Abstract
Objective
Negation is common in clinical documents and is an important source of poor precision in automated indexing systems. Previous research has shown that negated terms may be difficult to identify if the words implying negations (negation signals) are more than a few words away from them. We describe a novel hybrid approach, combining regular expression matching with grammatical parsing, to address the above limitation in automatically detecting negations in clinical radiology reports.
Design
Negations are classified based upon the syntactical categories of negation signals, and negation patterns, using regular expression matching. Negated terms are then located in parse trees using corresponding negation grammar.
Measurements
A classification of negations and their corresponding syntactical and lexical patterns were developed through manual inspection of 30 radiology reports and validated on a set of 470 radiology reports. Another 120 radiology reports were randomly selected as the test set on which a modified Delphi design was used by four physicians to construct the gold standard.
Results
In the test set of 120 reports, there were a total of 2,976 noun phrases, of which 287 were correctly identified as negated (true positives), along with 23 undetected true negations (false negatives) and 4 mistaken negations (false positives). The hybrid approach identified negated phrases with sensitivity of 92.6% (95% CI 90.9–93.4%), positive predictive value of 98.6% (95% CI 96.9–99.4%), and specificity of 99.87% (95% CI 99.7–99.9%).
Conclusion
This novel hybrid approach can accurately locate negated concepts in clinical radiology reports not only when in close proximity to, but also at a distance from, negation signals.
Introduction
With the increasing adoption of EHR (Electronic Health Record) 1 systems, huge amounts of patient data are stored electronically. These data contain valuable information for patient care, biomedical research and education. Data stored in a structured, standards-based format can facilitate fast and accurate retrieval. However, despite many efforts to acquire clinical data using structured data entry, 2,3 a large portion of the data are still acquired and stored in narrative clinical reports, using an unstructured format referred to as “free-text.” A large corpus of narrative clinical documents usually needs to be indexed either using words or concepts in a biomedical terminology to be used for information retrieval (IR). 4
Though word-based indexing using a vector space model 5 is simple and powerful, concept-based indexing using biomedical terminologies can improve the performance of biomedical IR over that of the vector space model. 6 The National Library of Medicine’s (NLM) Unified Medical Language System (UMLS) 7 provides comprehensive coverage of biomedical concepts and many researchers have studied a variety of approaches to concept-based indexing of clinical documents using the UMLS. 8–12
Negation is commonly seen in clinical documents 13 and may be an important source of low precision in automated indexing systems. 14 Because of the different semantics associated with affirmative biomedical concepts vs. when the same concepts are negated, it is essential to detect negations accurately to facilitate high performance document retrieval in clinical documents.
Generally speaking, negation is complex in natural languages, such as English. It has been an active research topic for decades. Researchers have approached this topic from both linguistic and philosophical perspectives. 15,16 In most cases, negation involves a negation signal, a negated phrase containing one or more concept(s), and optionally some supporting feature (pattern), which helps us locate the negated phrase. In the following example,
There is no evidence of cervical lymph node enlargement.
“no” is the negation signal used to denote that a following concept is negated; “cervical lymph node enlargement” is the negated phrase; while “evidence of” is the supporting phrase feature.
Background
Negation has been investigated by a number of researchers in the clinical domain. 14,17–19 Sager et al. implemented a comprehensive English grammar in the Linguistic String Project (LSP) Natural Language Processing (NLP) system to parse semantic phenomena including negations. 20 The system was later specialized for Medical Language Processing (MLP). 21 Inspired by the work of LSP, Friedman et al. developed the Medical Language Extraction and Encoding (MedLEE) system to encode clinical documents into a structured form using semantic grammar. 22 MedLEE encoded concepts including negated ones with various “certainty” modifiers, and thus was able to differentiate negated concepts from other concepts in retrieval. 17 A study in 2004 reported a recall of 77% and a precision of 89% in extracting UMLS concepts from sentences in discharge summaries, at a performance level comparable to that of individual experts. 23 Both above systems detect negations using grammar in NLP parsing, however, no evaluation was reported focusing on negation detection.
Mutalik et al. showed in Negfinder, 14 that a One-token Look-Ahead Left-to-right Rightmost-derivation (LALR(1)) parser could reliably detect negations in surgical notes and discharge summaries to achieve a sensitivity of 95.7% and a specificity of 91.8% without extracting syntactical structures of sentences and phrases as in full NLP parsing. It helped reduce the input complexity for the LALR(1) parser to replace words in text with UMLS concept IDs before negation tagging. Such a concept replacement process may impact the overall performance of negation detection; however, its performance was not reported. As pointed out by the authors, the limitations of such a single token look-ahead parser prevented it from detecting negated concepts correctly if the negation signal was more than a few words away from negated concepts.
