Abstract
Objective
This study evaluated a computerized method for extracting numeric clinical measurements related to diabetes care from free text in electronic patient records (EPR) of general practitioners.
Design and Measurements
Accuracy of this number-oriented approach was compared to manual chart abstraction. Audits measured performance in clinical practice for two commonly used electronic record systems.
Results
Numeric measurements embedded within free text of the EPRs constituted 80% of relevant measurements. For 11 of 13 clinical measurements, the study extraction method was 94%–100% sensitive with a positive predictive value (PPV) of 85%–100%. Post-processing increased sensitivity several points and improved PPV to 100%. Application in clinical practice involved processing times averaging 7.8 minutes per 100 patients to extract all relevant data.
Conclusion
The study method converted numeric clinical information to structured data with high accuracy, and enabled research and quality of care assessments for practices lacking structured data entry.
Introduction
Routine entry of clinical information in electronic patient records (“registration”) comprises an important data source for healthcare research and quality improvement. The 1990s saw creation of several European general practice registration networks. 1–7 Most such networks collect selected information from structured tables embedded in the electronic patient record (EPR) systems—for example, patients’ prescribing records, diagnostic codes, and demographic information.
Despite recognized potential and widespread pleas to register more structured clinical data, relevant, important clinical information remained scattered throughout different segments of the EPR. Much of that information, of great potential interest for research and quality improvement, only resided in the free text of patient records. 8–10 Physicians embed key information in free text instead of EPR structured tables due to time constraints during patient care, uncertainty about using codes, classification limitations, and inexperience and difficulties with the computer systems. 1,2,8,11
Many approaches to information retrieval exist. 12 In particular, Natural Language Processing (NLP) shows promising results for extracting and structuring clinical information from unstructured, “free text” medical records. 13,14 For example, NLP applications have classified medical problems lists, 15,16 extracted disease-related concepts from narrative reports, 17,18 and combined data from multiple discharge summaries. 19 NLP, however, has limitations making it less suitable for handling dense, telegraphic, ungrammatical clinical data that lack fixed structure and recognizable text formats. Furthermore, misspellings, personal idiosyncrasies, and transient local abbreviations make the words used to identify numeric data in free text highly variable and ambiguous. Thus, it is better to develop an extraction approach triggered by numeric values, per se, rather than their labels.
Case Description
Often, manual record abstraction occurs during the quality assessment of diabetes care. 20–22 Requisite clinical information usually includes measurements of blood pressure, weight, height, and laboratory results. 23–25 While administrative or centralized clinical databases contain some of these data, incomplete data registration in such systems often necessitates additional patient record review. 26 A need exists for automated capture of these data from EPRs. Such a method must be applicable across multiple sites lacking uniform data registration procedures.
Most general practices in The Netherlands use one of seven vendors’ major electronic patient record systems. The EPR information collected by general practitioners (GPs) can be stored either as structured tables or written free text fields. Text fields contain various notes entered by the GP or the GP’s assistants, including summaries of reports from outside sources. Sometimes clinicians embed electronically transmitted laboratory test results within free text fields, instead of the intended structured tables.
This study developed and evaluated a computerized extraction method to convert numeric clinical information stored anywhere in an EPR into structured data. There were no prerequisites for how and where the information was registered during general practice. The method addresses the following issues: 1) accuracy of data extraction for numeric clinical measurements relevant to diabetes care; 2) performance of the extraction method during clinical practice, and post-processing actions needed to optimize accuracy.
Methods
The study identified 13 numeric clinical measurements considered relevant for evaluating the quality of diabetes care. 23–25 These included measurements of systolic (SBP) and diastolic (DBP) blood pressure, weight, height, serum glucose (fasting, non-fasting, unspecified), glycosylated hemoglobin (HbA1c), and several measures of serum cholesterol (total, TC; high-density lipoprotein, HDL-C; and, low-density lipoprotein, LDL-C), triglycerides, and serum creatinine.
