Abstract
Objective
The goal of this research is to learn how the editorial staffs of bioinformatics and medical informatics journals provide support for cross-community exposure. Models such as co-citation and co-author analysis measure the relationships between researchers; but they do not capture how environments that support knowledge transfer across communities are organized.
Methods
In this paper, we propose a social network analysis model to study how editorial boards integrate researchers from disparate communities. We evaluate our model by building relational networks based on the editorial boards of approximately 40 journals that serve as research outlets in medical informatics and bioinformatics. We track the evolution of editorial relationships through a longitudinal investigation over the years 2000 through 2005.
Results
Our findings suggest that there are research journals that support the collocation of editorial board members from the bioinformatics and medical informatics communities. Network centrality metrics indicate that editorial board members are located in the intersection of the communities and that the number of individuals in the intersection is growing with time.
Conclusions
Social network analysis methods provide insight into the relationships between the medical informatics and bioinformatics communities. The number of editorial board members facilitating the publication intersection of the communities has grown, but the intersection remains dependent on a small group of individuals and fragile.
Until recently the medical informatics and bioinformatics communities led relatively separate existences. As a result of the independence with which the two communities functioned, each developed and cultivated diverging research agendas, as well as journals and publication outlets. However, the integration of molecular and genomic information into clinical practice and electronic medical records has spurned a convergence in education, 1 practice, 2 and research. 3 The burgeoning relationship was expressed in 2002 at the Annual Fall Symposium of the American Medical Informatics Association, where the Scientific Program Chair called for an integration under a community called “Bio*medical Informatics: One Discipline.” 4 Broadly speaking, the goal of the new society is to support a bridge between bioinformatics and medical informatics in the form of a biomedical informatics community.
New communities are built, in part, through partnerships of individuals within and between existing communities. 5 To build a new research community, it is necessary for researchers to interact, collaborate, and integrate ideas. Previous community-oriented research sought to identify relationships between various informatics communities by studying the co-citations and co-authors in published articles. 6-12 While citations illustrate who is reading and relating their research to whom, but it does not capture how one community is exposed to another. Similarly, author partnerships provide an indication of research groups that exist, but not how such groups come together or communicate with research communities at large.
One manner by which partnerships across communities are initiated by exposure to each community’s research, which implies that platforms to facilitate exposure are beneficial to new community building. Exposure platforms are diverse and range across informal meetings in the workplace, calls for interdisciplinary research grant proposals, and journals or conferences that enable researchers to share their findings with their peers. Within this context, program managers, conference program committee members, and journal editorial board (EB) members function as figureheads with the ability to facilitate exposure, communication, and interdisciplinary research.
In this article we evaluate how figureheads of medical informatics and bioinformatics, in the form of journal EB members (defined as also including editors and associate editors), are grouped together on editorial boards. We investigate the question, “How are members of the medical informatics and bioinformatics communities associated on various journals in an editorial capacity?” Moreover, “To what extent do these journals support an intersection of the communities?”
This research analyzes the intersection of figureheads from the medical informatics and bioinformatics communities using social networking analysis, i.e., associations based on groupings of people. 13 We utilize several methods from social network analysis and information retrieval to study the intersection as inferred by the editorial boards of bioinformatics and medical informatics research journals. More specifically, we utilize “centrality” metrics to evaluate where individuals and journals are situated in the biomedical intersection. This article extends our initial investigations 14 to account for the evolution of the intersection. The biomedical community is evolving, and to reflect this fact, we study a longitudinal setting by tracking changes in the network over the years 2000 through 2005. Our principal findings suggest that: (1) there exist EB members and journals in the intersection of the medical informatics and bioinformatics communities and (2) the number of editors in the intersection has grown with time.
Background
In this section, we review related research and alternative models by which biomedical communities have been studied. In general, prior research modeled interactions and relationships in the biomedical informatics community via co-citation and co-word analyses.
Investigations conducted by Morris 6 illustrated how the integration of techniques, research areas, and education concentrations constitute various subdisciplines within the medical informatics community. In his research, co-word analysis and multidimensional scaling of medical subject headings (MeSH) keywords from Medline abstracts were explored to discover subdomains within the overarching genre of medical informatics. Findings from this study revealed that medical informatics could be roughly partitioned into eight concepts: biochemistry, psychology, imaging methods, image diagnostics, immunology, molecular genetics, statistics, and the science and art of medicine. This partitioning further divided the findings from an earlier study by Morris and McCain, 7 in which it was shown that medical informatics as a discipline could be grouped into four general focus areas: biomedical engineering, biomedical computing, decision support systems, and education. This body of research supports the belief that distinct communities of medical informatics exist, but does not capture how such groups relate to each other, nor how they relate to the bioinformatics community.
Building on the previous work, Andrews 8 concentrated on the relationships of specific individuals in the medical informatics community. In this research, co-citation analysis was performed with the 50 most-cited American College of Medical Informatics fellows. The “importance” of specific individuals was inferred via unsupervised learning in the form hierarchical clustering. Co-citation analysis provides a characterization of how their work is related to that of others, which can capture cross-community interaction; however, it does not indicate whether there is a platform that encourages disparate communities to interact with each other.
In contrast to Andrews’ investigation of individual significance, Sittig 9 applied co-citation analysis to study the importance of medical informatics journals. Initial findings from this research revealed that, in comparison to other medical and scientific outlets, no specific medical informatics journal was a definitive best. Yet, in a follow-up study, Sittig 10 demonstrated that there exists a core set of publication outlets for the medical informatics community. Sittig extended this research into the biomedical informatics journals, where the resulting rankings implied that Methods of Information in Medicine was the most prominent. 11 In parallel research, Vishwanatham 12 reported similar results on the intersection of medical informatics and library sciences. All of the aforementioned studies focused on journals that in our findings are primarily outlets for the medical informatics community. As a result, they do capture how the bioinformatics community relates to such journals or how journals from their community support knowledge transfer to the medical informatics community.