NegEx, a regular expression–based algorithm developed by Chapman et al., 18 though simple to implement, has been shown to be powerful in detecting negations in discharge summaries 18 with a sensitivity (recall) of 77.8%, a positive predictive value (PPV, precision) of 84.5%, and a specificity of 94.5%. NegEx could identify a term as negated after it was mapped to a UMLS concept. The results were calculated using successfully mapped UMLS terms only. An improved NegEx (version 2) was reported to have generally lower performance in pathology reports without any customizations for the new document type. 24 This later study included text phrases not mapped to UMLS concepts and identified UMLS concept mapping as one major source of error.
More recently, Elkin et al. studied this problem using a negation ontology containing operators and their associated rules. 19 Operators were two sets of terms with one set starting negations and another set stopping the propagation of negations. Each sentence was first broken into text and operators, with text mapped to SNOMED CT concepts by Mayo Vocabulary Server using automated term composition (ATC). 25 Those concepts were assigned one of the three possible assertion attributes according to the negation ontology, with “negative assertion” being studied. This study expanded the previous studies by formally evaluating the coverage of concept mapping proceeding negation detection. The human reviewer in the study identified that 205 of 2,028 negative concepts were not mapped by SNOMED CT, revealing the terminology’s coverage of 88.7% of the negative concepts. The 205 unmapped negative concepts were not included in the gold standard in calculating the performance of negation assignment: the sensitivity (recall) of 97.2%, the PPV (precision) of 91.2%, and the specificity of 98.8%.
The published evaluations on negation detection used mainly lexical approaches without using the syntactical structural information of a sentence embedded in its parse tree generated through full NLP parsing. They were shown to be effective and reliable; however, determining the scope of a negation was noted as a challenge by several authors. 14,18 The above approaches could determine the scope of negations reliably when a negated concept is close to a negation signal, but unsatisfactorily when they are separated with multiple words not mapped to a controlled terminology. We have devised a novel approach including a classification scheme based on syntactical categories of negation signals and their corresponding natural language phrase patterns to support locating negated concepts both in close proximity to and at a distance from negation signals.
ChartIndex is an automated concept indexing system using a contextual indexing strategy 26 and an NLP approach for noun phrase identification 27 before concept mapping to improve indexing precision. It uses an open-source high-performance statistical parser, the Stanford Parser, 28 to generate a full parse tree of each sentence, which provides the syntactical information on sentence structure used by our negation detection approach. A classification of negations was first developed according to the syntactical categories of negation signals, and the phrase patterns required to locate negated phrases. One then uses a hybrid approach, combining regular expression matching and a grammatical approach, to locate negated phrases within a parse tree. The classifier first detects possible negation in a sentence, and classifies the negation into one of 11 categories by regular expression matching. The computer then extracts the negated phrases from the parse tree, according to grammar rules developed for that negation type. Regular expression matching is fast and sensitive in identifying the type of negations, while the grammatical approach helps locate negated phrases accurately within or outside the proximity of the negation signal.
In most previous studies, negative concepts were tallied only when they were in a controlled terminology, such as UMLS or SNOMED CT. It was plausible as the purpose of study was to assess a negation detection module placed after concept mapping process. To evaluate the impacts of concept mapping on negation detection, Mitchell et al. 24 allowed human reviewers to mark phrases not mapped to a controlled terminology and included unmapped phrases in the performance calculations. Similarly, we used phrases in the text without a concept mapping process in this study to evaluate the performance of negation assignment, independent of concept mapping.
Methods
Research Hypothesis
We tested the following two hypotheses in this experiment:
1 The structure information stored in parse trees helps identify the scope of negation.
2 For radiology reports, a negation grammar with good coverage could be derived from a small number of reports and the hybrid approach could achieve good performance using such a grammar.
For the purpose of this study, only complete negations (versus partial negations such as “probably not”) within a sentence are considered. Phrases were considered negated if they were indicated as “completely absent” in the clinical document. Normal findings and test results were not considered as negated based on discussions with physicians. In the following example given by Mutalik et al., “several blood cultures, six in all, had been negative,” “several blood cultures” were not considered as negated because it is a noun phrase representing a test, the results of which were normal. Negations within a word were not considered, as in the case of negative prefix or suffix, because they are often semantically ambiguous, moreover, the best way to represent these words may depend on the controlled terminologies used for concept encoding. For example, people would agree that “nontender” is a negation meaning “not tender,” however, “colorless” is itself a concept in SNOMED CT (263716002) to describe a color attribute: transparent. People may not agree on whether it is a true negation or how to represent it, either as “colorless” (263716002) or as negation of “colors” (263714004). Mutalik et al. gave more examples in their paper: 14 “A final issue is that many UMLS concepts themselves represent antonymous forms of other concepts, e.g., words beginning with “anti-,” “an-,” “un-,” and “non-.” Such forms are not necessarily negations. (Thus, an anti-epileptic drug is used when epilepsy is present; “non-smoker,” however, is a true negation.)” We decided to mark-up negated biomedical noun phrases instead of UMLS concepts, to focus on evaluating negation detection in this experiment. The concepts represented by these negated phrases, whether in a controlled terminology or not, are negated concepts.