Data Extraction Method
The computerized extraction method, triggered by numeric values in free text fields of the EPR, utilized nearby names and abbreviations used to label the measurement, including units and other specifications added to numeric values. A vocabulary of potential labels or specifications related to numeric values was generated by examining all unique words within two words from numbers embedded in text strings. Two authors independently reviewed this vocabulary and classified words as either belonging to a numeric measurement of interest or not. This vocabulary was used to select a text recognition algorithm that could correctly identify target words that varied in length, likelihood of different ways of spelling, and possible misinterpretation due to typing errors (see online-only Appendix, available at www.jamia.org). A character sequence algorithm was found to be most suitable for recognition of measurement labels, and therefore included in the data extraction method.
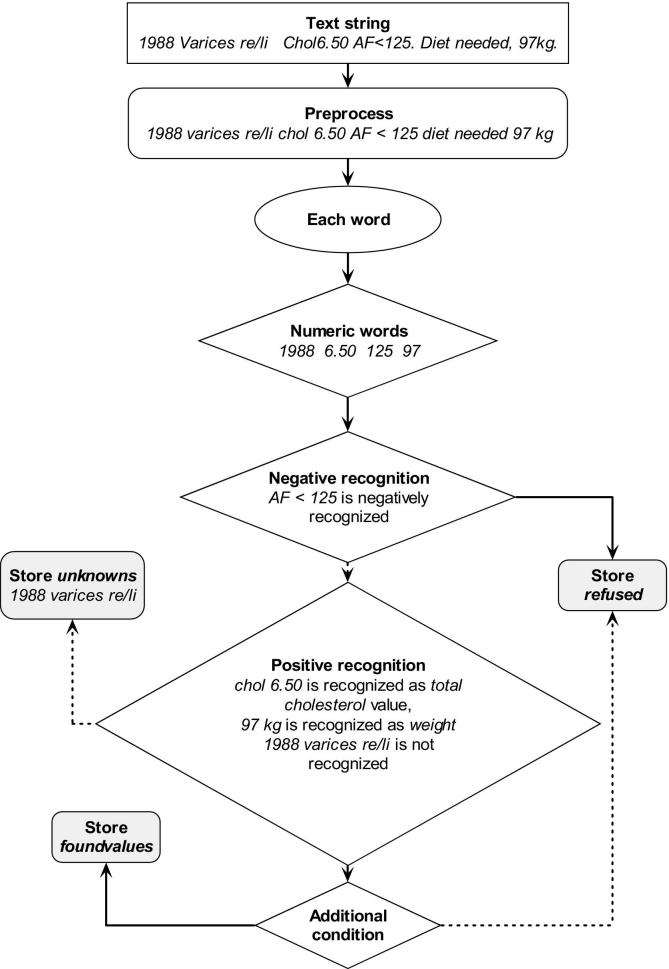
The data extraction method converted free text numeric measurement information into structured data (▶) as follows: (a) text strings, preprocessed to split compound strings and standardize character use, were split into substrings representing individual words; (b) each substring was classified as either containing or not containing relevant numeric values. Negative recognition (identification of irrelevant numerical values) involved evaluation of the four words neighboring a numeric substring, through comparison with a list of “negative definitions” for numeric values not of interest. Similarly, positive recognition involved comparison of neighboring words to the definitions made for each clinical measurement of interest. These comparisons included multiple definitions, so that, for example, “blood sugar” and “glucose” will both be recognized to represent the same clinical measurement. Unrecognized numeric values are stored together with their context words in a separate “unknowns” table which is monitored during the post-processing procedure. Authors incorporated this data extraction method into a software application that accesses two of the most commonly used EPR systems operating in general practice.
Figure 1.
Flow of the data extraction method (continuous lines are positive actions, interrupted lines are negative actions), with example text string.