To study the biomedical informatics intersection, we consider a set of journals derived from both communities. We use a social network analysis of the editorial boards to learn the extent to which there are editors and journals that support the intersection of the two communities.
Methods
This research assesses where the bioinformatics and medical informatics communities intersect from an editorial perspective. We use the groupings of editors on journals as a proxy for measuring the intersection of the two communities. Social networks of EB members are inferred from publicly available information in the form of editorial boards, which are accessible in print and on Internet archives. In the following section we provide a more detailed description of the data derived for this study and the methods we adopted for analysis.
Materials
For this study, we collected the editorial boards of approximately 40 journals that serve as publication outlets for the medical informatics and bioinformatics communities. To conduct a longitudinal analysis, we gathered editorial boards for each of the years 2000 through 2005. We do not include editorial boards for each journal for each year because not all journals included in this study existed during each year of study. Although no journal was discontinued during the time period of our study, and thus no journal was removed from the study once it was added, there are journals that were added as time moves forward. For example, the inaugural issue of the journal IEEE Transactions on Computational Biology and Bioinformatics (TCBB) was not published until 2004, so we include editorial boards for the years 2004 and 2005. A list of the journals used in this study, and the year that each journal was incorporated into the analysis, are shown in ▶. To summarize, there were 38 and 42 journals in 2000 and 2005, respectively.
Table 1.
Table 1 Journals Studied and Centrality Scores for the Year 2005
| Journal | First Year in Study | 1st Principal Component |
2nd Principal Component |
||
|---|---|---|---|---|---|
| Score | Rank | Score | Rank | ||
| Annals of Internal Medicine | 2000 | 0.04 | 40 | 0.00 | 29 |
| Applied Bioinformatics | 2002 | 0.58 | 35 | 12.41 | 10 |
| Artificial Intelligence in Medicine | 2000 | 75.61 | 5 | 2.97 | 15 |
| Bioinformatics | 2000 | 14.19 | 17 | 101.78 | 1 |
| Biosystems | 2000 | 0.78 | 34 | 6.08 | 12 |
| BMC Bioinformatics | 2000 | 2.63 | 28 | 46.11 | 3 |
| BMC Genomics | 2000 | 2.42 | 29 | 36.84 | 4 |
| BMC Medical Informatics and Decision Making | 2000 | 36.16 | 8 | 1.27 | 18 |
| BMC Medicine | 2000 | 7.23 | 21 | 0.14 | 25 |
| Briefings in Bioinformatics | 2000 | 6.15 | 22 | 31.44 | 6 |
| Computer Methods and Programs in Biomedicine | 2000 | 37.28 | 7 | −2.33 | 38 |
| Computerized Medical Imaging and Graphics | 2000 | 14.27 | 16 | 2.51 | 16 |
| Computers in Biology and Medicine | 2000 | 30.46 | 9 | 0.50 | 21 |
| Data Mining and Knowledge Discovery | 2000 | 4.36 | 26 | 1.42 | 17 |
| Evidence-based Healthcare and Public Health | 2000 | 0.85 | 32 | 0.05 | 26 |
| Handbook of Medical Informatics | 2000 | 144.79 | 2 | −5.82 | 39 |
| Health Informatics Journal | 2000 | 16.85 | 12 | −1.38 | 34 |
| Health Information and Libraries Journal | 2000 | 9.34 | 20 | −0.19 | 31 |
| IEEE Intelligent Systems | 2000 | 4.49 | 25 | 5.36 | 13 |
| IEEE Transactions on Computational Biology and Bioinformatics | 2004 | 2.23 | 30 | 33.63 | 5 |
| IEEE Transactions on Information Technology in Biomedicine | 2000 | 12.80 | 19 | 0.45 | 22 |
| In Silico Biology | 2000 | 0.82 | 33 | 12.73 | 9 |
| Informatics in Primary Care | 2000 | 20.35 | 10 | −1.85 | 36 |
| Informatics Review | 2000 | 17.45 | 11 | −1.54 | 35 |
| International Journal of Medical Informatics | 2000 | 134.52 | 4 | −19.05 | 40 |
| International Journal of Technology Assessment in Health Care | 2000 | 0.37 | 36 | 0.44 | 23 |
| Journal of Biomedical Informatics | 2000 | 72.08 | 3 | 10.27 | 7 |
| Journal of Clinical Monitoring and Computing | 2000 | 140.80 | 14 | 26.63 | 20 |
| Journal of Computational Biology | 2000 | 15.41 | 27 | 0.77 | 2 |
| Journal of Information Technology in Healthcare | 2000 | 3.57 | 23 | 63.90 | 33 |
| Journal of Medical and Biological Engineering and Computing | 2005 | 5.90 | 37 | −0.84 | 27 |
| Journal of Medical Internet Research | 2000 | 0.35 | 15 | 0.03 | 24 |
| Journal of Medical Systems | 2000 | 15.11 | 24 | 0.28 | 32 |
| Journal of Telemedicine and Telecare | 2000 | 4.80 | 41 | −0.83 | 30 |
| Journal of the American Medical Informatics Association | 2000 | 0.01 | 6 | 0.00 | 11 |
| Medical Decision Making | 2000 | 15.42 | 13 | 5.03 | 14 |
| Medical Informatics and the Internet in Medicine | 2000 | 14.12 | 18 | −2.16 | 37 |
| Methods of Information in Medicine | 2000 | 213.69 | 1 | −21.69 | 41 |
| Nucleic Acids Research | 2000 | 0.87 | 31 | 14.92 | 8 |
| Online Journal of Bioinformatics | 2002 | 0.14 | 39 | 1.20 | 19 |
| Technology and Health Care | 2000 | 0.29 | 38 | 0.02 | 28 |
In total, there were 1,617 distinct members in the union of the 2000 through 2005 editorial boards. There were on the order of 10,000 distinct editor-board records for the time period studied. In ▶, we show the number of journals and distinct EB members per year. To minimize the effect of larger boards, individuals who appeared on only one editorial board were removed from this study. This reduced the number of editors considered in each year by approximately 90%. All included journals contain at least one EB member from this reduced set.