Deriving Negation Grammar
The document collection used in our study was 1,000 de-identified radiology reports of six common imaging modalities from Stanford University Medical Center. After deriving a grammar-based classification scheme from a limited set of 30 reports (see below), the coverage of the classification was validated using another 470 reports. These 500 reports served as the training set. A negation detection module (NDM) was implemented using the above grammar, which was later tested on a set of 132 reports randomly selected from the remaining 500 reports.
To construct a negation grammar in radiology reports, we manually identified sentences with negations in 30 reports of all six modalities and marked-up negation signals, negated phrases, and negation patterns. We also studied the published literature to construct a more extensive list of possible negation patterns from a linguistics perspective. Instances and patterns were reviewed by a physician from a clinical application perspective to construct a preliminary classification of negations. Negations were classified based on the syntactical categories of negation signals, and the phrase patterns. The following is an example of one such grammar rule for an adjective-like negation type, where the negation signal is a determiner such as “no” or a preposition such as “without,” or an adjective such as “absent,” followed by a noun phrase to negate phrases. Determiners and prepositions are different from adjectives. However, the noun phrases, such as “evidence of” following “no,” “without,” or “absent,” determine the scope of the negated phrases in the same way. Therefore we categorize these three together to reduce the total number of grammar rules.
Adjective-like Negation : Phrasal : NounPhrase
Expression Pattern: N [JJ] NN⊻NNS {of⊻for⊻to suggest} NegdPhr
N → {no⊻without⊻absent}
JJ → {mammographic⊻significant}
NN → {evidence⊻feature⊻area⊻pattern⊻history⊻sign}
NNS → {features⊻areas⊻patterns⊻signs}
Grammar:
PP → IN0 NP
IN0 → {without}
NP → NP PP
NP → DT⊻JJ0 [JJ1] NN⊻NNS
NP → [JJ1] NN⊻NNS
DT → {no}
JJ0 → {absent}
JJ1 → {mammographic⊻significant}
NN → {evidence⊻feature⊻area⊻pattern⊻history⊻sign}
NNS → {features⊻areas⊻patterns⊻signs}
PP → IN NP
IN → {of⊻for}
NP → NegdPhr
Example: There is no evidence of cervical lymph node enlargement.
Note: N – Negation Signal, [JJ] – optional Adjective, NN – Noun singular, NNS – Noun plural, IN – Preposition, NegdPhr – Negated Phrase, NP – Noun Phrase, VP – Verb Phrase, PP – Prepositional Phrase, A⊻B – either A or B.
▶ shows a parse tree of the above sentence generated by the Stanford Parser, with each token of the sentence tagged with a Part-Of-Speech (POS) tag. A POS tag identifies the syntactic category of a sentence component, such as JJ for an adjective and NN for a noun. As shown in ▶ each grammar rule like the above was further translated into a structural rule to extract negated phrases within a parse tree: 1. Locate the noun phrase (NP) with a head from a small set of nouns such as “evidence” and modified by word “no,” “without,” or “absent;” 2. Locate the prepositional phrase (PP) headed by “of” or “for” following the above NP; 3. Extract the NP under the above PP, which contains the negated phrase (NegdPhr).
Figure 1.
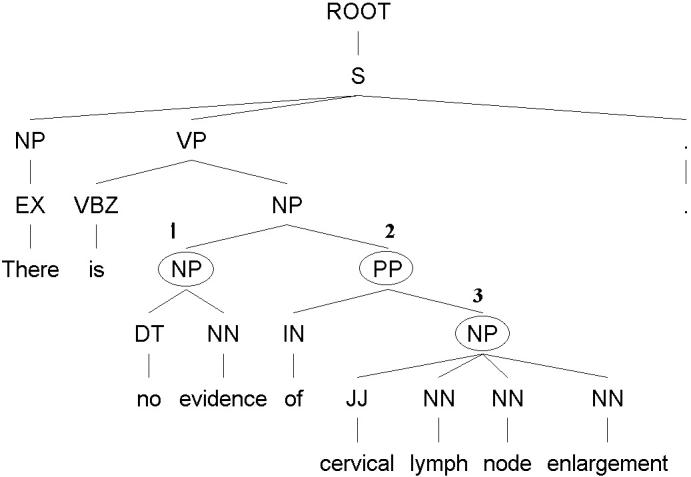
An example illustrating how to use a structural rule to extract negated phrases.
