Abstract
This article describes the algorithms implemented in the Essie search engine that is currently serving several Web sites at the National Library of Medicine. Essie is a phrase-based search engine with term and concept query expansion and probabilistic relevancy ranking. Essie’s design is motivated by an observation that query terms are often conceptually related to terms in a document, without actually occurring in the document text. Essie’s performance was evaluated using data and standard evaluation methods from the 2003 and 2006 Text REtrieval Conference (TREC) Genomics track. Essie was the best-performing search engine in the 2003 TREC Genomics track and achieved results comparable to those of the highest-ranking systems on the 2006 TREC Genomics track task. Essie shows that a judicious combination of exploiting document structure, phrase searching, and concept based query expansion is a useful approach for information retrieval in the biomedical domain.
A rapidly increasing amount of biomedical information in electronic form is readily available to researchers, health care providers, and consumers. However, readily available does not mean conveniently accessible. The large volume of literature makes finding specific information ever more difficult. Development of effective search strategies is time consuming, 1 requires experienced and educated searchers, 2 well versed in biomedical terminology, 3 and is beyond the capability of most consumers. 4
Essie, a search engine developed and used at the National Library of Medicine, incorporates a number of strategies aimed at alleviating the need for sophisticated user queries. These strategies include a fine-grained tokenization algorithm that preserves punctuation, concept searching utilizing synonymy, and phrase searching based on the user’s query.
This article describes related background work, the Essie search system, and the evaluation of that system. The Essie search system is described in detail, including its indexing strategy, query interpretation and expansion, and ranking of search results.
Background
The Essie search engine was originally developed in 2000 at the National Library of Medicine to support ClinicalTrials.gov, 5,6 an online registry of clinical research studies. From the beginning, Essie was designed to use synonymy derived from the Unified Medical Language System (UMLS) 7 to facilitate consumers’ access to information about clinical trials. The UMLS Metathesaurus contains concepts (meanings) from more than 100 medical vocabularies. Each UMLS concept can have several names, referred to as terms in this article.
Many consumers searching for medical information are unlikely to be familiar with the medical terminology found in medical documents and to use more common language in their queries. Most of the ClinicalTrials.gov documents about heart attacks do not contain the phrase “heart attack,” but instead use the clinical term “myocardial infarction.” Concept-based searching, which utilizes the UMLS-derived synonymy, has the potential to bridge this terminology gap.
One of the first retrieval systems that implemented automatic concept-based indexing and extraction of the UMLS concepts from users’ requests was SAPHIRE. 8 SAPHIRE utilized the UMLS Metathesaurus by breaking free text into words and mapping them into UMLS terms and concepts. Documents were indexed with concepts, queries were mapped to concepts, and standard term frequency and inverse document frequency weighting was applied. When measured with combined recall and precision, SAPHIRE searches performed as well as physicians using Medline, but not as well as experienced librarians. 9 The core functionality of mapping free text to UMLS concepts is generally useful and is freely available in the NLM MetaMap tool. 10
Other experiments with synonymy have produced mixed results. Voorhees 11 found a 13.6% decrease in average precision comparing effectiveness of conceptual indexing with baseline indexing of single words for 30 queries and 1,033 medical documents. Srinivasan 12 demonstrated an overall improvement of 16.4% in average precision, primarily due to controlled vocabulary feedback (expanding queries by adding controlled vocabulary terms). Aronson and Rindflesch 13 achieved 14% improvement in average precision through query expansion using automatically identified controlled vocabulary terms that were expanded using inflectional variants (gender, tense, number, or person) from the SPECIALIST lexicon 14 and synonyms encoded in the UMLS. Finally, concept-based indexing of Medline citations based on manual and semiautomatic indexing 15 is utilized in PubMed.
Similar to concept-based indexing, phrase indexing is believed to be useful in improving precision. 16 However, adding phrase indexing to otherwise good ranking schemes has not been demonstrated to improve performance dramatically. 17 Regardless of this ambiguity, phrases are necessary for identifying UMLS concepts. 18 Essie searches for phrases and maps them to UMLS concepts for synonymy expansion.
Queries can be further expanded with morphological variants (inflectional and derivational) of individual words. 19 Alternatively, queries and documents can be normalized using stemming. 19 In experiments with newswire text, Hull and Grefenstette 20 obtained approximately 5% improvement by stemming. Bilotti et al 21 revisited the exploration of stemming vs. morphological expansion, and found that morphological expansion resulted in higher recall.
Another factor affecting search is the strategy used to decide what constitutes a unit of text. 22 This tokenization determines what bits of text can be found. In many systems, one must specify whether certain characters, like hyphens, are part of a word or not part of a word. Such decisions frequently limit the ability of a system to deal properly with words and phrases that contain punctuation characters. The importance of tokenization in biomedical domains was demonstrated repeatedly in the Text REtrieval Conference (TREC) Genomics track evaluations. For example, much of Essie’s success in the 2003 evaluation can be attributed to tokenization. Further, the best average precision in the TREC 2005 evaluation was achieved by a system that broke text at hyphens, letter–digit transitions, and lower/upper case transitions. 23
Equally important to retrieving relevant information is the order of presentation of the search results. With the exception of Boolean systems, presentation order is typically determined using a scoring function that sorts documents in descending order of relevance. A survey of relevance ranking methods can be found in Singhal 24 and Baeza-Yates. 25
Essie adopts many of the ideas explored in earlier work. Essie implements concept-based searching by expanding queries with synonymy derived from UMLS concepts. Unlike SAPHIRE, Essie includes phrase searches of the original text and inflectional variants in addition to concepts; thus it is less reliant on concept mapping and should be more robust when concept mapping fails. Essie searches for phrases from the user’s query by preserving word adjacency as specified in the query rather than indexing terms from a controlled vocabulary. Queries are further expanded to include a restricted set of inflectional variants, as opposed to many search engines that rely on stemming. 25 Tokenization decisions in Essie are driven by characteristics of biomedical language 3 in which punctuation is significant. Phrase-based searching of synonymy, which equates dramatically different phrases, has forced a new approach to document scoring. Essie scoring is based primarily on where concepts are found in the document, rather than on their frequency of occurrence.
The Essie system was formally validated in the context of the TREC Genomics track. Essie participated in the 2003 and 2006 evaluations. The 2003 ad hoc retrieval evaluation was conducted on a document collection consisting of 525,938 Medline citations. The task was based on the definition of a Gene Reference Into Function (GeneRIF) 26 : For gene X, find all Medline references that focus on the basic biology of the gene or its protein products from the designated organism. Randomly selected gene names distributed across the spectrum of organisms served as queries (50 for training and 50 for testing of the systems). The available GeneRIFs were used as relevance judgments.