Table 2.
Table 2 Editorial Board Coverage by Distinct Individuals
| Year | Number of Journals | Number of Distinct EB Members | Number of Members on >1 Editorial Board | Coverage (%) |
|---|---|---|---|---|
| 2000 | 37 | 1,107 | 96 | 8.67 |
| 2001 | 37 | 1,112 | 112 | 10.07 |
| 2002 | 39 | 1,163 | 123 | 10.58 |
| 2003 | 39 | 1,178 | 116 | 9.85 |
| 2004 | 40 | 1,213 | 127 | 10.47 |
| 2005 | 42 | 1,248 | 136 | 10.90 |
| Average | 10.09 | |||
| Standard deviation | 0.79 |
There are several reasons why we choose to incorporate new journals with time. First, we justify our reasoning through organizational behavior theory. Prior research has shown tha communities are dynamic and thus are constantly in a state of evolution. 15–16 This notion is reflected by the fact that the set of members that are on a journal’s editorial board is not static. By incorporating new journals, we emulate the system for a community as it existed in each year. Second, the distinct set of EB members that exist for all journals is changing as well. By modeling the effect of new journals on the network, we reflect the evolution within existing communities as well as interaction between existing communities. For instance, as mentioned above, TCBB was founded to forge a stronger bond between the computational biology community and the bioinformatics communities. The decision to fill a perceived void culminated with the inaugural issue of TCBB in 2004.
Editorial Centrality
For this article, we use the following notation. Let J = {j 1, j 2, …, j m} be a set of journals. Let E = {e 1, e 2, …, e n} be the set of editors. Let B be an m × n Boolean matrix, which we call the editorial-board matrix, that represents which EB members are on which journals’ boards, such that b xy = 1 if journal j x has editor e y and 0 otherwise.
From the editorial-board matrix, we construct an n × n matrix representing editorial adjacencies A = B T B. We do not consider the number of boards an EB member is a member of, so all diagonal values aii are set to zero. Furthermore, we do not consider the number of times members appear on the same board, nor the prestige or ISI-Thompson Scientific journal impact factor ranking of a journal. As such, we threshold A into a Boolean matrix in which all cells with value ≥1 are set to 1.
We use the editorial-adjacency matrix to measure the centrality of each member. Specifically, we utilize Freeman’s betweenness centrality score. 17 If we consider the adjacency matrix as a graph, then betweenness can be likened to flow. Basically, the betweenness score for an EB member represents how often two members would have to pass through another EB member in order to meet. More formally, betweenness is measured via shortest paths in the network. Let p i j be the set of nodes in the shortest path from member e i to member e j. Then, the betweenness centrality score for a member e k is calculated as:
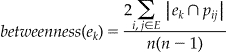 |
By this construction, the betweenness score for a member e k is directly proportional to the number of times e k exists in all shortest paths of the editorial network.
Journal Centrality
The betweenness centrality measure provides an indication of which EB members are situated within the biomedical intersection of bioinformatics and medical informatics. However, betweenness does not provide a model that allows us to determine the centrality of a journal with respect to the intersection. Yet, the editorial adjacency matrix can be converted into an alternative representation that allows us to relate journals via an EB member’s network positioning.
Specifically, we measure a journal’s contribution to the intersection using the principal components derived from the editorial adjacency matrix. Formally, the singular value decomposition of the editorial adjacency matrix yields
where U and V are orthonormal n × n matrices and Λ is an n × n matrix with non-zero eigenvalues along the diagonal. Each column of the V matrix is an eigenvector and can be thought of as distinct concepts derived from the relationships in A. Each row of U = {u i1, … u in} represents the degree that member i is explained by concepts 1 through n (i.e., vectors v 1, v 2, …, v n of V). The diagonal values λ1, λ2, …, λn of Λ represent the significance of each concept in terms of how much variance each eigenvector accounts for in the network.
Given the decomposition, we measure how well each journal is represented by each concept, or weighted vector of EB members. We project the editorial boards matrix B onto the new components space using the following operation:
In this form, matrix X is similar to matrix U, except each row vector in matrix X represents how similar a journal is to each concept derived from the editor adjacency matrix.