Using a PERL script, 1,384 sentences containing possible negations were extracted from 6,590 sentences in the remaining 470 reports of the training set. We then marked-up the 1384 sentences using the draft classification and repeated the review process to improve the grammar. To further validate the comprehensiveness of the classification, we inspected 1,600 sentences marked by the script as not containing negations. This resulted in 14 instances of negations being found, none of which revealed any new syntactical patterns, but it did add two additional negation signals to our list.
In the above process, a comprehensive syntactical classification of negations in radiology reports was obtained. The classification has been shown in ▶, with negations firstly classified based on the syntactical category of negation signals as an adjective-like (such as “no, absent” and a preposition such as “without”), adverb (such as “not”), verb (such as “deny”), and noun (such as “absence”) respectively. To locate negated phrases, negations are further classified based on phrase patterns, including words critical in defining negation patterns and the negation patterns themselves. The first two columns of the table contain the syntactical categories of negation signals and phrase patterns, which support locating negated phrases not in close proximity with negation signals from the output of an NLP parser. The third column contains examples for each category, where negation signals are bolded, negation patterns are italic, and negated phrases are underlined.
Table 1.
Table 1 The Classification of Negations, Based on the Syntactical Category of Negation Signal, and Phrase Patterns
| Negation Signal Type | Phrase Pattern | Example |
|---|---|---|
| Adjective-like (+ prepositions) | Sentence | The left submandibular gland is surgically absent. |
| Adjective phrase | IV access was unable to be established. | |
| Noun phrase | Three views of the left shoulder demonstrate no evidence of fracture. | |
| Simple | There is absent flow signal throughout the right common carotid. | |
| Adverb | “Be” adjective | The previously identified isoechoic nodule is not present on the current examination. |
| “Be” verb-past-participle | The right ovary is not seen. | |
| “Do” | There does not appear to be any significant osteolysis. | |
| Verb | Passive verb | The fusiform aneurysm has been excluded. |
| Active verb | The patient refused IV access. | |
| Noun | The lack of mass effect argues against neoplastic process. | |
| Double negation (not a negation) | We cannot exclude malignancy. |
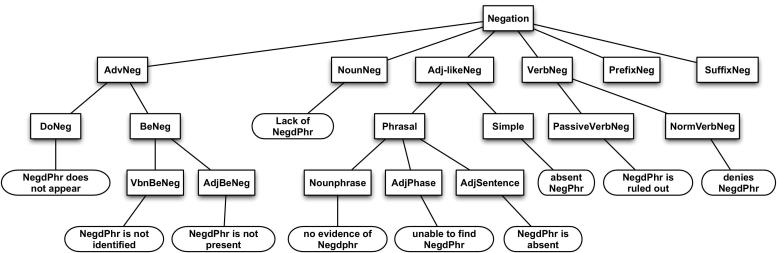
▶ shows the relationship between categories more intuitively in a tree. The first-level child nodes are the syntactical categories of negation signals, verb, adverb, adjective, and noun. The SuffixNeg and PrefixNeg are negations marked by suffixes and prefixes, which were not considered in this study. The nodes under them are sub-categories based on phrase patterns. Finally, the leaf nodes contain an example of the negation category with NegdPhr representing a negated phrase. Please refer to Appendix A (available as an online data supplement at www.jamia.org) for further details. There are no negated phrases in double negation, thus, it was not shown in the figure.
Figure 2.
Negation classification tree. Negation classes are shown in rectangular boxes. Negation examples are shown in the leaf oval boxes.
Regular expressions were developed using negative signals such as “no” and important text features such as “evidence” in the above example. They matched negation patterns broadly with high sensitivity, while were able to classify negations in the above categories when applied in order. After a negation was found, the NDM then traversed the parse tree to locate negated phrases with high specificity. The NDM usually started from the node containing the negative signal. Using the grammar corresponding to the negation type, the NDM extracted the tree node(s) of the negated noun phrase. As noted by Chapman et al., some nouns such as “change” are not real negations when appearing as the head of a negated noun phrase. There are also conjunctions such as “but,” prepositions such as “besides,” and adverbs such as “other than” to reduce the negation scope within a composite noun phrase. Therefore, the extracted noun phrases were scanned for pseudo-negations and decreased scope to generate a final version of negated phrases. Multiple negations (defined by having more than one negation signal) in one sentence were processed multiple times in word order according to each individual negation type. Double negations were frequent in the corpus with restricted text patterns. They were screened out using regular expressions before counting multiple negations.
