search Tool [23] for nucleotides (blastn) and translated to proteins (tblastx). For inclusion in Tables S1 and 4 we applied an arbitrary maximum expect value of 10−4 and an arbitrary minimum of 70% identity at the nucleotide sequence level or two or more alignments to the same locus/function in different plant species.
Phylogenetic analyses of DArT data and DNA sequence data
Because the standard DArT protocol (DNA not sheared, not subtracted) yielded the most consistent data across plates and also produced the greatest number of DArT markers of any of the treatments, only DArT markers produced using the standard treatment were used in phylogenetic analyses. Asplenium samples Avi288 and Avi289 were excluded due to substrate ambiguity (Table 1). To infer relationships, a binary matrix was generated in which absence (0) or presence (1) of DNA fragments was scored (Table S1). Both DArT data sets (Asplenium and Garovaglia) were analysed using principal component analyses (PCA) and principal coordinate analyses (PCoA) with Euclidean distances using the software packages PAST [24] and MVSP (Kovach Computing, Pentraeth, UK). In the case of Garovaglia data, a hierarchical PCoA procedure was performed in which the most distant specimens were excluded step-by-step to obtain a higher resolution in the scatter plots for the more closely related specimens. Phylogeny reconstructions were carried out using either distance-based approaches with LogDet and Nei-Li distance corrections or character based approaches, for example Bayesian inference of phylogeny with the MK1 model, maximum parsimony with equally weighted and unordered characters. NeighborNet analyses were employed to explore putative alternative relationships. Evolution of substrate specificity was reconstructed using the consensus topology based on all trees sampled from the plateau phase of the Bayesian analyses carried out with MrBayes. Both maximum parsimony and maximum likelihood approaches were applied to reconstruct changes in substrate preferences. These analyses were carried out with the appropriate software MacClade [25], Mesquite [25], MrBayes [26], PAUP* 4.0 [27], Splitstree [28], Tracer [29], and Treecon [30]. DNA sequence data were assembled and aligned using the Lasergene software package (DNASTAR, Madison, WI, USA) and MacClade. Phylogenetic hypotheses based on sequence data were generated using PAUP and MrBayes.
Results
The frequencies of polymorphic markers detected on the Asplenium and Garovaglia discovery arrays were 6% and 15% respectively, resulting in 444 and 905 polymorphic markers recovered respectively (Table 5). The reproducibility of the two DArT genotyping arrays was successfully validated by independent assays from the same DNA (unpublished data). The polymorphism information content [18] for each marker is shown in Table S1; with average PIC (0.21 for Asplenium, 0.25 for Garovaglia), lower than expected for randomly chosen bi-allelic loci (0.50) and lower than in previous DArT studies in barley (0.38) [7] and cassava (0.42) [10].
Table 5. Summary of quantitative evaluation of Asplenium and Garovaglia DArT markers (for individual markers see Table S1).
| Asplenium | Garovaglia | |||||
| no SSH | SSH | total | no SSH | SSH | total | |
| sheared | no | yes | - | no | yes | - |
| clones tested for polymorphism | 3072 | 3072 | 6144 | 3840 | 2304 | 6144 |
| polymorphic (DArT) markers recovered | 126 | 318 | 444a | 420 | 485 | 905 |
| frequency of polymorphism | 0.04 | 0.08 | 0.06 | 0.11 | 0.21 | 0.15 |
| average reproducibility | 97.85 | 97.63 | 97.71 | 98.73 | 99.3 | 99.03 |
| average call rate | 99.68 | 99.61 | 99.64 | 99.54 | 99.75 | 99.65 |
| average PIC | 0.24 | 0.19 | 0.21 | 0.27 | 0.23 | 0.25 |
| DArT markers sequenced (attempted) | 36 (49) | 38 (47) | 74 (96) | 36 (49) | 38 (47) | 74 (96) |
| ave. length of sequenced DArT markers | 539 | 176 | 344 | 480 | 182 | 327 |
| DArT marker length range | 292–852 | 82–565 | 82–852 | 279–789 | 37–649 | 37–789 |
| frequency of redundancy | 0.2 | 0.31 | 0.26 | 0.14 | 0.47 | 0.31 |
For distribution and phylogenetic analyses, 84 of the 444 Asplenium DArT markers that displayed an identical scoring pattern across all samples analysed were excluded (see Materials and Methods).
To examine taxonomic specificity of DArT markers and to evaluate DArT protocol variables, especially enrichment by suppression subtractive hybridization (SSH), Asplenium and Garovaglia DArT markers (Table S1) were classified according to their distribution among species. Asplenium DArT markers were distributed as follows: A. trichomanes private (65%), A. viride private (16%), A. trichomanes/A. viride shared (13%) and A. trichomanes/A. viride putatively shared (present in A. trichomanes, “X” in A. viride, 5%). Garovaglia DArT marker presence was distributed as follows: G. powellii private (30%), G. elegans ssp. dietrichiae private (33%), G. elegans ssp. elegans and/or G. elegans fo. latifolia private (19%) unassignable by taxonomy (18%).