Results
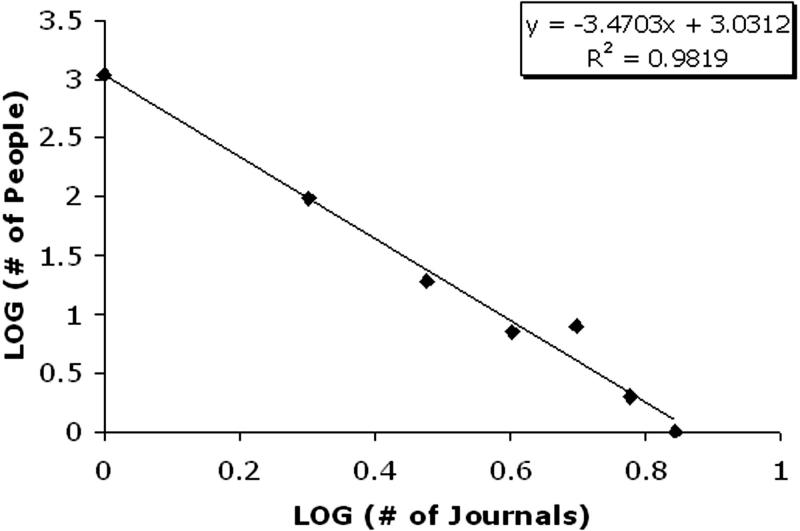
Before analyzing the intersection of the communities, we first characterize how members are distributed across editorial boards. As illustrated in ▶, approximately 10% of members are on more than one editorial board. However, the distribution of the EB members to journals is not uniform. Rather, the distribution is highly skewed and follows a power law. The distribution of the number of boards per member is illustrated for the 2005 dataset in ▶, where it can be verified that the distribution follows a log-log linear relationship. There is one EB member, Jochen Moehr, who is on seven, the largest number, of the editorial boards. After Moehr, there are two members, Enrico Coiera and Jeremy Wyatt, who are on six, the next largest number, of the editorial boards. Yet, there are nine members who are on five editorial boards. In our studies, we find that a similar power law trend holds true for each year in the study.
Figure 1.
Frequency of number of journals per editorial board member in the year 2005.
Editorial Centrality
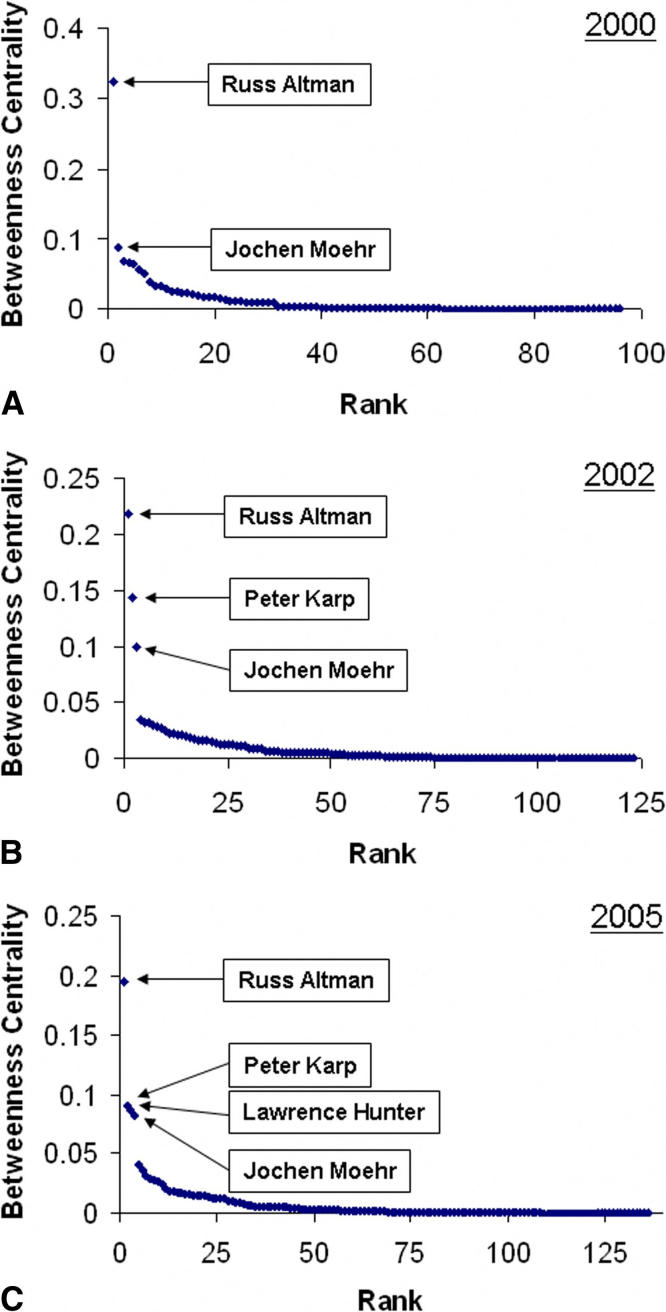
We depict the distribution of betweenness centrality scores for EB members for the years 2000, 2002, and 2005 in ▶. First we note that although Moehr, Coiera, and Wyatt are on the most editorial boards, according to the betweenness metric, Russ Altman occupies the most central position in the system in every year of our study. Moreover, Altman’s betweenness centrality is consistently at least two times larger than the next most central individual. For instance, in 2005 Altman’s centrality score (0.1949) is 2.25 times larger than the next most central individual, Peter Karp (0.0871). In contrast to rank by the number of boards a member is on, Moehr is found at the fourth-ranked betweenness position.
Figure 2.
Betweenness centrality score distribution, ordered by rank for the years (A) 2000, (B) 2002, and (C) 2005.