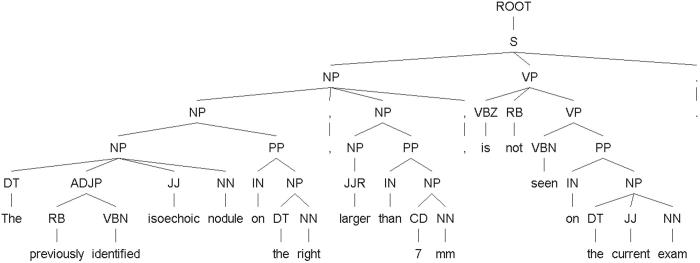
An example of a parse tree output from the Stanford Parser is shown in ▶. This sentence may pose a difficulty for most negation detection algorithms not using the structural information in the sentence’s parse tree, because the negated phrase “The previously identified isoechoic nodule on the right” and the negation signal “not” are separated by the phrase “larger than 7 mm.” However, with the help of the parse tree, it is clear that, syntactically, the verb phrase (VP) “is not seen …” negated its subject, a noun phrase (NP) composed of two noun phrases, “The previously identified isoechoic nodule on the right” and “larger than 7 mm.” Thus, we were able to tag both of them as a negated composite noun phrase, with the help of the parse tree in ▶ and the grammar developed in the previous step.
Figure 3.
A parse tree of a negation case where negated phrase “The previously identified isoechoic nodule” is at a distance from negative signal “not.”
Evaluation
One hundred thirty-two reports of all six modalities were randomly selected from the remaining 500 reports in our document collection. These reports were pre-tagged by NDM with negated phrases indicated. We adopted a simplified Delphi approach 29,30 in establishing a gold standard for the test set with the help of four physicians. Because the physicians were given pre-tagged reports, thus, their decisions on negations might be susceptible to the influence of the NDM’s pre-tagging, producing a “priming bias.” We used 12 of the 132 reports to evaluate priming bias; the remaining 120 reports were split into four groups of 30 reports, each being evaluated by different pairs of physicians. The physicians inspected pre-tagged reports to identify and correct miss-tagging. The markups agreed upon by both physicians were included in the gold standard. Those marked differently by the two physicians were collected and sent back to them showing the tagging by the other physician without any discussions between them. This gave physicians a chance to correct trivial mistakes made in the first inspection cycle and also allowed them to give the results a second thought after seeing each other’s opinion. More than half of the disagreements were resolved when the results were collected for the second round. In cases where the physicians still gave different markups and could not reconcile their difference through discussions, a third physician served as a judge to decide the correct markup, which completed the gold standard.
Using the gold standard, we measured the recall and precision of negation detection, together with the inter-rater agreement ratio and priming bias.
Results
We assessed the priming bias by comparing the markups made by a physician using 12 reports with and without computer pre-tagging. There was more than a month between the two experiments to minimize the effect of the physician remembering his previous markups. Initial results showed a Kappa value of 80% (95% CI 68.7–84.4%). The physician failed to detect several straight-forward negations in the reports without computer markups. The physician was given a second chance to review his own markups, which enabled him to identify most of the negations missed the first time. This second round process was also similar to the process in the construction of the gold standard before calling in a judge for an unresolved disagreement. The second round result had a Kappa value of 92.9% (95% CI 84.1–96.7%), which indicated a high level of agreement between this physician’s markups with and without computer pre-tagging and limited priming bias. This also demonstrated that the two-round process significantly improved the reliability of negation tagging for individual physicians.
Four physicians were assigned as four pairs with each pair inspecting 30 pre-tagged reports in the test set of 120 clinical radiology reports. Their agreement on negated phrases is shown in ▶.
Table 2.
Table 2 Agreement Between Physicians on Negated Phrases Using Pre-tagged Reports
| Pair 1 | Pair 2 | Pair 3 | Pair 4 | Mean | |
|---|---|---|---|---|---|
| Agreement | 92.2% | 90.6% | 89.5% | 94.9% | 91.8% |
▶ shows the final results using the 120 radiology reports. The sensitivity of negation detection, using our hybrid approach, was 92.6% (95% CI 90.9–93.4%). The positive predictive value (PPV) was 98.6% (95% CI 96.9–99.4%), and the specificity was 99.8% (95% CI 99.7–99.9%).
Table 3.
Table 3 Results on 120 Test Reports in Terms of True Positives (TP), False Positives (FP), False Negatives (FN), True Negatives (TN), Sensitivity, Positive Predictive Value (PPV), and Specificity
| TP | FP | FN | TN | Sensitivity | PPV | Specificity | |
|---|---|---|---|---|---|---|---|
| CT | 57 | 0 | 5 | 553 | 91.9% | 100.0% | 100.0% |
| Mammo | 47 | 1 | 1 | 314 | 97.9% | 97.9% | 99.7% |
| MR | 76 | 0 | 6 | 518 | 92.7% | 100.0% | 100.0% |
| PROC | 38 | 1 | 4 | 749 | 90.5% | 97.4% | 99.9% |
| RAD | 17 | 1 | 5 | 208 | 77.3% | 94.4% | 99.5% |
| US | 52 | 1 | 2 | 320 | 96.3% | 98.1% | 99.7% |
| All | 287 | 4 | 23 | 2,662 | 92.6% | 98.6% | 99.8% |
CT = computed tomography; Mammo = mammogram; MR = magnetic resonance imaging; PROC = radiology procedure; RAD = radiograph; US = ultrasound.