The 2006 Genomics track collection consists of 162,259 full-text documents subdivided into 12,641,127 paragraphs. The task for participating systems was to extract passages providing answers and context for 28 questions formed from four genomic topic templates. 27 Each question contains terms that define: (1) biological objects (genes, proteins, gene mutations, etc.), (2) biological processes (physiological processes or diseases), and (3) a relationship between the objects and the processes. Relevance judges determined the relevance of passages to each question and grouped them into aspects identified by one or more Medical Subject Headings (MeSH) terms. Document relevance was defined by the presence of one or more relevant aspects. The performance of submitted runs was scored using mean average precision (MAP) at the passage, aspect, and document level.
Essie approached the task as document retrieval, indexing each paragraph as a document and applying the standard Essie retrieval strategies to queries created for each question. The goal was to use Essie “as is” in a new retrieval task to explore the applicability of the underlying algorithms. The results of the evaluation are presented in the Validation section.
A detailed description of the Essie algorithms for tokenization, morphological variation, concept expansion, and document scoring follows.
System Description
Overview
The Essie search system consists of two distinct phases: indexing and searching. The indexing phase identifies and records the position of every token occurrence in the corpus. The searching phase uses query expansions to produce a set of search phrases, each of which is a sequence of tokens. Tokens are matched against the indexes to identify potentially relevant documents. Finally, probability of relevance is quantified by document scoring.
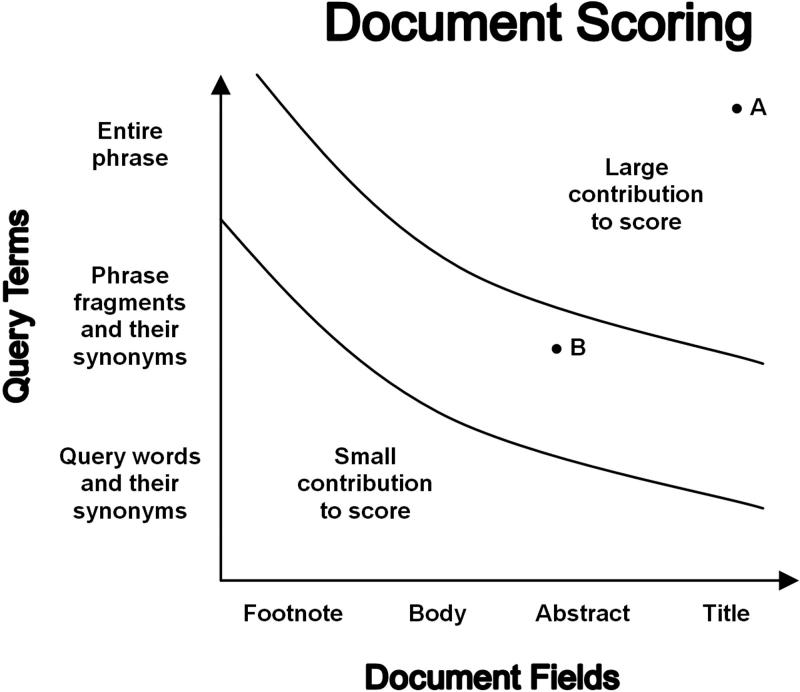
The scoring algorithm accounts for (1) the relative values of the different phrases produced by query expansion and (2) the relative values of the locations in the document where phrases were found. Essie’s scoring algorithm can be summed up as preferring “all the right pieces in all the right places” (▶). “The right pieces” are phrases from the query, with large penalties for dropping words or subphrases. There is less penalty for breaking word adjacency and small adjustments for using synonyms and word variants. “The right places” are the valuable fields of a structured document, such as the title. Further, summary and key word fields are preferred over general text, addendums, and ancillary information.
Figure 1.
Abstract diagram of Essie’s scoring algorithm. Term occurrences are weighted by: (1) the similarity to the user’s query, and (2) the importance of the field where they are found. For example, in a search for “heart attack,” a document with “heart attack” in the title (point A) would score higher than a document with “myocardial infarction” in the abstract (point B).
For example, for the user query “heart attacks in older adults,” the following variants might be weighted as:
• Heart attacks in older adults (weight: 1.0)
• Myocardial infarction AND older adults (weight: 0.18)
• Heart AND attack AND seniors (weight: 0.04)
Within a Medline citation, one might establish a weighting of the various document areas as:
• Title: 0.9
• Abstract: 0.5
• Affiliation: 0.1
Thus, a document with “heart attacks in older adults” in the title has “the right pieces” in “the right places” and results in such a document having a high rank.
Indexing
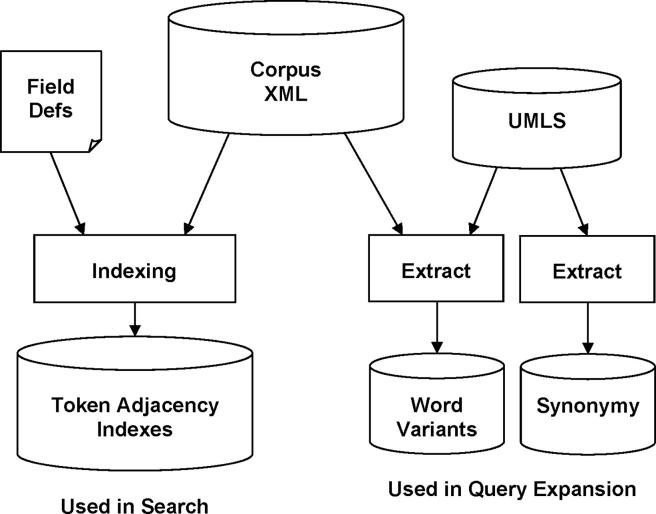
Before searches can be performed, the corpus of source documents must be tokenized and indexed (▶). This process produces token adjacency indexes, which consist of position information about every token occurrence in the corpus.
Figure 2.
Index building and related preprocessing. Token adjacency indexes are derived from the corpus and support efficient searches for arbitrary phrases. Word variants are extracted primarily from the Unified Medical Language System (UMLS) (additional compound words and plurals are mined from the corpus), and are used in term expansion. Synonymy is extracted from the UMLS and is used for concept expansion.
Essie uses a fine-grained tokenization strategy. Every sequence of letters, every sequence of digits, and individual punctuation characters are treated as separate tokens. For example, “non-hodgkin’s lymphoma” consists of the six tokens:
1 non
2 –
3 hodgkin
4 ’
5 s
6 lymphoma
Position information is kept for every occurrence of every token so that adjacency can be determined. An occurrence of the term “non-hodgkin’s lymphoma” is found whenever the six constituent tokens are found in adjacent token positions.
Because of the fine-grained tokenization, it is possible to perform exact literal searches, including punctuation, and to distinguish between nearby term variants. The decision about which variants to include in a search is made at search time and is not built into the underlying indexes. This allows flexibility in the handling of difficult terms that incorporate hyphens, apostrophes, and other punctuation.