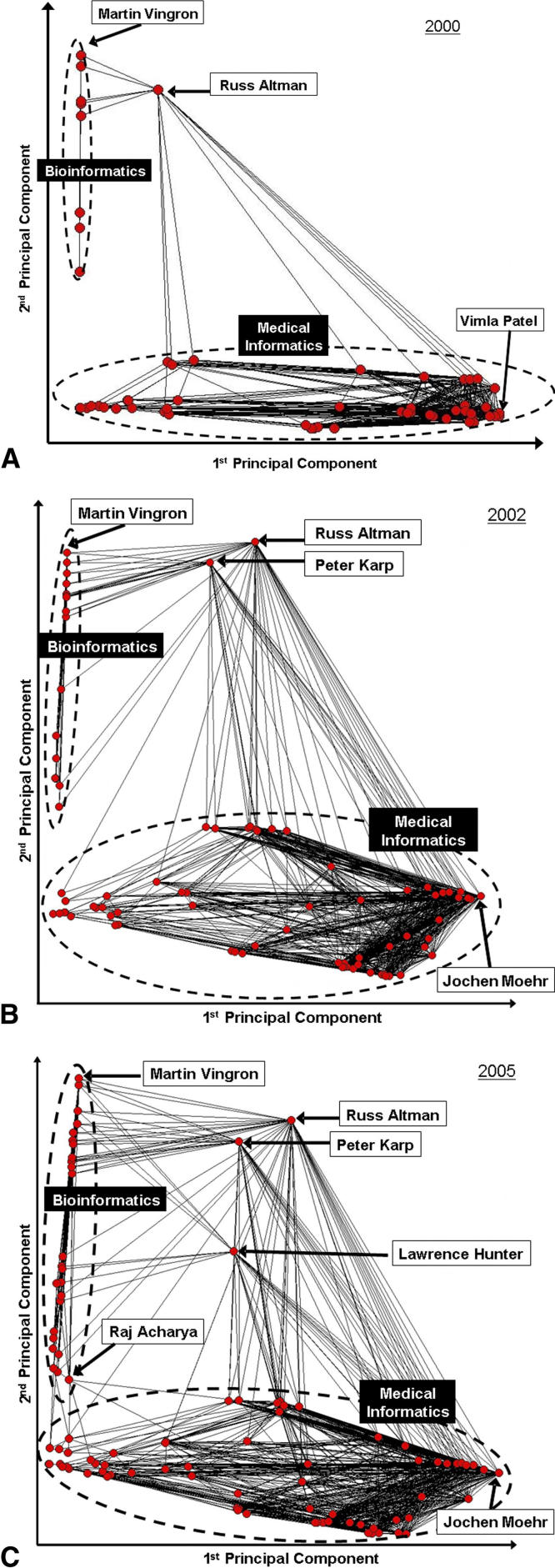
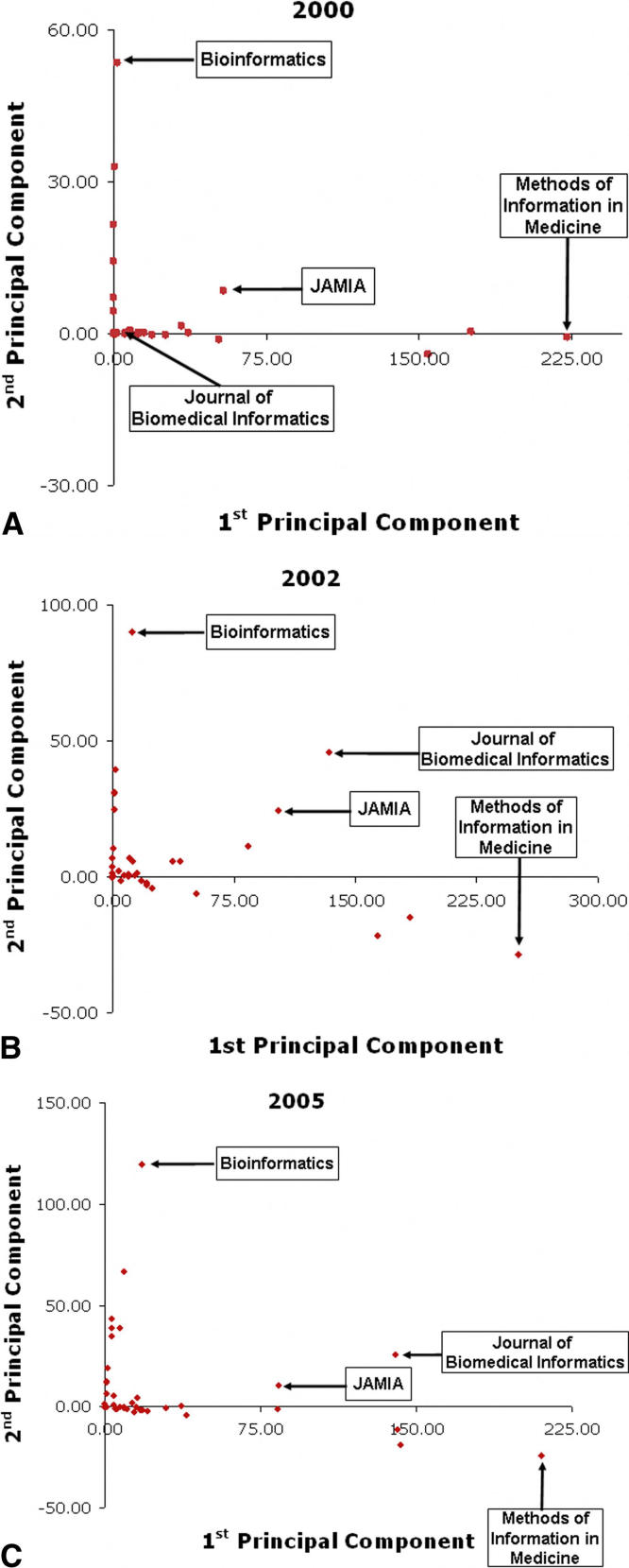
By considering the principal components representation of the editorial adjacency matrix, we gain insight into why EB members like Altman have higher centrality. In our investigations we found that the first two principal components of the matrix account for most of the variance in the data. In ▶, we depict the social network of the EB members for the years 2000, 2002, and 2005 as plotted against the first two principal components of the network. In each year, the network can be partitioned into three clusters. The largest group clusters along the first principal component and captures EB members who tend to be found on journals that are more traditionally oriented to the medical informatics community. The second group clusters along the second principal component and captures EB members who tend to be found on journals that are more traditionally for the bioinformatics community. The third group falls between the first two groups and characterizes individuals who function as “boundary spanners”—EB members who are crucial to the linkage of disparate organizations. 18,19 It was noted by Thompson 18 that such persons facilitate the integration of knowledge from disparate communities and facilitate information exchange.