The fraction of Asplenium DArT markers in each of these categories varied according to treatment: DArT markers derived from the pooled Asplenium metagenome using the standard DArT protocol (no shearing, no SSH) (Asplenium plates 1–6, n = 103) were distributed as follows: A. trichomanes private (58%), A. viride private (12%), A. trichomanes/A. viride shared (20%) and A. trichomanes/A. viride putatively shared (10%). By contrast, DArT markers derived solely from A. viride genomic DNA driver Avi284b (Asplenium plates 7–8, n = 22) were enriched for A. viride private (50%) and A. trichomanes/A. viride shared (50%) polymorphisms. In the reverse treatment, A. viride Avi284b genomic DNA was subtracted from the pooled Asplenium metagenome (Asplenium plates 11–12 & 15–16, n = 193) and the resulting dataset was enriched for A. trichomanes private polymorphisms (86%). Lastly, when A. viride Avi284b genomic DNA was subtracted from a sub-metagenome containing only A. viride genomic DNA samples except Avi284b (Asplenium plates 9–10 & 13–14, n = 35), the proportion of A. trichomanes private polymorphisms was reduced (9%) and A. viride private polymorphisms increased (71%).
The fraction of Garovaglia DArT markers in each category also varied according to treatment: DArT markers derived from the pooled Garovaglia metagenome using the standard DArT protocol (no shearing, no SSH, Garovaglia plates 1–8, n = 262) were distributed as follows: G. powellii private (15%), G. elegans ssp. dietrichiae private (32%), G. elegans ssp. elegans and/or G. elegans fo. latifolia private (31%) or unassignable by taxonomy or geography (23%). By contrast, DArT markers derived solely from G. elegans genomic DNA driver Gel6504 (Garovaglia plates 15–16, n = 55) were enriched for G. elegans ssp. dietrichiae private (35%) and G. elegans ssp. elegans and/or G. elegans fo. latifolia private (42%) polymorphisms. In the reverse treatment, G. elegans Gel6504 genomic DNA was subtracted from the pooled Garovaglia metagenome (Garovaglia plates 9–14, n = 413) and the resulting dataset was enriched for G. powellii private polymorphisms (44%).
To detect DArT marker specificity by substrate or geography, we examined the distribution of DArT markers in substrate-specific or geographically defined Asplenium and Garovaglia specimens. In the complete unsubtracted Asplenium data set (Asplenium plates 1–6), the A. viride private marker class included 1 global limestone-specific DArT marker (marker ID Asplenium6F4), 4 serpentine-specific DArT markers, and 7 non-substrate-specific DArT markers; the A. trichomanes/A. viride shared marker class included 6 DArT markers absent from global limestone and 1 DArT marker absent from serpentine (marker ID Asplenium3O24). In the complete unsubtracted Garovaglia data set (Garovaglia plates 1–8, Table S1), 33% of DArT markers are specific to Australian G. elegans samples while 19% are specific to Papua New Guinean G. elegans samples.
The genotype of G. elegans sample Gel6434 contains both G. powellii- and G. elegans (Australia)-private DArT markers, potentially consistent with a hybrid origin. Perplexingly, this accession was not a known or suspected hybrid. Re-examination of the cushion from which this accession was dissected revealed that it was a mixed-taxon cushion containing individuals of both G. elegans ssp. dietrichiae and G. powellii. Gel6434 was therefore retroactively excluded from the marker distribution analyses of all of the other markers reported above.
The copy number and genomic origin of any marker system, and the potential for redundancy between markers, is an important consideration for evolutionary applications. Only approximately 15% of Asplenium DArT markers hybridized to their respective un-amplified, labelled metagenomes (unpublished data) indicating that approximately 85% of Asplenium DArT markers are low-copy sequences. The level of redundancy between supposedly independent DArT markers on each genotyping array, as well as between SSH treatments, was assessed by sequencing a selection of 74 Asplenium and 74 Garovaglia DArT markers, (Table S1, summarized in Table 4). This revealed that 10.7% of Asplenium DArT markers and 16.2% of Garovaglia DArT markers shared over 99% identity at the nucleotide sequence level to other DArT markers (Table 5). Redundancy dropped to 3.3% and 5.6% respectively when SSH-derived DArT markers were removed. Similar redundancy estimates, including lower redundancy in un-subtracted libraries, are reported for wheat [11], tomato, sorghum and sugarcane (DArT P/L, unpublished).
Table 4. Predicted locus/function for sequenced DArT markers with >70% identity to one or more GenBank sequences or (aand) >1 plant species hit for the same function.
| DArT marker | length | predicted function or locus |
| Asplenium | ||
| 2 B8 | 442bp | expressed gene (similarity to cDNA, mRNA and/or protein)a |
| 2 G20 | 581 | expressed gene (possible retrotransposon) |
| 2 H15 | 691 | cyclophilin-like protein (CYP20)a (redundant with Asplenium 8 E23) |
| 2 K16 | 708 | Hcr2 |
| 2 M10 | 852 | expressed gene (similarity to cDNA, mRNA and/or protein)a |
| 3 I22 | 704 | expressed gene (similarity to cDNA, mRNA and/or protein) |
| 4 A18 | 537 | GGPP synthase |
| 5 H4 | 460 | methyltransferasea |
| 7 E12 | 627 | eukaryotic translation initiation factor 3B (EIF3B)a |
| 7 M10 | 640 | cyc07 and/or rps3 a |
| 7 O5 | 527 | expressed gene (similarity to cDNA, mRNA and/or protein) |
| 8 E23 | 698 | cyclophilin-like protein (CYP20)a (redundant with Asplenium 2 H15) |
| 8 H10 | 340 | glutamine synthetasea |
| 12 D11 | 101 | Chloroplast-encoded trnK-psbA-trnHa |
| 12 G13 | 146 | glyceraldehyde-3-phosphate dehydrogenasea |
| 12 H16 | 124 | Mitochondrial pseudogene rpl2 |
| 13 F23 | 173 | carbohydrate transporter |
| 16 C20 | 123 | expressed gene (similarity to cDNA, mRNA and/or protein)a |
| Garovaglia | ||
| 1 D19 | 637 | expressed gene, protein binding functiona (redundant with Garovaglia 5 C20) |
| 1 E5 | 382 | structural maintenance of chromosomes I (SMCI)a |
| 1 K11 | 279 | kinase |
| 3 D6 | 333 | expressed gene (similarity to cDNA, mRNA and/or protein) |
| 4 M15 | 448 | protein kinasea |
| 4 P4 | 292 | expressed gene (similarity to cDNA, mRNA and/or protein) |
| 5 C20 | 635 | expressed gene, protein binding functiona (redundant with Garovaglia 1 D19) |
| 6 H21 | 598 | expressed gene (similarity to cDNA, mRNA and/or protein) |
| 7 H9 | 693 | biotin synthase |
| 9 K13 | 649 | Chloroplast-encoded16s rRNA genea |
| 12 F16 | 155 | polyribonucleotide nucleotidyltransferase (3'RNAse)a |
| 13 B20 | 138 | histidine kinasea |
| 14 O1 | 118 | expressed gene (similarity to cDNA, mRNA and/or protein) |
| 14 O18 | 201 | sugar transportera |
DArT markers are identified by discovery array plate number and location (row, column) in Table S1.