Two additional datasets are generated during the indexing process. The word variants dataset, used for term expansion, is derived from the corpus and the UMLS SPECIALIST Lexicon. The synonymy dataset, used for concept expansion, is derived from the UMLS. Both datasets are essentially lookup tables that match a single word or term to a set of words or terms. Details about the contents of these datasets are given in the Query Expansion section below.
Search
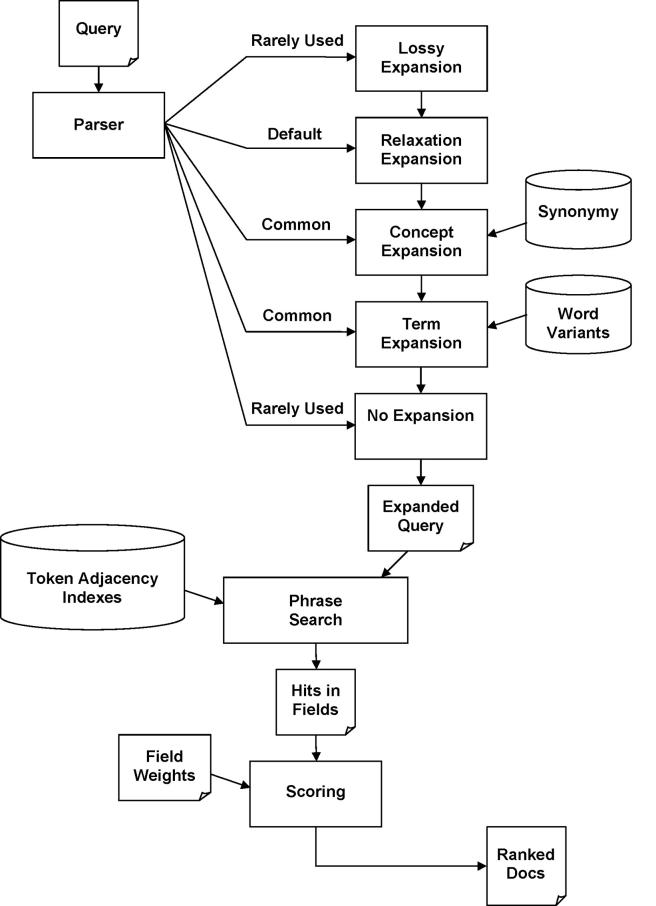
Search processing (▶) uses the datasets derived during indexing (▶). First, the query is parsed to extract search syntax operators and search texts. Then, search texts are expanded, typically with relaxation expansion (breaks text into fragments), which includes concept expansion (adds synonymy) and term expansion (adds word variants). Expansion results in a large set of phrases, all of which are searched as sequences of tokens. Hits in the corpus are collected, and the documents containing them are scored, ranked, and returned.
Figure 3.
Search processing. Queries are parsed to extract search syntax and search texts. Syntax operators can control query expansion, but the default is relaxation expansion, which extends concept and term expansion. Expansion results in a large set of variations of the original search text, all of which are searched as phrases. Hits in the corpus are collected, and the documents containing them are scored, ranked, and returned.
There are more than 50 syntax operators to control a wide range of search functionality. The syntax includes parentheses, AND, OR, and NOT, and additional operators to control query expansion options. Details of the syntax are peripheral to the discussion of the search system and are explained only as needed.
Query Expansion
Query expansion is used to increase recall, with as little degradation in precision as possible. Five query expansion levels have been developed, each an extension of the previous level (▶). For levels of expansion from no expansion to concept expansion, the query is treated as if it consists of a single multiword term, such as “non-hodgkin’s lymphoma.” Relaxation expansion and lossy expansion deal with complex multiple concept queries such as “non-hodgkin’s lymphoma in children with early onset diabetes.” Query expansion defaults to relaxation expansion, but can be controlled with operators that are part of the Essie search syntax.
Table 1.
Table 1 Five Levels of Query Expansion; Each Level Extends the Expansion of the Previous One
| Expansion | Key Feature |
|---|---|
| No expansion | Exact literal search, token-for-token match |
| Term expansion | Include word variants such as plurals and possessives |
| Concept expansion | Also include synonyms of search text |
| Relaxation expansion | Break up search text and add synonyms for all fragments |
| Lossy expansion | Allow some fragments to be missing |
Term expansion
We use the word “term” to refer to both single-word and multiword phrases that identify a concept that may or may not be in the UMLS. Term expansion extends a strict literal search by including word variants for plurals, possessives, hyphenation, compound words, and alternative spellings. Essie does not use stemming or non-noun (verb, adjective, adverb) inflectional variation. Experiments with these and other forms of inflection resulted in an unacceptable loss of precision, as in {numb, number, numbest, numbs, numbers, numbing, numbering}. Many useful variants that are lost due to limited inflectional variation are recovered through the use of synonymy included via concept expansion.
Essie derives most of its word variants from the SPECIALIST Lexicon. Additional variants are derived from the corpus during the indexing phase. Heuristics are used to recognize or generate plurals for new words when they are long enough (five or more characters) and occur often enough (four or more occurrences in two or more documents). Authors and searchers alike use apostrophes freely, so Essie is tolerant of incorrect apostrophe usage. Compound words are derived from the corpus when multiple forms of a hyphenated word occur, as in doubleblind, double-blind, and double blind.
Variation is applied word by word, and can result in a large number of variants for long queries. A few of the variants for non-hodgkin’s lymphoma are:
• non-hodgkin’s lymphoma
• non hodgkins’ lymphomae
• nonhodgkins lymphomas
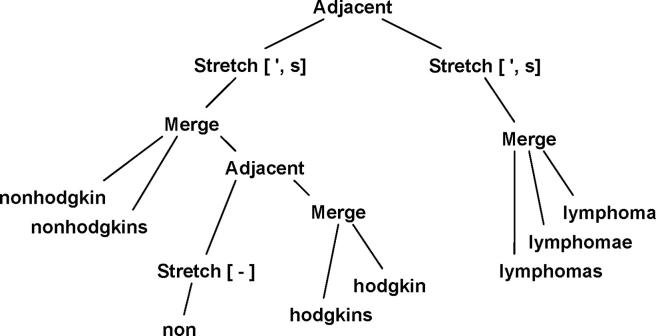
Rather than search each variant as an exact literal search, queries are converted into expression trees containing token occurrences and primitive operations such as merge, adjacent, and stretch. The leaf nodes of the tree represent lists of occurrences of individual tokens. Merge nodes combine lists of individual occurrences into a single list. Adjacent nodes combine adjacent occurrences into a single occurrence of a longer multitoken phrase. Stretch nodes extend a token occurrence to include additional optional tokens to the right. ▶ illustrates the expression tree for non-hodgkin’s lymphoma.