Figure 3.
First two principal components of the editorial adjacency graph for the years (A) 2000, (B) 2002, and (C) 2005. Nodes correspond to members, and lines correspond to existing edges in the editorial adjacency graph.
Network Evolution and Fragility
Next, we considered how the network of EB members has evolved. We first looked into how many persons keep the network connected as a single component. We characterize this set of individuals as the minimal set of nodes that disconnect the network into at least two components. We summarize our findings in ▶. We find that the number of EB members who connect the network has monotonically increased from 2000 through 2005. In the year 2000, there was only one EB member who was in the intersection. By the year 2005, there were six individuals who supported the intersection. Thus it appears that the biomedical informatics intersection is growing stronger with time.
Table 3.
Table 3 Minimal Set of Individuals Necessary to Disconnect the Social Network
| Year | Number of EB Members | Members Ranked by Betweenness Centrality |
|---|---|---|
| 2000 | 1 | 1) Altman, Russ |
| 2001 | 2 |
|
| 2002 | 2 |
|
| 2003 | 2 |
|
| 2004 | 5 |
|
| 2005 | 6 |
|
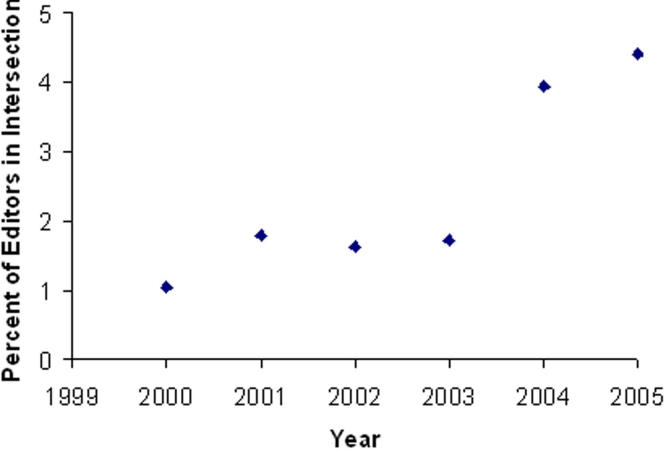
However, this increase is misleading because the number of people in the network has grown as well. To determine whether the growth in the intersection size was due to the growth of the network, we analyzed the ratio of people in the intersection to people in the network. We plot this ratio as a function of year in ▶. To evaluate whether the overall increasing trend is statistically significant, we performed a statistical hypothesis test to compare the intersection between two populations: Population A, which consists of EB members in the social network from the year 2000; and Population B, which consists of EB members in the social network for the year 2005. We then partitioned EB members into two classes: Class I, persons who are in the intersection (i.e., the minimal set of persons necessary to split the network into at least two components); and Class II, persons who are not in the intersection. To account for the small sample sizes, we applied a Fisher’s exact test and found that the p value was < 0.03. This result indicates that the proportion of EB members in the intersection has increased by a statistically significant proportion.
Figure 4.
Percent of members in intersection to total number of EB members in the network.
The previous analysis confirms the intersection is growing, but how robust is it? To answer this question, we considered how the current state of the biomedical informatics intersection is affected if boundary spanners are removed from the system. In other words, what would happen to the organization of the network when we remove the EB member with the largest betweenness score? To answer this question, we dropped the most central individual from the editorial adjacency matrix and recomputed the betweenness centrality. When Altman is dropped from the system, we find that the rank ordering of EB members on betweenness centrality does not change the ranks 2 through 6 in the original data rank of ▶. More concretely, when Altman is removed, persons ranked 2 through 6 rotated up one ranking into positions 1 through 5. Translating this result into the names that are depicted in ▶, Karp became the most significant, followed by Lawrence, followed by Moehr. A similar effect is observed when we remove both Altman and Karp (ranks 1 and 2). However, if we remove the Altman, Karp, and Hunter (ranks 1, 2, and 3), we observe a significant shift in terms of centrality. As a consequence, it appears that the cross-community relationship is not very fault tolerant in terms of EB members in the system. Specifically, the person at original rank 5, Raj Acharya, jumps ahead of Moehr into the first position. Moreover, EB members from the original ranks of 9 through 35 move into ranks 2 through 4. Despite the fact that the system is not fault tolerant for cross-community relations, it does remain a connected environment.
Table 4.
Table 4 Betweenness Centrality Scores in 2005 before and after the Removal of the Highest Ranking Central Editors
| EB Member Name (Last, First) | Original Network |
With Top Three Ranks Removed |
||
|---|---|---|---|---|
| Betweenness Centrality | Rank | Betweenness Centrality | Rank | |
| Altman, Russ | 0.1949 | 1 | n/a | n/a |
| Karp, Peter | 0.0900 | 2 | n/a | n/a |
| Hunter, Lawrence | 0.0871 | 3 | n/a | n/a |
| Moehr, Jochen | 0.0827 | 4 | 0.0366 | 5 |
| Acharya, Raj | 0.0423 | 6 | 0.2421 | 1 |
| Huang, HK | 0.0275 | 10 | 0.1678 | 2 |
| Ratib, Osman | 0.0142 | 21 | 0.0936 | 3 |
| Schwartz, Alan | 0.0078 | 36 | 0.0796 | 4 |
n/a, not applicable.