Discussion
This study used a methodology that allowed us to assess negation detection in clinical documents, without compounding the evaluation with a concept mapping process. This is possible because our hybrid approach is able to extract negated phrases according to the syntactical structure of sentences within parse trees, and thus, does not rely on concept mapping to group words together before detecting negations. As shown in ▶, the approach is more intuitive in understanding complex sentences and is able to locate the negated phrase according to the understanding of a whole sentence.
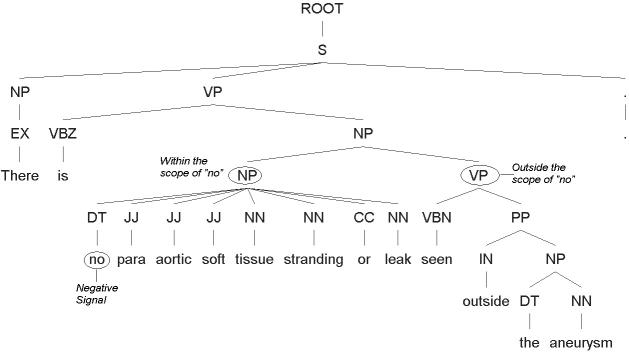
In this approach, a negated phrase is usually well-scoped because the syntactical role of the negation signal is well-defined by the parse tree. As shown in ▶, the NDM is able to tag “para aortic soft tissue stranding or leak” as negated but not “the aneurysm” using the grammar for negation type 4 in Appendix A (available as an online data supplement at www.jamia.org). This is because the negative signal “no” clearly has its scope defined by the noun phrase (NP) node over “no para aortic soft tissue stranding or leak,” while “the aneurysm” is an NP sitting under a VP which is outside the scope of “no.”
Figure 4.
An example showing how the scope of negation is determined by the syntactical structure specified by the parse tree above. “Para aortic soft tissue stranding or leak” is negated while “the aneurysm” is not.
This approach achieved excellent precision, which is not surprising because the negation grammar imposes patterns combining syntactical, lexical, and parse tree information and is therefore highly selective in identifying negated phrases.
Error Analysis
▶ shows the classification of errors made by the computer program on 132 radiology reports.
- A False negatives and positives caused by errors in parse trees. There were a total of 30 errors made by the program. The Stanford parser generated parse trees, allowing us to scope negations based on the understanding of sentences both syntactically and semantically. However, the grammar might not work as expected if parse trees contain errors. Parsing error was the most important source of error, consisting of 16 errors (53.3% of total errors). There were three types of parsing errors:
- 1 Confused a gerund with a present participle, two False Negatives (FNs), 7.7% of total FNs.
- 2 Confused a VBN with a VBD, two FNs, 7.7% of total FNs.
- 3 Other parsing errors, typically attaching preposition phrases or noun phases at the wrong places in parse trees, nine FNs (34.6%) and three FPs (75%).
-
B False negatives caused by the incomplete negation grammar. The grammar certainly does not contain all negation signals and phrase patterns allowed in English and it is likely to be incomplete when processing large numbers of new documents. When the grammar did not have a negation signal specified, it did not recognize a negation at all. For example,
“radiographs labeled from specimen # 1 fail to demonstrate any definite abnormality.”
Table 4.
Table 4 Source of Errors Calculated on 132 Radiology Reports
| Source of Errors | False Negatives | False Positives | ||
|---|---|---|---|---|
| Verb: gerund or present participle | 2 | 7.7% | 0 | 0.0% |
| Verb: past particle or past tense | 2 | 7.7% | 0 | 0.0% |
| Other | 9 | 34.6% | 3 | 75.0% |
| Missing negation signal | 2 | 7.7% | 0 | 0.0% |
| Missing phrase pattern | 9 | 34.6% | 0 | 0.0% |
| Irregular use and other | 2 | 7.7% | 1 | 25.0% |
| Total | 26 | 100% | 4 | 100% |
Our grammar did not include “fail” as a possible negation signal, and thus missed this case entirely. There were two new negation signals identified in the 132 reports tested, “fail” and “resolution,” which accounted for two FNs (7.7%).
It was actually more commonly seen that the grammar did not contain the corresponding phrase pattern of a known negation signal to extract negated phrases. Here is an example,
No focal lucency or endosteal scalloping is noted to suggest multiple myeloma.