Figure 4.
A search expansion tree. Leaf nodes load lists of occurrences (aka hits) for tokens as found in the token adjacency indexes. Adjacent and merge nodes build up multitoken phrase hits. The stretch operation extends hits to include optional extra tokens on the right. Evaluation of the entire tree produces hits for the term expansion of “non-hodgkin’s lymphoma.”
The advantage of using expression trees is that search times scale linearly with search text length, even though the number of variants grows exponentially. A drawback is that the trees will generate grammatically incorrect variants such as a phrase where every noun is in its possessive form. However, inclusion of such variants in the search causes no degradation of precision because no ambiguity is introduced and incorrect forms are simply not found.
Concept expansion
Concept expansion is implemented by expanding query terms with synonyms derived from the UMLS. In order to map into the UMLS, both the query terms and the UMLS terms are converted into a standard normal form. For example, all of the forms of non-hodgkin’s lymphoma, mentioned above, normalize to “nonhodgkin lymphoma.”
Normalization is implemented as the inverse of the term expansion described above. Whereas term expansion expands nouns with their singular and plural forms, term normalization converts both singular and plural nouns to a standard form, which happens to be the singular. Likewise, normalization converts to a preferred spelling, stripped hyphens, stripped possessives, and the shortest form of compound words. In addition, all letters are lowercased and runs of white space are converted to single blanks. The result is that any term variant produced by term expansion will map to an identical normal form.
Normalized terms from the query are mapped to normalized terms from the UMLS. This mapping may find a match of one or more UMLS concepts, each with a list of synonyms. All the synonyms for all the matched concepts are then expanded with term expansion and treated as additional variants of the original query term.
Deriving useful synonymy from the UMLS has a number of complexities. The UMLS contains many ambiguous terms. For example, 22% of the 3,764 concepts in the semantic neighborhood of “heart” were found to be ambiguous. 28 In addition, many short words are also acronyms. The term “cold” can refer to an illness, a temperature, or chronic obstructive lung disease. Further, the integration of over 100 source vocabularies into the UMLS leads to incorporation of some obscure terms, abbreviations, and foreign language synonyms, which can be difficult to identify and exclude. Many problematic terms are excluded from the synonymy via exception lists and heuristics based on characteristics such as term length and punctuation. There is also some limited human review, but this process is labor intensive, often requires subjective judgments, and is prone to error. The cleaned-up synonymy continues to produce bad hits, but appears to do more good than harm. While we continue to improve the synonymy, a weighting penalty discounts hits due to synonymy.
Mapping user queries to concepts is only partially successful. Variability in word order and the complexity of natural language inevitably lead to incomplete concept mapping, even when using a resource such as the UMLS with over one million concepts and five million terms. Because of the uncertainty in concept mapping and the potential ambiguity of synonyms, Essie gives occurrences of terms that exactly match the user’s query the highest weighting and discounts synonymous terms. Thus a search result set includes documents that contain the terms specified in the user’s query as well as synonyms identified through concept mapping.
Relaxation expansion
Up to this point the discussion has focused on simple queries that consist of a single search term, such as “amyotrophic lateral sclerosis.” Compound queries containing multiple terms, such as “heart attacks in elderly” or “secondary infections in low income patients with AIDS,” will not map directly into synonymy and will typically not be found in a corpus exactly as specified in the query.
A simple brute-force strategy has been developed to deal with compound queries. As discussed above, concept expansion treats the entire query as a phrase, expands it with synonymy and word variants when possible, but still requires strict adjacency between tokens (words). With relaxation expansion, the query is relaxed by breaking adjacency requirements. A compound phrase is broken into fragments by inserting AND operators, which still require the presence of both arguments in the document but not their adjacency. When adjacency is broken, any leading or trailing stop words on the resulting fragments are dropped while embedded stop words are retained. Additional details and an example are provided in the Relaxation expansion section.
A relaxation expansion search performs a concept expansion search for all combinations of all fragments that span the original phrase. Thus a relaxation expansion of a query such as “heart attacks in elderly” will also search for “heart attacks” AND “elderly” and will perform concept expansion on each of those fragments, so that the results will include “myocardial infarction” AND “older adults.” A scoring penalty is applied for each broken adjacency requirements, as discussed in the scoring section below.
Lossy expansion
Relaxation expansion is not sufficient to handle long natural language queries such as “What is the latest research on stage IV breast cancer and that new drug Herceptin?” The most relevant document might contain discussion of breast cancer and Herceptin, but may not mention latest research or new drugs. To handle this situation, Essie provides a lossy expansion, which permits words and fragments from the query to be dropped, with a substantial scoring penalty.
With lossy expansion, all possible subsets of query fragments are searched, weighted, and combined using an OR operator. Thus a lossy expansion of a query such as “heart attacks in elderly” will include searches for: (1) heart AND elderly, (2) myocardial infarction, and (3) attack AND adults. We believe that lossy expansion is outperformed by relaxation expansion with interactive query modification, and is only valuable when there is no opportunity for user interaction.
Scoring
Underlying the Essie scoring algorithm is the notion that phrases are qualitatively different from individual words. Essie treats phrases as atomic units of meaning, and any occurrence of that phrase is relevant to some degree. This strategy is a consequence of incorporating synonymy from the UMLS, which implies that an occurrence of “ALS” (single-word term) is somehow as relevant as an occurrence of “amyotrophic lateral sclerosis” (three-word term). Essie’s approach differs from techniques where relevancy is based on individual word frequency, such as cosine similarity measures and the inverse document frequency (IDF) formalism. 25
Essie uses a scoring method in which scores range from zero to one and are treated as the probability of relevance. Currently, the probabilities are based on heuristics and intuition and are not likely to be statistically correct. However, treating scores as probabilities provides a mathematical framework for their manipulation. Essentially, every occurrence of a search term (or its synonyms) contributes a chance that the document is relevant. The scoring algorithm consists of various rules (▶) for aggregating single-occurrence probabilities into an overall document probability.
Table 2.
Table 2 Operations Used in Scoring
| Operator | Description | Usage | Scoring | Primary Use |
|---|---|---|---|---|
| AND | Probability both occur | A AND B | PA * PB | Weighting |
| OR | Probability either occurs | A OR B | PA + PB − PA * PB | Aggregating |
| NOT | Probability A does not occur | NOT A | 1 − PA | Exclusion |
Scores are treated as probabilities, PA and PB, of independent events, A and B. If finding a search term in the title gives a probability of 0.8 that the title is relevant, then finding a second occurrence yields an aggregate probability of 0.8 + 0.8 − 0.82 = 0.96.