The main reason that Moehr does not have a substantial increase in centrality, but remains in the top five ranks, is because his centrality score is derived from his editorial associations within medical informatics. Removal of Altman, Karp, and Hunter, who are central to cross-community paths in the network, do not affect Moehr’s centrality within the medical informatics subnetwork.
The clusters help explain why the removal of Altman does not destabilize the betweenness score rankings. Altman, Karp, and Hunter occupy positions in the biomedical informatics intersection. We could remove Karp, but not Altman or Hunter, with similar results. Once both are removed, Acharya and Huang are pushed into central positions. However, the latter do not facilitate the intersection as well, which manifests in our results as a significant increase in the system’s average betweenness score from 0.0086 (standard deviation of 0.022) to 0.011 (standard deviation of 0.018). A t-test confirms that the means of the distributions are significantly different at the 95% confidence interval. Therefore, we conclude that Altman, Karp, and Hunter are the most central in the intersection of medical informatics and bioinformatics EB members.
Journal Centrality and Migration
Next, we turned our attention to the relationship between journals and the intersection. Based on the clusters discovered in the decomposition of the editorial adjacency matrix, we considered projections of the journal editorial boards along the first two eigenvectors. The resulting journal projections for the years 2000, 2002, and 2005 are depicted in ▶. The values for the projections of the year 2005 are provided in ▶. Akin to our observations of EB member centrality, we notice that journals cluster along each of the two components. Along the first principal component, we find well-known medical informatics journals. From most to least prominent, i.e., from right to left, we find Methods of Information in Medicine, Journal of Biomedical Informatics, International Journal of Medical Informatics. This finding lends support to the earlier observations of Sittig and Vishwanatham that Methods of Informatics in Medicine is a significant journal within the medical informatics community. The journal’s negative value with respect to the second principal component suggests that this journal is negatively correlated with the bioinformatics community.
Figure 5.
Journals projected onto the first two eigenvectors of the editorial adjacency matrix for the years (A) 2000, (B) 2002, and (C) 2005.
We find a similar result within the bioinformatics community. Along the second principal component, we find bioinformatics journals, from most prominent to least prominent, i.e., from top to bottom: Bioinformatics, Journal of Computational Biology and BMC Bioinformatics.
In the year 2000, we find that there is one journal that marginally resides between the communities. This journal is the Journal of the American Medical Informatics Association (JAMIA). By 2002, we find that the Journal of Biomedical Informatics (JBI) leapfrogs JAMIA into the most prominent position and has relatively large values along both principal components. As of the year 2005, it appears that the only journal in the intersection of both communities is JBI. Out of 42 journals, this journal ranks second in the medical informatics concept and seventh in the bioinformatics component. The value of JBI along the medical informatics component is two-thirds that of the Methods of Information in Medicine, but it is only 0.3 that of Bioinformatics. Thus, it appears that JBI does a significantly better job of exposing medical informatics to bioinformatics research than vice versa. JAMIA remains the next closest journal in the intersection, with a value of one-third that of JBI’s in the second principal component.
Discussion
In this section, we elaborate on our findings and discuss some of the limitations of our study.
Summary of Findings and Historical Confirmation
The first message of this research is that our analysis supports the belief that there is an intersection of the medical informatics and the bioinformatics communities. Keep in mind, our findings do not imply that the EB members who are the most central in the network are the most influential individuals in their respective communities. Furthermore, it should be made evident that the above journal rankings are not an indication of the prestige of the journals from a research perspective. The journal rankings are an indication of how EB members from different communities are grouped together. Thus, our results measure different aspects of journals than metrics such as the ISI impact factor. From an editorial perspective, our findings provide an indication that the medical informatics and bioinformatics communities are coalescing to facilitate knowledge exchange.
A second message is based on an historical study of the Journal of Biomedical Informatics (JBI). Specifically, our findings lend support to a conscious organizational restructuring effort undertaken by the JBI management. Prior to 2001, JBI was named Computers and Biomedical Research. Then, in 2001 the management renamed the journal the Journal of Biomedical Informatics, and in an editorial by the editor-in-chief 20 it was made evident that they “made a number of changes to update and reorient the journal … simultaneously seeking to fill a niche not clearly identified as a central focus by the other journals that publish papers in biomedical informatics research” and “The Journal of Biomedical Informatics (JBI) is intended to complement rather than to compete with the other major journals in biomedical informatics. In particular, we wish to emphasize papers that elucidate methodologies that generalize across biomedical domains and that help to form the scientific basis for the field.”
The movement of JBI into a central position within the principal components space of our network analysis illustrates that the management achieved their goals of reorganization in terms of EB member associations. As of 2005, the JBI moved closer to the first principal component, and thus became more populated with EB members in the medical informatics community. It is possible that this movement away from bioinformatics editorial membership is due to the evolution of the bioinformatics community, particularly through the establishment of new journals such as TCBB, or through reorganizations of existing journals, which affected the editorial social network. Although journals such as JBI and JAMIA have migrated closer to the medical informatics community since 2002, our findings suggest that information exposure across the medical informatics and bioinformatics communities, as represented by boundary spanning EB members, is growing stronger.
Limitations and Extensions
There are both technical and ideological limitations to our study. We outline several issues and propose how our work can be extended to resolve these limitations.