BLAST searches of 74 Asplenium and 74 Garovaglia DArT marker sequences against GenBank recovered 18 (24%) and 14 (19%) significant alignments respectively, and included known mitochondrial (i.e. rpl2 mitochondrial pseudogene), chloroplast (i.e. psbA and 16s) and nuclear (i.e. Hcr2 and cyc07/rps3) loci as well as proteins (e.g. CYP20, EIF3B and G3PDH) and predicted expressed genes (Table 4).
PCO and PCoA analyses of the Asplenium and Garovaglia DArT data sets derived from the standard DArT procedure (not subtracted) recovered a cumulative explanatory percentage for axes 1 and 2 of 72.7% for Asplenium and 73.5% for Garovaglia, the latter increasing to 100% in the exclusively Papua New Guinean sample subset, but not in the exclusively Australian subset. These DArT data enabled reconstruction of intraspecific structure in A. viride revealing phylogeographic and substrate specificity patterns (Figures 1 & 2). These patterns were not detectable in our analyses of cpDNA or nrDNA sequences, due to insufficient polymorphisms to generate a fully resolved phylogeny (data not shown). The rps4-trnS IGS contained just two polymorphic positions, while the trnL-F region had one unambiguous polymorphic nucleotide position. In both cpDNA regions, one polymorphic site separates samples Avi169 and Avi272 from the remaining samples of A. viride. Variation in the nuclear pgiC data set was similarly low, with seven unambiguous polymorphic sites each of which was specific to only one specimen (e.g. Avi69, Avi245, Avi284a).
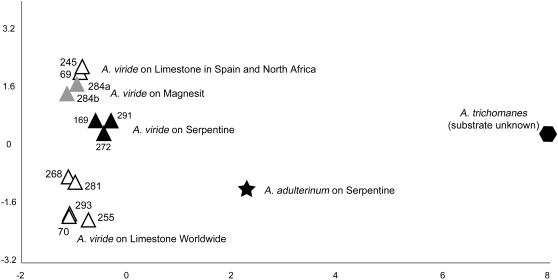
Figure 1. Two dimensional PCO scatter plot of the Asplenium viride DArT marker data set (triangles) plus a single specimen of A. adulterinum (star) and A. trichomanes (hexagon).
Color of symbol corresponds to the substrate on which the sample was growing: limestone = white, serpentine = black and magnesit = gray. Numbers correspond to the sample number (Table 1).
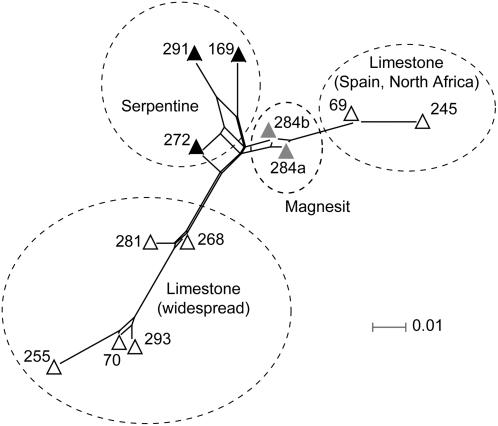
Figure 2. Splitgraph obtained by a NeighborNet analysis with LogDet distances for the Asplenium viride DArT marker set.
Sample numbers, symbols and symbol colors as in Figure 1. The dotted ovals mark putative groups found in phylogenetic analyses (Figure 3).
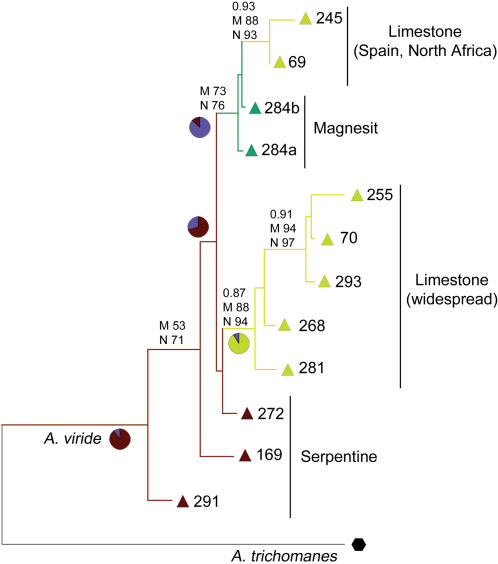
All analyses of the Asplenium DArT marker data indicate one group with a widespread limestone genotype and a second group containing a north-African/Iberian limestone genotype with the two magnesit samples from Austria as putative relatives (Figures 1, 2, 3). In PCO analyses, the three A. viride samples occurring on serpentine are closely associated, and form a putative group in some phylogenetic reconstructions, forming a basal grade at the base of the remaining A. viride groups in others. The three samples of serpentine A. viride were collected in three different regions of Europe: Scandinavia (Avi291), Scotland (Avi169) and Austria (Avi272). The Asplenium DArT marker data set suggests that serpentine was the ancestral substrate of A. viride with two independent colonizations onto limestone (Figure 3).