Here “multiple myeloma” is considered negated as well as “focal lucency or endosteal scalloping,” through the phrase pattern “no … noted to suggest.”
Nine FNs (34.6%) were caused by incomplete phrase patterns.
There were also errors caused by irregular use of the language and other reasons. This accounted for two FNs (7.7%) and one FP (25%).
For example,
“No perinephric collections are identified or renal masses.” “Renal masses” should be tagged as negated in the above, however, this sentence was hardly correct grammatically.
Limitations
The first limitation was the comprehensiveness of such a manually derived negation grammar. Even though we had manually validated our grammar on a larger set of 470 reports and initial results during the grammar development showed very good coverage, the syntactical and lexical negation grammar was shown to be not as comprehensive as expected during the test. The sensitivity (recall) of 92.6% (95% CI 90.9–93.4%) is significantly lower than the PPV (precision) of 98.6% (95% CI 96.9–99.4%). One important reason was that reviewers have slightly different definitions of negations even though they agree with each other most of the time. For a total of 29 false negatives initially brought up in the first round on the set of 120 reports, reviewers did not reach agreement on 12 of them after seeing the markups by the other physician. Because none of the reviewers were involved in the grammar development, the grammar used in the NDM was thus not as “comprehensive” as the total of four reviewers. However, we should note that even though this limitation is reflected in the coverage of the negation grammar in this work, it would also apply to other approaches to the evaluation of negation detection, as differences in the understanding of negations among developers and reviewers do exist and they negatively impact the measured performance of the system independent of the negation detection approach used.
More importantly, the performance of extracting negated phrases from a full parse tree was obviously limited by the parsing performance of the NLP parser. As documented in our previous study, 27 the noun phrase identification is harder for longer maximal noun phrases due to the ambiguity of English. For example, the parser may not be able to attach a modifying prepositional noun phrase at the right level of a parse, resulting in a noun phrase identification error. In such cases, different words of a biomedical phrase may be separated far apart in a parse tree, and thus missed by the NDM. Our radiology report corpus had many incomplete sentences in sections including the Impression section. These fragmented sentences were mostly well-formed long NPs with preposition phrase attachments or other structures. The presence of incomplete sentences indeed negatively impacted the parsing performance, to a much lesser degree compared to the surgical pathology reports we had. We had expected the parsing performance would cap the performance of negation detection at a lower level. However, the parser was able to get most low-level structures right in a parse tree, even when it did not get the sentence completely right. It occurred much more often for negation instances to be located in a relatively small part of the parse tree using simple syntactical structures. We envision that it is a long-term gradual process to improve the parsing performance of NLP parsers.
The NDM does not handle affixed negations or negations across sentence boundaries. As discussed previously, the definition and utility of detecting negations within a word may depend on applications and the controlled terminologies used for encoding. It is possible to develop a lexical scanner to scan each word token for negative affixes to work with the concept mapping process. Tokens stripped off negative affixes usually represent more basic concepts thus, may be more likely to be mapped into controlled terminologies. Therefore, it could be an important extension to study affixed negations.
Another important limitation was that our study was done on radiology reports only. Chapman et al. documented that radiology reports contained only two thirds of frequently used negation phrases found in non-radiology reports. 30 Therefore, the approach described in this study should be further validated using other types of narrative clinical documents.
Conjoined noun phrases can be very difficult to parse due to ambiguity. We took a practical approach and our grammar handles conjoined noun phrases based on the output of the Stanford Parser and made no efforts in parsing out each smaller noun phrase.
We are integrating the NDM developed in this work into the ChartIndex concept indexing system, which will provide further data in evaluating its performance. Moreover, we plan to expand the NDM to work on other types of clinical documents.
Conclusion
We devised a novel hybrid approach, combining regular expression matching with grammatic parsing, to detect negations. A negation classification and grammar were developed based on the syntactical categories of negation signals and their corresponding phrase patterns. Regular expression matching provided fast classification of negation cases, and enabled the NDM to extract negated phrases from parse trees using corresponding structural grammar rules. The structure grammar rules developed using linguistic principles were more powerful than previous approaches in detecting negated concepts at a distance from negation signals. Our results show that the classification and grammar provided good coverage and this hybrid approach can accurately locate negated concepts in clinical radiology reports.
Footnotes
We are very grateful to the reviewers, Nigam Shah, MD, Todd Ferris, MD, Mia Levy, MD, and Cesar Rodriguez, MD, for their hard work and great patience. We also thank Dr. Wendy Chapman for publishing the algorithm of NegEx online and promptly answering our questions on NegEx.