Single-term scoring
Working with a corpus of structured data in XML format provides ready access to document fields, such as title, abstract, conditions, treatment, key words, etc. The first step of the scoring algorithm is the determination of field scores from the number of term occurrences in each field. A value of 0.8 was empirically derived as the default prior probability that a field is relevant given one occurrence of a search term. Multiple occurrences of a term within one field are aggregated (via OR operator per ▶) so that two occurrences yields a probability of 0.96, and three, 0.992. Clearly this is a crude estimate for absolute probabilities, but only the relative values of field scores matter for relevance ranking.
Essie searches for terms, their synonyms, and a conservative set of variants of the terms and synonyms. Because of language complexities and expansion algorithm deficiencies, variants and synonyms may change meaning from the user’s intent. To address this uncertainty, all variant occurrences incur a weighting penalty of 0.9, and all synonym occurrences, 0.8. Variants of synonyms are included at no additional penalty beyond the 0.8. These numerical values are empirical and promote face validity—users indicate a preference for documents with the original search terms over documents with variants and synonyms.
Because scores are probabilities, and cannot be larger than one, all weights are penalties in the range of zero to one. Weights can also be viewed as probabilities. For example, the 0.8 weight penalty for using a synonym is an estimate that our synonymy is correct (i.e., does not distort relevance) 80% of the time.
Some fields are more significant than others, as confirmed recently in TREC Genomics track evaluations. 29 Based on the informativeness of titles in biomedical literature, 30 one expects that an occurrence of a term in the title is more significant than an occurrence in the abstract. In addition, because results are generally listed by title, the presence of the search term in the title is critical for face validity. These considerations led to introduction of weighting factors for the fields, with a significant bias toward titles. These document field weights are adjustable in configuration files. To date, the most effective choice of weights generally results in field weighting being the dominant factor in determining document score. Field weights can be thought of as a-priori probabilities that the document is relevant given that the field is relevant.
Probabilities that the fields are relevant are determined first; then each field score is weighted by the field weight. Finally, scores from all fields are aggregated to produce the probability that the entire document is relevant to the query. Generally, highly relevant documents have score contributions from several fields and an aggregated score of more than 0.90 out of 1.00.
The primary use of document scores is to rank the most relevant documents first. The actual value of the probabilities need not be correct, or even approximately correct, so long as ranks are preserved. Given a corpus annotated with relative relevancy scores, it should be possible to rigorously determine appropriate values for field weights rather than the current ad-hoc approach for assigning weights. Such a corpus would be significantly more difficult to construct than existing corpora with binary relevance judgments. To our knowledge, such a corpus does not exist.
One might expect that field length would have a significant impact on relevancy. A logarithmic dependency on field length was implemented, but resulted in a negligible improvement in precision. This can be readily understood if you break the field length into two components: (1) the average length of the field, and (2) the length variation with respect to the average. The average length variation will produce a weighting that applies to the field but does not vary from document to document. This weighting is already part of the user-chosen field weighting discussed above. So, while it is true that abstracts are generally longer than titles, the greater length is already accounted for by the higher weight given to titles. The length variation from document to document within a particular field is often reasonably small. For example, title lengths are 88 ± 39 characters in ClinicalTrials.gov records and 92 ± 39 characters in Medline citations. Because the adjustment is applied as a logarithm of the field length, this turns into roughly a 10% variation in score for an occurrence of a search term. These occurrence scores are then aggregated over all occurrences of all search terms in all fields in each document. In sum, the field length adjustments had a minimal effect on the overall document score and resulted in a negligible improvement in precision.
Some fields contain texts that are shorter than a sentence, such as a key word. In these cases, a partial match is common such as “heart disease” in a field containing “ischemic heart disease.” Essie has parameters that will weight partial matches lower, effectively boosting the scores where there is an exact match that covers the entire field, such as “heart disease” in “heart disease.” It is also possible to weight a partial match at the beginning or end of a field higher than a partial match in the middle of a field. By default, a full match is given weight 1.0, a match at the beginning or end of a field is given weight 0.9, and a partial match in the middle is given weight 0.8.
In summary, occurrences are weighted and aggregated (▶) to produce a score for each field (▶). Field scores are then weighted and aggregated to produce a score for each document. The net result is a score that is a probability that the document is relevant to the given search term. For single-term searches with concept expansion or less, no more is required, and the scores are ranked and documents returned.
Table 3.
Table 3 Summary of Scoring Algorithm for a Single-term Search with Concept Expansion or Less
|
Relaxation expansion
Relaxation expansion breaks adjacency requirements by inserting AND operators within the query search text. A relaxation expansion will search for all combinations of all fragments that span the original text. The more the text is fragmented by inserting AND operators, the larger the weighting penalty applied. Longer texts are deemed more likely to consist of multiple terms, and are penalized less. The weighting factor is determined by:
Weighting Penalty = αβ
where α = 0.02, β = NANDs / (NWords − 1).
For example, a search of the compound query text “heart attacks in elderly” with relaxation expansion is rendered in the Essie search syntax as:
WEIGHT [1.00] (heart attacks in elderly) OR
WEIGHT [0.14] (heart attacks AND elderly) OR
WEIGHT [0.14] (heart AND attacks in elderly) OR
WEIGHT [0.02] (heart AND attacks AND elderly)
Each of the fragments is searched with concept expansion (i.e., expanded with synonymy). Fragments that are medical terms, such as “heart attacks,” will map into UMLS concepts, be expanded with synonymy, find occurrences in documents, and contribute to the final score. Fragments that do not make much sense, such as “attacks in elderly,” are unlikely to occur in documents, will not map into UMLS concepts, and generally do not contribute to the final score.
Due to the substantial weighting penalties applied for breaking adjacency, documents containing the original, unbroken query will be ranked at the top. Documents found by fragments are ranked lower. The only time documents found by fragments have the highest rank is when there are no documents containing the original unbroken query. Thus, relaxation expansion increases recall without a significant degradation in precision of the “best” documents.
Lossy expansion
When using lossy expansion, Essie allows query fragments to be dropped, although severe scoring penalties apply. In the current implementation, dropping a single word has a penalty equal to breaking three adjacency requirements from relaxation expansion. Dropping a multiword fragment costs as much as dropping the words individually. The previous example with lossy expansion becomes:
- WEIGHT [1.0]
- (WEIGHT [1.00] (heart attacks in elderly) OR
- WEIGHT [0.14] (heart attacks AND elderly) OR
- WEIGHT [0.14] (heart AND attacks in elderly) OR
- WEIGHT [0.02] (heart AND attacks AND elderly)) OR
- WEIGHT [0.01]
- WEIGHT [1.00] (heart attacks) OR
- WEIGHT [0.14] (heart AND attacks) OR
- WEIGHT [0.14] (heart AND elderly) OR
- WEIGHT [1.00] (attacks in elderly) OR
- WEIGHT [0.14] (attacks AND elderly)) OR
- WEIGHT [0.0001]
- WEIGHT [1.00] (heart) OR
- WEIGHT [0.14] (attacks) OR
- WEIGHT [0.14] (elderly))
Due to the substantial weighting penalties applied for dropping words, documents containing all of the original query terms will have highest rank. Lossy expansion increases recall without significant degradation in “precision at ten,” because the results from lossy expansion will have a lower rank than results from concept expansion and relaxation expansion.