Figure 3. Consensus phylogram obtained using Bayesian inference of phylogeny analyses for the A. viride DArT marker data set rooted with A. trichomanes.
Posterior probabilities (0.XX), maximum parsimony bootstrap values (M XX), and bootstrap values for a neighbor joining distance analyses with Nei-Li distances (N XX) are given above branches if they are >0.75 for posterior probabilities or >50% for bootstrap values. Sample numbers and symbols as in Figure 1. Color of symbol corresponds to the substrate on which the sample was growing: limestone = yellow, serpentine = dark red and magnesit = green. Branch color corresponds to the character state reconstructed using a maximum parsimony approach. The pie charts represent the likelihood of substrate preference for branches with a putative switch between limestone, magnesit, or serpentine substrates, shown as either limestone versus non-limestone or serpentine versus non-serpentine.
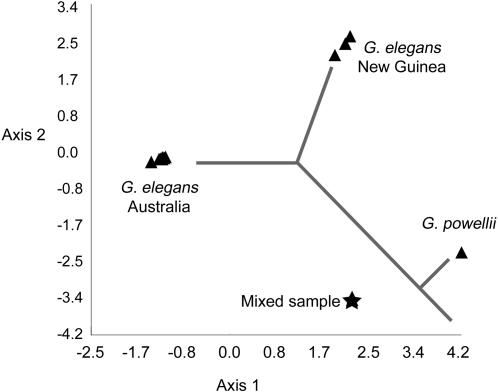
DNA sequence data from chloroplast (Asplenium trnL-F, rps4-trnS; Garovaglia trnG) and nuclear (Asplenium pgiC) loci largely corroborated the results of the above analyses of Asplenium and Garovaglia DArT data (Figures 2 & 4 and unpublished data). The analysis of Garovaglia chloroplast DNA sequence data resulted in a phylogram with a clear separation of Papua New Guinean and Australian specimens (Figure 4). The same clusters were recovered using Garovaglia DArT markers. Neither cpDNA nor the highly variable DArT markers recovered any phylogeographic patterns within Australia. In a hierarchical PCO analysis procedure (step by step exclusion of more distantly related samples), no increase in the cumulative percentage explained by axes 1 and 2 was observed.
Figure 4. Two-dimensional PCO scatter plot of the Garovaglia elegans DArT marker data set superimposed with the phylogenetic tree obtained based on cpDNA.
Garovaglia powelli is included to root the phylogenetic tree. Triangles correspond to single species samples, whereas the star corresponds to a mixed sample that includes individuals of G. elegans and G. powellii.
In both Asplenium and Garovaglia, DArT markers enabled the detection of additive variation caused by hybridization (A. adulterinum) or a mixed-taxon specimen (Garovaglia). The genotype of A. adulterinum, a known allopolyploid A. viride x trichomanes hybrid, contains DArT markers that are private to both A. trichomanes and to the widespread limestone A. viride (Table S1). The position of A. adulterinum in the PCO analysis is intermediate between that of A. trichomanes and widespread limestone A. viride (Figure 1). Chloroplast DNA sequence data corroborate these DArT-based results: A. adulterinum exhibits an A. viride chloroplast haplotype (100% sequence identity with the common cp haplotype of A. viride), consistent with the known maternal inheritance of chloroplast DNA in Asplenium and that A. viride is always A. adulterinum's ovule parent [31], [32]. The genotype of the mixed-taxon Garovaglia sample Gel6434 contains DArT markers private to both G. powellii and the Australian G. elegans group.
Discussion
This study demonstrates that the DArT protocol can be utilized to reproducibly detect largely low-copy genomic variation in sample sets from two lineages of seed-free land plants, suggesting that DArT may be useful across a broader taxonomic spectrum than that addressed in previous studies focusing on cultivated angiosperm species [6]–[11]. Asplenium and Garovaglia DArT data were useful for making biological inferences, for example, phylogeographic structure in plants with a high dispersal capacity, naturally occurring hybridization, exploration of chimeric environmental samples, and the reconstruction of ecological differentiation. Here we discuss procedures for both detecting and analysing DArT data for evolutionary applications, practical considerations for investigators, and the evolutionary hypotheses suggested by these results.
Procedures for detecting DArT markers for evolutionary studies
The frequencies of polymorphism detected on the Asplenium (6%) and Garovaglia (15%) discovery arrays are similar to those reported for previous studies in cultivated angiosperms and their wild relatives: barley (2.9–10.4%) [12], cassava (9–14%) [10], mouse-ear cress (7.3%) [6]. The number of polymorphic DArT markers obtained from Asplenium (444) and Garovaglia (905) is a function of number DArT clones screened in the discovery array step, and the frequency of polymorphism therein. Because marker redundancy was estimated as less than 10% between DArT markers from un-subtracted metagenomic libraries, it is clear that several times as many DArT markers can be developed for the Asplenium or Garovaglia metagenomes through screening additional clones/arrays before saturation of marker libraries is reached.