References
- 1.McDonald C. The barriers to electronic medical record systems and how to overcome them J Am Med Inform Assoc 1997;4:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poon AD, Fagan LM. PEN-Ivory: the design and evaluation of a pen-based computer system for structured data entry Proc Annu Symp Comput Appl Med Care 1994:447-451. [PMC free article] [PubMed]
- 3.Bell DS, Greenes RA. Evaluation of UltraSTAR: performance of a collaborative structured data entry system Proc Annu Symp Comput Appl Med Care 1994:216-222. [PMC free article] [PubMed]
- 4.Hersh WR. Information Retrieval: A Health and Biomedical Perspective. 2nd ed.. New York, NY: Springer-Verlag; 2003.
- 5.Salton G. Automatic Text Processing: The Transformation, Analysis, and Retrieval of Information by Computer. Boston, MA: Addison-Wesley Longman Publishing Co., Inc; 1989.
- 6.Aronson AR, Rindflesch TC, Browne AC. Exploiting a large thesaurus for information retrieval. 1994. Proceedings of RIAO.
- 7.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System Methods Inf Med 1993;32:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: the MetaMap program Proc AMIA Symp 2001:17-21. [PMC free article] [PubMed]
- 9.Friedman C, Liu H, Shagina L, Johnson S, Hripcsak G. Evaluating the UMLS as a source of lexical knowledge for medical language processing Proc AMIA Symp 2001:189-193. [PMC free article] [PubMed]
- 10.Hersh WR, Mailhot M, Arnott-Smith C, Lowe HJ. Selective automated indexing of findings and diagnoses in radiology reports J Biomed Inform 2001;34(4):262-273Aug. [DOI] [PubMed] [Google Scholar]
- 11.Nadkarni P, Chen R, Brandt C. UMLS concept indexing for production databases: a feasibility study J Am Med Inform Assoc 2001;8:80-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elkin PL, Bailey KR, Chute CG. A randomized controlled trial of automated term composition Proc AMIA Symp 1998:765-769. [PMC free article] [PubMed]
- 13.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. Evaluation of negation phrases in narrative clinical reports Proc AMIA Symp 2001:105-109. [PMC free article] [PubMed]
- 14.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS J Am Med Inform Assoc 2001;8(6):598-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horn L. A natural history of negation. Chicago, Ill: University of Chicago Press; 1989.
- 16.Ernest T. The phrase structure of English negation. The Linguistic Review. 9:109–144.
- 17.Friedman C, Hripcsak G, Shagina L, Liu H. Representing information in patient reports using natural language processing and the extensible markup language J Am Med Inform Assoc 1999;6(1):76-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries J Biomed Inform 2001;34(5):301-310. [DOI] [PubMed] [Google Scholar]
- 19.Elkin PL, Brown SH, Bauer BA, Husser CS, Carruth W, Bergstrom LR, et al. A controlled trial of automated classification of negation from clinical notes BMC Med Inform Decis Mak 2005;5(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sager N. Natural Language Information Processing: A Computer Grammar of English and Its Applications. Reading, MA: Addison-Wesley; 1981.
- 21.Sager N, Friedman C, Lyman MS. Medical Language Processing: Computer Management of Narrative Data. Reading, MA: Addison-Wesley; 1987.
- 22.Friedman C, Alderson P, Austin J, Cimino J, Johnson S. A general natural-language text processor for clinical radiology J Am Med Inform Assoc 1994;1:161-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing J Am Med Inform Assoc 2004;11(5):392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell KJ, Becich MJ, Berman JJ, Chapman WW, Gilbertson J, Gupta D, et al. Implementation and evaluation of a negation tagger in a pipeline-based system for information extract from pathology reports Medinfo 2004;11(Pt 1):663-667. [PubMed] [Google Scholar]
- 25.Elkin PL, Bailey KR, Chute CG. A randomized controlled trial of automated term composition Proc AMIA Symp 1998:765-769. [PMC free article] [PubMed]
- 26.Huang Y, Lowe HJ, Hersh WR. A pilot study of contextual UMLS indexing to improve the precision of concept-based representation in XML-structured clinical radiology reports J Am Med Inform Assoc 2003;10(6):580-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Lowe HJ, Klein D, Cucina RJ. Improved identification of noun phrases in clinical radiology reports using a high performance statistical natural language parser, augmented with the UMLS Specialist Lexicon J Am Med Inform Assoc 2005;12(3):275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein D, Manning CD. Accurate Unlexicalized Parsing. 2003. In Proc of the 41st Meeting of the Association for Computational Linguistics.
- 29.Friedman CP, Wyatt JC. Evaluation Methods in Medical Informatics. New York, NY: Springer-Verlag; 1997.
- 30.Hripcsak G, Wilcox A. Reference standards, judges, comparison subjects: roles for experts in evaluating system performance J Am Med Inform Assoc 2001;9:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]