Dropping fragments in lossy expansion raises the question of the relative value of each of the fragments. Because all fragments are searched, the number of times each fragment occurs in each document and in the corpus as a whole is known. We have experimented with using an inverse document frequency measure to evaluate the value of each fragment with weights based on the logarithm of the number of occurrences found for each fragment. The effect on ranking was sufficiently small that the experiments were deemed inconclusive, and this strategy has not been adopted.
Validation
Evaluating a search system in the biomedical domain is difficult. There are few corpora, little agreement about what constitutes a reasonable query, and few relevance judgments. Studies on human judgments of relevancy show wide disagreement. The TREC Conference offers some assistance in this area, but the numbers of queries and relevance judgments in the biomedical domain are quite small.
TREC 2003
TREC 2003 Genomics track used mean average precision (MAP—average precision at each point a relevant document is retrieved) as the official evaluation metric. 29 Essie (formerly referred to as SE) achieved the high score of 0.42 MAP, ranking first among a total of 25 groups that submitted 49 official runs for scoring. In fact, all strategies for reranking Essie results failed to improve the MAP for the NLM submissions to TREC. Runs from the University of California at Berkeley, 0.39 MAP, and the National Research Council of Canada, 0.39 MAP, ranked next highest in this evaluation. 29
Several lessons were learned from participating in TREC 2003. Most important was the realization that it is critical to understand the structure and content of the data. Recognizing and mapping of organism name in the query to the MeSH descriptors in the corpus documents was the biggest contribution to MAP. Next in importance was field weighting. Weighting the title larger than the abstract was significantly better than treating all fields equally. Tokenization and term variation was important, especially for gene names. Essie’s strategy of indexing all tokens, including punctuation, allowed for the recognition of compound word forms, such as JAK2, JAK-2, and JAK 2. Further, complex terms like d-ala(2) are lost if individual characters and punctuation are treated as stop words. Synonymy did not contribute significantly to MAP scores in TREC 2003 because the queries already contained many useful synonyms.
TREC 2006
The 2006 Genomics TREC task was to extract relevant passages from full-text HTML journal articles. To accomplish this task, the NLM team followed the strategy of: (1) extracting “<p>” delimited paragraphs, (2) relevancy ranking the paragraphs with Essie, and then (3) trimming the best paragraphs down to the few best sentences. This last step dropped leading and trailing sentences that did not contain query terms, which should have minimal impact on the Essie relevance judgments. Because the source text was HTML, it was difficult to extract useful document structure. In the end, all text was treated equally, and Essie field weighting played no significant role. Essie was used “as is,” with no modifications to ranking and retrieval algorithms.
Three runs of Essie searches were conducted and submitted for evaluation: automatic, manual, and interactive. In the automatic run, an Essie query was created for each TREC question by extracting the biological processes and objects terms. 31 The inaccuracies in the automatic recognition of the biological objects and processes were corrected in the queries for the manual search. In the interactive run, the queries were created by a domain expert interacting with the system. This run most closely models how Essie is used in practice. Details of query formulation and analysis of the results are provided in the NLM 2006 TREC report. 31
Although no attempt was made to find different aspects pertaining to a question, the interactive Essie run achieved the second highest aspect MAP = 0.405, behind the University of Illinois team that achieved MAP = 0.441, out of a total of 92 submitted runs. The three runs submitted by the University of Illinois team ranked above the interactive run in document MAP as well (▶). In the passage retrieval, Essie ranked 9th, possibly due to the fact that we discarded the references sections of the full-text papers, but in fact many of those were judged to be relevant.
Table 4.
Table 4 Text REtrieval Conference (TREC) 2006 Comparison for Passage, Aspect, and Document Mean Average Precision (MAP) for Essie Automatic, Manual, and Interactive Searches (Total of 92 Submitted Runs)
| System | Passage MAP | Aspect MAP | Document MAP |
|---|---|---|---|
| Essie interactive | 0.0827, 9th | 0.4051, 2nd | 0.4730, 4th |
| Essie manual | 0.0470, 25th | 0.2664, 12th | 0.3648, 23rd |
| Essie automatic | 0.0406, 34th | 0.1922, 33rd | 0.3421, 36th |
| Best passage run (Tsinghua University) | 0.1486, 1st | 0.3040, 7th | 0.4335, 7th |
| Best aspect and document run ∗ | 0.0750, 10th | 0.4411, 1st | 0.5439, 1st |
∗ University of Illinois at Chicago.
Essie was used “as is” to perform a nearby retrieval task, relevant passages in unstructured text, as opposed to relevant documents with structured text. The good results indicate that the underlying retrieval and relevancy ranking algorithms have some merit for the biomedical domain. Further evaluation is ongoing.
Production Use
Essie has been used as the search engine for ClinicalTrials.gov 6 since October 2001. Other groups at the NLM are also utilizing Essie for their projects. The Genetics Home Reference 32 uses Essie to serve consumer information about genetic conditions and the genes that are responsible for them. The NLM Gateway 33 searches simultaneously in multiple library resources, and uses Essie directly for certain data sets as well as indirectly in some of the systems it queries. Another user of Essie is Medline Database On Tap, 34,35 which provides Medline access through wireless handheld devices at the point of care. Each of these projects has structured data in XML form, benefits from using synonymy derived from the UMLS, and has chosen Essie as its search system.
Discussion
Implementation Considerations
The strategies used by Essie are computationally expensive and resource intensive. The token adjacency index can be up to ten times the size of the document set. The Essie Medline implementation makes use of servers that have a considerable amount of random-access memory (64 GB) to reduce the use of slower disk access. Described below are some specific techniques used to make implementation of the algorithms more practical.
The index for each token occurrence carries context information for some number of tokens to its left and right. The number of context tokens included varies depending on the number of occurrences of the token. For rare tokens, the context may include as many as ten tokens on either side. For common tokens, the context will contain only two tokens on either side. Including adjacent tokens in the context makes the index considerably larger, but allows many search optimizations and provides good locality of reference for increased computational efficiency. For example, in the term “non-hodgkin’s lymphoma,” context information can be used to check for an adjacent “non,” hyphen, apostrophe, and letter “s,” so that only the occurrences for tokens “hodgkin” and “lymphoma” need to be loaded from disk. The adjacency of the rest of the tokens can be determined from the contexts.