That the average PIC values (0.21 for Asplenium, 0.25 for Garovaglia) are lower than previous DArT studies in barley (0.38) [7] and cassava (0.42) [10], and is likely due to the choice of sample sets with a strong structure, and clearly defined outgroups. All DArT markers private to the outgroups exhibited low PIC values as expected, and thus the high frequency of such markers impacted strongly on average PIC value.
Because the aim of this study was to pilot DArT in new taxonomic contexts and apply it to different classes of questions, a range of methodologies were compared to inform similar future studies. The distributions of DArT markers derived from sample-specific vs. pooled metagenomes exhibited expected shifts in marker distribution. For example, the DArT markers derived from one A. viride sample (Avi284b, Asplenium plates 7 & 8) or the A. viride sub-metagenome (Asplenium plates 9, 10, 13 & 14) were enriched for markers private to and shared with A. viride compared to the baseline marker set derived from the complete pooled Asplenium metagenome. Similarly, the distributions of DArT markers derived from subtracted vs. un-subtracted libraries exhibited expected shifts in marker distribution. For example, when Avi284b genomic DNA was subtracted from the A. viride sub-metagenome (all A. viride samples except Av284b), the number of A. viride private polymorphisms was enriched as expected. Thus restricting the starting metagenome and/or performing SSH allows investigators to intentionally enrich for DArT markers private to any one or more input samples in cases where such an enrichment is desired. However, when the maximum number of random markers is preferred, such as for unbiased evolutionary analyses, SSH is unnecessary for or even detrimental to the detection and typing of DArT markers in randomly derived sample sets.
Procedures for analysing DArT markers to reconstruct the evolutionary history of plant species
The phylogenetic analyses of Asplenium and Garovaglia DArT data largely corroborated results generated using direct sequencing, and often improved upon the resolution provided by these methods. Considering the small sample set used in these pilot studies, our result holds much promise for the use of DArT to address evolutionary-genetic hypotheses when large sample sets are employed. The two case studies were designed to demonstrate the utility of DArT markers for studies in which highly variable markers are required for evolutionary interpretation and in which relationships are poorly resolved using DNA sequence data due to a low level of variation. Various other marker systems, isozymes and arbitrarily amplified dominant (AAD) markers for example, have been employed to overcome this problem [4], [33], [34]. DArT markers share several features with AAD markers but also differ substantially in other respects. DArT markers and AAD markers have the same binary data structure, that is, presence or absence of markers or bands and a dominant inheritance. Here we discuss some of the relevant distinguishing features of DArT data in comparison with AAD markers, and recommend how DArT data should be handled in phylogenetic or phylogeographic studies.
Non-independence of markers and false homology of bands are common problems associated with AAD markers. These drawbacks restrict the application of AAD markers to very closely related samples (e.g. within species) and often necessitate a dense sampling of the lineages to infer evolutionary history [4]. Because DArT markers are detected by DNA-DNA hybridization rather than fragment size, DArT markers were expected to suffer only a very low level of erroneous homology assessment/assignment. This assumption was validated by Wenzl et al. [12] through genetic mapping of over 2000 DArT markers in many mapping populations of barley, where fewer than 2% of the markers mapped to more than one locus in the genome. The high percentage (mostly above 70%) accumulation for axis 1 and axis 2 in all PCO and PCoA analyses of DArT markers in both case studies provides additional support that this expectation is justified. DNA sequencing of DArT markers confirmed that redundancy between different DArT markers is low. Some DArT markers, however, are still likely to be erroneously scored as independent if they originated from partially overlapping genomic regions. DArT marker DNA sequences suggest that very few DArT markers are mistakenly treated as independent and further comfort is derived from the observation that all three plant genomes contributed to the DArT data set.
A related problem is the assumption of homology when DArT markers are absent and data are analysed in a parsimony framework [35]. Similar to other presence/absence data (AFLP, ISSR), homology assessment is restricted to the hypothesis that the state of presence is the result of presence of homologous markers but the state of absence may be the result of alternative processes [35]. Currently, it is not possible to evaluate the putative misleading effects of such a bias. A further restriction of presence/absence data is the limited number of character states (two). However, this problem is likely to be offset by the large number of DArT markers available [35], which exceeds several hundred for both case studies presented here.
Distance-based methods are putatively less affected by these problems because some similarity scores take data structure and heritage (which in DArT is likely dominant) into account. Thus, Nei-Li similarity score [36] is probably the most appropriate for DArT data, for the same reasons given for AFLP data, i.e. presence/absence data structure and dominant inheritance [37]. This is an advantage of using the DArT technique because existing software tools designed for analyses of AAD markers can be easily adjusted to analyse DArT data. However, users should keep in mind the existence of modified versions of these distance measures [37]. The selection of tree building algorithms, for example UPGMA and Neighbor-Joining (NJ) is also important in the context of distance-based approaches. UPGMA is likely to result in incorrect topology as the result of the imbalanced distribution of variation among putative branches in the tree. Recently developed approaches such as NeighborNet and split decomposition analyses as implemented in Splitstree [28] are likely to be more powerful approaches for phylogenetic reconstruction based on DArT data transformed into appropriate distance matrices. Importantly, DArT procedures hold the potential to generate co-dominant markers by taking into account the strength of the signal for each DArT marker. This improvement could potentially overcome several restrictions limiting the analyses of AAD markers and current (dominant) DArT markers.