First, we made several simplifications regarding the associations between EB members that may bias our results. A first simplification is that we disregarded the prestige of the journal that EB members are associated with. Not all journals are equal in their ability to publish high-quality research, and require different levels of effort and oversight from EB members. To address this issue, it may be useful to incorporate journal rankings based on citation analysis, such as that published by ISI–Thompson Scientific. A second simplification is that we do not characterize the strength of an edge between two EB members in the network. This is because we threshold the editorial adjacency matrix, which discounts the number of times members collocate. A social network that explicitly includes this feature may be desirable. In addition, the study did not include members who served on only one editorial board. Some journals prefer that their EB members do so.
Second, our research does not address biomedical informatics in general. The investigations in this article address: (1) the extent to which forums that expose one community to the other exist, and (2) how editorial boards bring researchers from disparate communities to a common editorial platform. Editorial board associations indicate associations of researchers and how a research community gains exposure from another community. Thus, editorial boards do not imply the extent of collaboration across communities. To gain an understanding of the collaborations that exist across communities, we believe that different models, such as the social networks derived from co-authorships, are necessary to provide insight into cross-community collaboration.
Conclusion
This research investigated how researchers from bioinformatics and medical informatics are associated on editorial boards to facilitate a biomedical informatics intersection. We extracted the editorial boards of approximately 40 journals over a five-year period, 2000 through 2005, and constructed social networks as inferred from the groupings of EB members. We decomposed the networks according to editorial centrality metrics, which indicated that the number of members who intersect the communities, in terms of centrality, has significantly grown. Based on editorial centrality, we discovered there exist journals that are situated at the editorial intersection of the communities. Although the size of the editorial intersection has grown, as of 2005 it was fragile and heavily dependent on a small group of EB members. Our research provides insight into the forums that allow exposure between the medical informatics and bioinformatics communities. However, our results are preliminary in that they do not model the biomedical community in general. In future research, we intend to study cross-community interaction using researcher relationships, such as co-authorship networks.
Footnotes
Funded in part by National Science Foundation Integrative Graduate Education and Research Traineeship grant 9972762 in Computational Analysis of Social and Organizational Systems. The views expressed in this manuscript are those of the authors and do not necessarily reflect those of the federal government or the management of the journals used in this study.
The authors acknowledge helpful discussions with Edoardo Airoldi, George Davis, and Latanya Sweeney. The authors thank all of the editors and publishers that provided archived journals and lists of editorial boards, with special note to Nancy Lorenzi, Randy Miller, and Ted Shortliffe. Social network graphs were produced with the software program UCINET.
References
- 1.Hersh W. The full spectrum of biomedical informatics education at Oregon Health and Science University Yearbook of Medical Informatics 2007;46:80-83. [PubMed] [Google Scholar]
- 2.Martin-Sanchez F, Iakovidis I, Norager S, et al. Synergy between medical informatics and bioinformatics: facilitating genomic medicine for future healthcare Technical Information Series 2003GRC192, GE Global Research. 2003.
- 3.Altman R. Bioinformatics in support of molecular medicine Proc AMIA Annu Fall Symp 1998:53-61. [PMC free article] [PubMed]
- 4.Kohane IS, ed. Bio*Medical Informatics: Proceedings of the 2002 AMIA Annual Symposium. 2002.
- 5.Coates T. Building New Communities: Learning from Weblogs. Proc 5th International Conference on Virtual Communities. 2002.
- 6.Morris TA. Structural Relationships within Medical Informatics Proc AMIA Annu Fall Symp 2000:590-594. [PMC free article] [PubMed]
- 7.Morris TA, McCain KW. The structure of medical informatics journal literature J Am Med Inform Assoc 1998;5:448-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews JA. An author co-citation analysis of medical informatics J Med Libr Assoc 2003;91:47-56. [PMC free article] [PubMed] [Google Scholar]
- 9.Sittig DF, Kaalaas-Sittig J. A citation analysis of medical informatics journals Proc Medinfo ’95 1995;8:1452-1456. [PubMed] [Google Scholar]
- 10.Sittig DF. Identifying a core set of medical informatics serials: an analysis using the MEDLINE database Bull Med Libr Assoc 1996;84:200-204. [PMC free article] [PubMed] [Google Scholar]
- 11.Sittig DF. Use of fuzzy set theory to extend Dhawan’s journal selection model: ranking the biomedical informatics serials Bull Med Libr Assoc 1999;87:43-49. [PMC free article] [PubMed] [Google Scholar]
- 12.Vishwanatham R. Citation analysis in journal rankings: medical informatics in the library and information science literature Bull Med Libr Assoc 1998;86:518-522. [PMC free article] [PubMed] [Google Scholar]
- 13.Wasserman S, Faust K. Social Network Analysis: Methods and Applications. Cambridge: Cambridge University Press; 1994.
- 14.Malin B, Carley K, Sweeney L. Where’s the “Biomedical” in Biomedical Informatics: A Social Network Analysis of the Bioinformatics and Medical Informatics Intersection. Data Privacy Laboratory Working Paper LIDADP-20. Pittsburgh, PA: Carnegie Mellon University; 2005. June.
- 15.Romanelli E. The evolution of new organizational forms Annu Rev Sociol 1991;17:79-103. [Google Scholar]
- 16.Carley K. Organizational adaptation Ann Operations Res 1997;75:25-47. [Google Scholar]
- 17.Freeman LC. A set of measures of centrality based on betweenness Sociometry 1977;40:35-41. [Google Scholar]
- 18.Thompson JD. Organizations in Action. Englewood Cliffs, NJ: Prentice-Hall; 1967.
- 19.Mintzberg H. The Nature of Managerial Work. Englewood Cliffs, NJ: Prentice-Hall; 1973.
- 20.Shortliffe T. Editorial J Biomed Inform 2001;34:2-3. [Google Scholar]