In relaxation expansion, the query is broken into fragments by inserting AND operators between words. The number of possible fragments increases quadratically with the length of the query. Fortunately, searching for long terms that are not in the corpus typically fails with no occurrences after examining just a few tokens. The cost of searching for all of the fragments in long queries effectively grows linearly.
In lossy expansion, some query fragments can be dropped, and all possible subset combinations of fragments are searched. The number of possible combinations of fragments increases exponentially with the length of the query. Fortunately, any combination containing a fragment with zero occurrences can be skipped. This effectively reduces the number of combinations that must be searched and makes it feasible to handle the combinations of a long query.
Despite these optimizations, the explosive nature of the expansions makes the implementation vulnerable to failure when given a very long query. The worst queries occur when a user types in a long document title as his or her search query. In this case, every fragment and combination exists in the corpus, requiring tremendous computation to fully evaluate all the possibilities. To handle this case, fragments and combinations are evaluated in order of most value and searches are abandoned after exceeding a predetermined amount of computation.
Practical Usage
Interactive systems primarily use relaxation expansion, not lossy expansion. The relaxation algorithm retrieves fewer documents but with reasonable assurance of relevance. The lossy algorithm retrieves many more documents, but the lowest-ranked documents are rarely relevant. For this reason, the lossy algorithm is generally used for information retrieval experiments where there is no human feedback loop to enable query refinement. Relaxation expansion works well for interactive systems, where the users have an opportunity to refine their search if they are unsatisfied with the results.
Given that the relaxation implementation has checked for the presence of every fragment and word in the user’s query, it is possible to take this information and present it to the user in the form of search suggestions. These suggestions present the productive fragments of the user’s query in various combinations and eliminate words and phrases from the user’s query that are not found in the corpus. Suggestions are always available if the user looks for them, but in the special case in which a user query fails to find any documents, suggestions are prominently displayed. So, whenever the original search is overspecified, the user is offered several nearby queries that are less restrictive and that will yield documents.
Current Limitations
Essie’s search strategy works well in the domain for which it was designed. The current implementation of Essie is limited to a domain that satisfies a number of constraints:
• A corpus consisting of documents that are structured in XML with sufficient structure to distinguish between more valuable and less valuable sections of a document
• A relatively homogeneous corpus in which the value and size of the structured fields is fairly uniform
• A relatively static corpus that enables sufficient time for the extensive indexing required by the algorithms
• Documents in the biomedical domain in which multiword terms are used extensively and in which the UMLS provides an appropriate basis for synonymy
Future Directions
The weights and penalty values used in Essie were derived in a fairly ad-hoc way. Values were adopted that seemed reasonable after performing a number of searches and examining the ordering of search results. Frequently, deciding which of two documents should have a higher rank based on a given query is quite arbitrary. We continue to tune the weights and penalty values. One interesting possibility for future work would be to identify or build an appropriate training set and then use machine learning techniques to optimize the weights and penalties.
In addition to the ranking of documents based on direct relevance to a user’s query, Essie’s document scoring algorithm makes it possible to include other ranking criteria. For example, it is possible to establish general filters to adjust rankings based on general criteria such as documents about “treatments,” “diagnosis,” “genetics,” or other topics. This strategy has been used in a number of other systems. 36–39 Evaluation of the utility of such re-ranking strategies is currently underway.
Another possible area for future work is the incorporation of a feedback loop from the top-ranked documents. 40,41 This strategy would augment the existing system by identifying the most significant phrases and words from the best documents and then adding those phrases and terms back into the search query.
Essie is a work in progress, and we will continue to address limitations as they become apparent. In particular, we are constantly evaluating the effectiveness of the ranking algorithm. Every corpus indexed has led to new insights and subsequent refinements.
Conclusion
Essie was developed for the ClinicalTrials.gov project at the NLM. The goal of Essie’s indexing, searching, and ranking algorithms is to reduce the burden on the user of developing sophisticated query strategies when searching a biomedical corpus. Preliminary evaluations conducted as part of the TREC Genomics track demonstrate the effectiveness of the Essie retrieval and ranking algorithms. The system is in active use on ClinicalTrials.gov as well as several other projects at the Lister Hill National Center for Biomedical Communications. Essie is an active research project with ongoing development and enhancements.
Essie utilizes a fine-grained tokenization algorithm that preserves punctuation information along with concept and phrase searching. Phrases and words from the user’s query augmented with appropriate variants and synonyms are “all the right pieces.” The document structure is exploited to distinguish the high-value areas of a document from the low-value areas to define “all the right places.” Essie’s ranking algorithm can be summarized as giving preference to documents with “all the right pieces in all the right places.”
Footnotes
This article is written by an employee of the US Government and is in the public domain. This article may be republished and distributed without penalty.
The views expressed in this paper do not necessarily represent those of any U.S. government agency, but rather reflect the opinions of the authors.
The authors thank Dr. Alexa McCray, Dr. Deborah Zarin, and the research community at the Lister Hill Center for their support and encouragement.
References
- 1.Alper BS, White DS, Ge B. Physicians answer more clinical questions and change clinical decisions more often with synthesized evidence: A randomized trial in primary care Ann Fam Med 2005;3:507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward D, Meadows SE, Nashelsky JE. The role of expert searching in the Family Physicians’ Inquiries Network (FPIN) J Med Libr Assoc 2005;93:88-96. [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: A description based on the theories of Zellig Harris J Biomed Inform 2002;35:222-235. [DOI] [PubMed] [Google Scholar]
- 4.McCray AT, Tse T. Understanding search failures in consumer health information systems AMIA Annu Symp Proc 2003:430-434. [PMC free article] [PubMed]
- 5.McCray AT, Ide NC. Design and implementation of a national clinical trials registry J Am Med Inform Assoc 2000;7:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. ClinicalTrials.gov. 2006. Available at: http://clinicaltrials.gov. Accessed July 26, 2006.
- 7.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System Methods Inf Med 1993;32:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hersh W, Greenes R. SAPHIRE—An information retrieval system featuring concept matching, automatic indexing, probabilistic retrieval, and hierarchical relationships Comput Biomed Res 1990;23:410-425. [DOI] [PubMed] [Google Scholar]
- 9.Hersh W, Hickam DH, Haynes RB, McKibbon KA. Evaluation of SAPHIRE: An automated approach to indexing and retrieving medical literature Proc Annu Symp Comput Appl Med Care 1991:808-812. [PMC free article] [PubMed]
- 10.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: AMIA Annu Symp Proc 2001:17-21. [PMC free article] [PubMed]
- 11.Voorhees EM. Natural language processing and information retrievalIn: Pazienza MT, editor. Information Extraction: Towards Scalable, Adaptable Systems. New York: Springer; 1999. pp. 32-48.