In general, character-based approaches are a more powerful tool to assess phylogenetic patterns and some authors therefore argue for their superiority [35]. Most parsimony analyses are now commonly used in addition to distance based approaches to evaluate AAD data sets, and these analyses should also be employed to analyse DArT data sets. More recently, Bayesian inference of phylogeny was made available by implementing a restriction site (binary) model based on an F81-like model into MrBayes 3.01 [26]. This model can also be used to analyse DArT marker data sets due to the underlying assumptions for the restriction sites model, non-observable absence data for example, fitting the structure of DArT data.
Practical advantages of DArT in evolutionary research
The results reported here provoke the question: what are the practical advantages and disadvantages of using DArT (as opposed to other methods) to detect genomic variation for analysis in an evolutionary or phylogenetic framework? DArT offers seven key advantages over other widely used methods for detecting molecular variation: First, DArT is a sequence-independent discovery tool that requires no preliminary sequence information such as the identification of candidate loci or time-intensive development and optimization of primers. Thus DArT is likely to find variable sequences that other sequence-dependent technologies might miss. Second, DArT recovers a high level of variation, and offers the potential for very high throughput analysis of both markers and specimens. Third, the presence of a DArT marker is determined by DNA-DNA hybridization rather than fragment length and hence DArT suffers less from ambiguous homology assessments than other finger-printing methods utilising AAD markers, especially RAPDs, ISSRs and AFLPs [4]. Indeed, independent segregation of DArT markers in mapping experiments in barley [12] as well as in other species like wheat and sorghum (unpublished data) provides strong evidence for this point. Fourth, DArT is highly reproducible; DArT marker scoring is consistent among independent replicates (this study and [7]), indicating that DArT may be a more reliable genotyping method than other AAD markers [4]. Fifth, redundancy between DArT markers is both quantifiable and low (this study and [7]). Sixth, DArT markers are very easily sequenced (no gel isolation required), allowing similarity searches against sequence databases and internal redundancy estimates. Twenty-four percent of Asplenium and 19% of Garovaglia DArT marker DNA sequences yielded GenBank alignments including known mitochondrial, chloroplast and nuclear loci, proteins and predicted expressed genes (Table 4). Amongst these were matches to rpl2, psbA and rps3 genes, and the locus encoding G3PDH, all of which are routinely used in sequence-based evolutionary studies in plants [38]–[44]. This demonstrates that DArT is capable of identifying known polymorphic loci, and suggests that the sequences of the hundreds of other unknown polymorphic fragments recovered by DArT may facilitate primer design for the amplification of novel target loci for direct sequencing. Finally, DArT is able to detect both genetic hybrids and chimeric environmental samples, demonstrating its utility for studies exploring the origin of hybrids and their specific genetic structure.
Evolutionary insights derived from this study
Analyses of Asplenium and Garovaglia DArT data enabled the reconstruction of intraspecific structure in A. viride and G. elegans and revealed substrate specificity and/or phylogeographic patterns which, for A. viride, were not detectable using chloroplast or nuclear DNA sequences. All analyses of the Asplenium DArT data indicated a widespread limestone genotype group and a second limestone genotype group occurring in northern Africa and the Iberian Peninsula, incorporating two magnesit samples from Austria as putative relatives (Figures 1, 2, 3). The phylogeographic and substrate specificity patterns detected in analyses of A. viride DArT data (Figures 1 and 2) provoke at least two alternative scenarios of substrate evolution in A. viride: (1) A. viride originated on serpentine and twice colonized limestone habitats or (2) the extant serpentine populations of A. viride are relicts able to survive on a less favorable substrate. Both scenarios are consistent with the lack of alleles private to serpentine populations of A. viride (unpublished data). For Garovaglia, both DArT and cpDNA data clearly separated the Australian from the Papua New Guinean samples (Figure 4). When contrasted against the geographical distinction between Papua New Guinean and Australian G. elegans (ssp. dietrichiae, endemic to Australia) specimens, the lack of micro-biogeographic structure detected in Australian ssp. dietrichiae clade (Figures 1 and 2) suggests a high rate of gene exchange among the Australian populations (which may be a reflection of its nanandrous breeding system) or a low rate of gene exchange between Australian and New Guinean populations. Lastly, DArT enabled the detection of additive variation caused by genetic hybridization (A. adulterinum) or a mixed-taxon specimen (G. elegans Gel6434).
In conclusion, while DArT has been previously used to complement existing technologies in crop breeding and genomics [10], our studies show that the abundant non a priori comparative molecular data generated by DArT also holds real potential for use in a wide range of high-throughput evolutionary studies. These include the detection of biological correlations and phenomena including similarity, hybridization, mixed environmental samples, ecological differentiation and geographical distribution, as well as other studies of non-model organisms including gene discovery, QTL mapping, population and conservation genetics, speciation and environmental forensics.
Supporting Information
DArT markers, specimen genotypes and sequence-associated data. This supplementary table contains 1) detailed information about each of the 1349 DArT markers recovered for this study including marker-specific library preparation data and statistics, 2) DArT genotyptes of Asplenium and Garovaglia specimens, and 3) sequence-associated data from each of the 148 sequenced DArT markers including GenBank accession numbers, fragment lengths, blastn and tblastx identifications, % identity (closest match), and number different species hit for same function.