- 12.Srinivasan P. Retrieval feedback in Medline J Am Med Inform Assoc 1996;3:157-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aronson AR, Rindflesch TC. Query expansion using the UMLS Metathesaurus Proc AMIA Annu Symp Proc 1997:485-489. [PMC free article] [PubMed]
- 14.Browne AC, Divita G, Aronson AR, McCray AT. UMLS language and vocabulary tools AMIA Annu Symp Proc 2003:798. [PMC free article] [PubMed]
- 15.Aronson AR, Mork JG, Gay CW, Humphrey SM, Rogers WJ. The NLM Indexing Initiative’s Medical Text Indexer Medinfo 2004;11(pt 1):268-272. [PubMed] [Google Scholar]
- 16.Harman D. In: Harman DK, editor. Overview of the Fourth Text REtrieval Conference (TREC-4). Gaithersburg, MD: National Institute of Standards and Technology (NIST); 1995. pp. 1-24Proceedings of the Fourth Text REtrieval Conference (TREC-4). NIST Special Publication 500–236.
- 17.Mitra M, Buckley C, Singhal A, Cardie C. In: Devroye L, Chrisment C, editors. An analysis of statistical and syntactic phrases. Montreal, Canada: McGill University; 1997. pp. 200-214Conference Proceedings of RIAO-97. June.
- 18.Rindflesch TC, Aronson AR. Ambiguity resolution while mapping free text to the UMLS Metathesaurus Proc Annu Symp Comput Appl Med Care 1994:240-244. [PMC free article] [PubMed]
- 19.Manning CD, Schütze H. Foundations of Statistical Natural language Processing. Cambridge, Massachusetts: MIT Press; 2003.
- 20.Hull DA, Grefenstette G. A Detailed Analysis of English Stemming Algorithms. Grenoble,France: Xerox Research Centre Europe; 1996. Report No. 1996–023.
- 21.Bilotti MW, Katz B, Lin J. What works better for question answering: Stemming or morphological query expansion?. Sheffield, England: ACM Press; 2004. pp. 1-7In: SIGIR 2004: Proceedings of the Information Retrieval for Question Answering Workshop, 2004 July 25–29.
- 22.Fox C. Lexical analysis and stoplistsIn: Frakes WB, Baeza-Yates R, editors. Information Retrieval. Upper Saddle River, NJ: Prentice Hall; 1992. pp. 102-130.
- 23.Huang X, Zhong M, Si L. York University at TREC 2005: Genomics track. Gaithersburg, MD: National Institute of Standards and Technology (NIST); 2005. In: Proceedings of the Fourteenth Text REtrieval Conference, 2005 Nov 15–18.
- 24.Singhal A. Modern information retrieval: A brief overview IEEE Data Eng Bull 2001;24:35-43. [Google Scholar]
- 25.Baeza-Yates R, Ribiero-Neto A. Modern Information Retrieval. New York: Addison Wesley; 1999.
- 26.Mitchell JA, Aronson AR, Mork JG, Folk LC, Humphrey SM, Ward JM. Gene indexing: characterization and analysis of NLM’s GeneRIFs AMIA Annu Symp Proc 2003:460-464. [PMC free article] [PubMed]
- 27.Hersh WR, Cohen A, Roberts P, Bhupatiraju RT. TREC 2006 genomics track overview. Gaithersburg, MD: National Institute of Standards and Technology (NIST); 2006. In: Proceedings of the Fifteenth Text REtrieval Conference, 2006 Nov 14–17.
- 28.Bodenreider O, Hole WT, Humphreys BL, Roth LA, Srinivasan S. Customizing the UMLS Metathesaurus for your applications AMIA Annu Symp Proc 2002;T13:158. [Google Scholar]
- 29.Kayaalp M, Aronson AR, Humphrey SM, et al. Methods for accurate retrieval of Medline citations in functional genomics. Gaithersburg, MD: National Institute of Standards and Technology (NIST); 2003. pp. 441-450In: Proceedings of the Twelfth Text REtrieval Conference, 2003 Nov 18–21.
- 30.Demner-Fushman D, Hauser S, Thoma G. The Role of Title, Metadata and Abstract in Identifying Clinically Relevant Journal Articles AMIA Annu Symp Proc 2005:191-195. [PMC free article] [PubMed]
- 31.Demner-Fushman D, Humphrey SM, Ide NC, et al. Finding relevant passages in scientific articles: Fusion of automatic approaches vs. an interactive team effort. Gaithersburg, MD: National Institute of Standards and Technology (NIST); 2006. In: Proceedings of the Fifteenth Text REtrieval Conference, 2006 Nov 14–17.
- 32. Genetics Home Reference. 2006. Available at: http://ghr.nlm.nih.gov. Accessed July 26, 2006.
- 33. NLM Gateway. 2006. Available at: http://gateway.nlm.nih.gov. Accessed July 26, 2006.
- 34. Medline Database on Tap. 2006. Available at: http://mdot.nlm.nih.gov/proj/mdot/mdot.php. Accessed July 26, 2006.
- 35.Hauser SE, Demner-Fushman D, Ford GM, Thoma G. Preliminary comparison of three search engines for point of care access to Medline citations AMIA Annu Symp Proc 2006;945. [PMC free article] [PubMed]
- 36.Cimino JJ, Aguirre A, Johnson SB, Peng P. Generic queries for meeting clinical information needs Bull Med Libr Assoc 1993;81:195-206. [PMC free article] [PubMed] [Google Scholar]
- 37.Haynes RB, Wilczynski N, McKibbon KA, Walker CJ, Sinclair JC. Developing optimal search strategies for detecting clinically sound studies in Medline J Am Med Inform Assoc 1994;1:447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratt W, Hearst MA, Fagan LM. A knowledge-based approach to organizing retrieved documents. Orlando, Florida: AAAI Press (American Association for Artificial Intelligence); 1999. In: AAAI ‘99: Proceedings of the 16th National Conference on Artificial Intelligence.
- 39.Demner-Fushman D, Lin J. Knowledge extraction for clinical question answering: Preliminary results. Pittsburgh, PA: AAAI Press (American Association for Artificial Intelligence); 2005. pp. 1-10In: Proceedings of the AAAI-05 Workshop on Question Answering in Restricted Domains, 2005 Jul 9–13.
- 40.Harman D. In: Belkin NJ, Ingwersen P, Pejtersen AM, editors. Relevance feedback revisited. Copenhagen, Denmark: ACM Press; 1992. pp. 1-10SIGIR 1992: Proceedings of 15th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, 1992 June 21–24.
- 41.Ruthven I, Lalmas M. A survey on the use of relevance feedback for information access systems Knowledge En Rev 2003;18:95-145. [Google Scholar]