(0.50 MB XLS)
Acknowledgments
We acknowledge Heinar Streimann and Jacques de Sloover for collecting Garovaglia specimens in Papua New Guinea and the curators of the New York Botanical Garden (NY) for giving us permission to use these specimens, Drs Adolf and Oluna Ceska, Dr Curtis Bjjoerk, Dr Fred Rumsey, Dr Mark Carine and Mr Stephan Jessen for support collecting Asplenium specimens, and Meredith Murphy Thomas for technical assistance. We also thank many members of Diversity Arrays Technology Pty Ltd team for help in execution of this project.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work has been financially supported by The Natural History Museum Research Fund, NERC grant GR3/12073 in support of specimen collection and Marie Curie Postdoctoral Fellowship MEIF-CT-2003-501682 to Niklas Pedersen.
References
- 1.Avise JC. Sunderland, MA: Sinauer Associates; 2004. Molecular markers, natural history, and evolution. [Google Scholar]
- 2.Ranz JM, Machado CA. Uncovering evolutionary patterns of gene expression using microarrays. Trends in Ecology & Evolution. 2006;21:29–37. doi: 10.1016/j.tree.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Gilad Y, Borevitz J. Using DNA microarrays to study natural variation. Current Opinion in Genetics & Development. 2006;16:553–558. doi: 10.1016/j.gde.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Bussell JD, Waycott M, Chappill JA. Arbitrarily amplified DNA markers as characters for phylogenetic inference. Perspectives in Plant Ecology Evolution and Systematics. 2005;7:3–26. [Google Scholar]
- 5.Kilian A, Huttner E, Wenzl P, Jaccoud D, Carling J, et al. The fast and the cheap: SNP and DArT-based whole genome profiling for crop improvement. In: Tuberosa R, Phillips RL, Gale M, editors. In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution. Bologna, Italy: Avenue Media; 2003. pp. 443–461. [Google Scholar]
- 6.Wittenberg AHJ, van der Lee T, Cayla C, Kilian A, Visser RGF, et al. Validation of the high-throughput marker technology DArT using the model plant Arabidopsis thaliana. Molecular Genetics and Genomics. 2005;274:30–39. doi: 10.1007/s00438-005-1145-6. [DOI] [PubMed] [Google Scholar]
- 7.Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, et al. Diversity Arrays Technology (DArT) for whole-genome profiling of barley. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9915–9920. doi: 10.1073/pnas.0401076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaccoud D, Peng K, Feinstein D, Kilian A. Diversity Arrays: a solid state technology for sequence information independent genotypinG. Nucleic Acids Research. 2001;29:1–7. doi: 10.1093/nar/29.4.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lezar S, Myburg AA, Berger DK, Wingfield MJ, Wingfield BD. Development and assessment of microarray-based DNA fingerprinting in Eucalyptus grandis. Theoretical and Applied Genetics. 2004;109:1329–1336. doi: 10.1007/s00122-004-1759-9. [DOI] [PubMed] [Google Scholar]
- 10.Xia L, Peng KM, Yang SY, Wenzl P, de Vicente MC, et al. DArT for high-throughput genotyping of Cassava (Manihot esculenta) and its wild relatives. Theoretical and Applied Genetics. 2005;110:1092–1098. doi: 10.1007/s00122-005-1937-4. [DOI] [PubMed] [Google Scholar]
- 11.Akbari M, Wenzl P, Caig V, Carling J, Xia L, et al. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theoretical and Applied Genetics. 2006;113:1409–1420. doi: 10.1007/s00122-006-0365-4. [DOI] [PubMed] [Google Scholar]
- 12.Wenzl P, Li HB, Carling J, Zhou MX, Raman H, et al. A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits. BMC Genomics. 2006;7:206. doi: 10.1186/1471-2164-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang SY, Pang W, Ash G, Harper J, Carling J, et al. Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Theoretical and Applied Genetics. 2006;113:585–595. doi: 10.1007/s00122-006-0317-z. [DOI] [PubMed] [Google Scholar]
- 14.Lovis JD, Reichstein T. Über das spontane Entstehen von Asplenium adulterinum aus einem natürlichen Bastard. Die Natuerwissenschaften. 1967;55:117–120. doi: 10.1007/BF00624239. [DOI] [PubMed] [Google Scholar]
- 15.Doyle J, Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 16.Trewick SA, Morgan-Richards M, Russell SJ, Henderson S, Rumsey FJ, et al. Polyploidy, phylogeography and Pleistocene refugia of the rockfern Asplenium ceterach: evidence from chloroplast DNA. Molecular Ecology. 2002;11:2003–2012. doi: 10.1046/j.1365-294x.2002.01583.x. [DOI] [PubMed] [Google Scholar]
- 17.Diatchenko L, Lau YFC, Campbell AP, Chenchik A, Moqadam F, et al. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME. Optimizing Parental Selection for Genetic-Linkage Maps. Genome. 1993;36:181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- 19.Taberlet P, Gielly L, Pautou G, Bouvet J. Universal Primers for Amplification of 3 Noncoding Regions of Chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 20.Schneider H, Ranker T, Russell SJ, Cranfill R, Geiger JMO, et al. Origin of the endemic fern genus Diellia coincides with the renewal of Hawaiian terrestrial life in the Miocene. Proceedings of the Royal Society B-Biological Sciences. 2005;272:455–460. doi: 10.1098/rspb.2004.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa H, Watano Y, Kano K, Ito M, Kurita S. Development of primer sets for PCR amplification of the PgiC gene in ferns. Journal of Plant Research. 2002;115:65–70. doi: 10.1007/s102650200010. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen N, Newton AE. Phylogenetic and morphological studies within the Ptychomniales, with emphasis on the evolution of dwarf males. In: Newton AE, Tangney R, DeLuna E, editors. Pleurocarpous Mosses: Systematics and Evolution. Boca Raton, FL: CRC Press (Taylor & Francis Group); 2007. pp. 367–392. [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammer O, Harper D, Ryan P. PAST: Palaeontological Statistics Software package for education and data analysis. Palaeontologia Electronica. 2001;4:9. [Google Scholar]
- 25.Maddison WP, Maddison DR. Interactive Analysis of Phylogeny and Character Evolution Using the Computer-Program Macclade. Folia Primatologica. 1989;53:190–202. doi: 10.1159/000156416. [DOI] [PubMed] [Google Scholar]
- 26.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 27.Swofford DL. Paup-a Computer-Program for Phylogenetic Inference Using Maximum Parsimony. Journal of General Physiology. 1993;102:A9–A9. [Google Scholar]
- 28.Bryant D, Moulton V. Berlin: Springer; 2002. NeighborNet: An agglomerative method for the construction of planar phylogenetic networks. In: Algorithms in Bioniformatics: Second International Workshop, WABI 2002, Rome, Italy, September 17–21, 2002. Proceedings. pp. 375–391. [Google Scholar]
- 29.Rambaut A, Drummond A. Oxford, UK: University of Oxford; 2004. TRACER, version 1.2. [Google Scholar]
- 30.Vandepeer Y, Dewachter R. Treecon-a Software Package for the Construction and Drawing of Evolutionary Trees. Computer Applications in the Biosciences. 1993;9:177–182. doi: 10.1093/bioinformatics/9.2.177. [DOI] [PubMed] [Google Scholar]
- 31.Vogel JC. University of Cambridge; 1995. Multiple origins of polyploids in European Asplenium (Pteridophyta). Ph.D. thesis. [Google Scholar]
- 32.Vogel JC, Russell SJ, Rumsey FJ, Barrett JA, Gibby M. On hybrid formation in the rock fern Asplenium x alternifolium (Aspleniaceae, Pteridophyta). Botanica Acta. 1998;111:241–246. [Google Scholar]
- 33.Pelser PB, Gravendeel B, van der Meijden R. Phylogeny reconstruction in the gap between too little and too much divergence: the closest relatives of Senecio jacobaea (Asteraceae) according to DNA sequences and AFLPs. Molecular Phylogenetics and Evolution. 2003;29:613–628. doi: 10.1016/s1055-7903(03)00139-8. [DOI] [PubMed] [Google Scholar]
- 34.Koopman WJM. Phylogenetic signal in AFLP data sets. Systematic Biology. 2005;54:197–217. doi: 10.1080/10635150590924181. [DOI] [PubMed] [Google Scholar]
- 35.Simmons MP, Zhang LB, Webb CT, Muller K. A penalty of using anonymous dominant markers (AFLPs, ISSRs, and RAMS) for phylogenetic inference. Molecular Phylogenetics and Evolution. 2007;42:528–542. doi: 10.1016/j.ympev.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Nei M, Li WH. Mathematical Model for Studying Genetic Variation in Terms of Restriction Endonucleases. PNAS. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Archibald JK, Mort ME, Crawford DJ, Santos-Guerra A. Evolutionary relationships within recently radiated taxa: comments on methodology and analysis of inter-simple sequence repeat data and other hypervariable, dominant markers. Taxon. 2006;55:747–756. [Google Scholar]
- 38.Laroche J, Bousquet J. Evolution of the mitochondrial rps3 intron in perennial and annual angiosperms and homology to nad5 intron 1. Molecular Biology and Evolution. 1999;16:441–452. doi: 10.1093/oxfordjournals.molbev.a026126. [DOI] [PubMed] [Google Scholar]
- 39.Olsen KM. SNPs, SSRs and inferences on cassava's origin. Plant Molecular Biology. 2004;56:517–526. doi: 10.1007/s11103-004-5043-9. [DOI] [PubMed] [Google Scholar]
- 40.Stefanovic S, Olmstead RG. Testing the phylogenetic position of a parasitic plant (Cuscuta, Convolvulaceae, Asteridae): Bayesian inference and the parametric bootstrap on data drawn from three genomes. Systematic Biology. 2004;53:384–399. doi: 10.1080/10635150490445896. [DOI] [PubMed] [Google Scholar]
- 41.Howarth DG, Baum DA. Genealogical evidence of homoploid hybrid speciation in an adaptive radiation of Scaevola (Goodeniaceae) in the Hawaiian Islands. Evolution. 2005;59:948–961. [PubMed] [Google Scholar]
- 42.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koehler-Santos P, Lorenz-Lemke AP, Muschner VC, Bonatto SL, Salzano FM, et al. Molecular genetic variation in Passiflora alata (Passifloraceae), an invasive species in southern Brazil. Biological Journal of the Linnean Society. 2006;88:611–630. [Google Scholar]
- 44.Dane F, Liu JR, Zhang CK. Phylogeography of the bitter apple, Citrullus colocynthis. Genetic Resources and Crop Evolution. 2007;54:327–336. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DArT markers, specimen genotypes and sequence-associated data. This supplementary table contains 1) detailed information about each of the 1349 DArT markers recovered for this study including marker-specific library preparation data and statistics, 2) DArT genotyptes of Asplenium and Garovaglia specimens, and 3) sequence-associated data from each of the 148 sequenced DArT markers including GenBank accession numbers, fragment lengths, blastn and tblastx identifications, % identity (closest match), and number different species hit for same function.
(0.50 MB XLS)