Post-Processing Procedures
Most data of interest were originally manually entered into an EPR system by clinical personnel. Data entry errors were possible, and errors in numerical text identification might occur. A stepwise, semi-automated post-processing procedure was developed to identify and correct potential errors in extracted data, including checking for numeric values outside specified ranges, identifying unexpected peaks in clinical measurement time-series, and locating specific information in the “unknowns” table. Identified potential errors were visually reviewed by a trained registration worker, who could correct a numeric value or its variable allocation, delete erroneous data, or add missed data. All such actions were logged, together with reasons for modifications. Reasons were classified into three categories: (1) data entry errors in the GP practice (e.g., typing errors), (2) problems introduced by the automated data extraction system (e.g., false positives, cutoff or goal values instead of actual clinical measurements, dates instead of values), and (3) general data problems occurring in electronic databases (e.g., unit conversions or bogus values introduced by EPR system conversions).
Evaluation
Extraction method accuracy was assessed by comparison with a gold standard dataset of manually abstracted EPRs. Sixty randomly selected complete patient records from six GPs served as the gold standard. A single reviewer identified all values for any of the 13 selected measurements reported during one year. Verification involved double coding of 30 gold standard EPRs by an independent general practitioner. For these 30 records, agreement was excellent between both reviewers (kappa = 0.98). Sensitivity was measured as the proportion of correct (gold standard) measurements found with the automated data extraction method; and positive predictive value was determined as the proportion of gold standard (correct) measurements compared to the total number of extracted measurements—calculated before and after post-processing.
The evaluation in clinical practice utilized the data extraction software on two EPR systems in ten GP practices. Data were extracted for all 767 patients previously identified with type 2 diabetes in these practices. The two EPR systems (Promedico and MicroHIS) comprised two-thirds of the market share in the study region. The GP practices for this test were different from those contributing to the gold standard. During the evaluation, the data extraction software logged the time it required for data retrieval and interpretation. Additional effort to perform post-processing procedures was timed by activity logging in the central database maintenance software.
The regional Scientific Advisory Group of the General Practitioners Association approved the anonymous data collection procedure for this project.
Example
For the first dataset of 60 EPRs, 80% of the observations for the 13 selected measurements occurred only in free text segments of the EPRs; for 5 out of the 13 selected measurements this was above 90%. Between 3 and 12 unique labels were associated with each selected clinical measurement. During clinical practice evaluations, median rates of observations available only in free text fields ranged from 74% to 99%. Rates of structured registration varied considerably among GP practices. The number of unique labels within and between GP practices varied widely. For example, we found a total of 39 unique labels for “unspecified glucose value” in the GP practices, with each individual GP practice using 5 to 21 of them.
The data extraction software sensitivity exceeded 84% (compared to the gold standard) for all clinical measurements of interest except unspecified glucose. False positive results generated by the method caused low positive predictive values for height and HDL-cholesterol, but good PPVs for other measurements (▶). Applying the post-processing procedure improved the sensitivity for six of the clinical measurements, and increased the PPV to 100% for all measurements (▶).
Table 1.
Table 1 Accuracy of Data Extraction of 60 EPRs from 6 GP Practices
| Clinical Measurement | N | Sensitivity |
Positive Predictive Value |
||
|---|---|---|---|---|---|
| BP | AP | BP | AP | ||
| Diastolic Blood Pressure | 165 | 93.9 | 93.9 | 100.0 | 100.0 |
| Systolic Blood Pressure | 166 | 95.2 | 95.2 | 96.9 | 100.0 |
| Weight | 72 | 100.0 | 100.0 | 100.0 | 100.0 |
| Height | 16 | 100.0 | 100.0 | 19.3 | 100.0 |
| Glucose fasting | 64 | 84.4 | 89.1 | 91.5 | 100.0 |
| Glucose non-fasting | 89 | 94.4 | 97.8 | 100.0 | 100.0 |
| Glucose unspecified | 145 | 66.2 | 68.7 | 85.7 | 100.0 |
| HbA1c | 154 | 98.7 | 100.0 | 99.3 | 100.0 |
| Total cholesterol | 57 | 98.2 | 98.2 | 100.0 | 100.0 |
| HDL cholesterol | 42 | 95.2 | 95.2 | 51.3 | 100.0 |
| LDL cholesterol | 38 | 94.7 | 94.9 | 97.3 | 100.0 |
| Triglycerides | 38 | 100.0 | 100.0 | 100.0 | 100.0 |
| Creatinine | 55 | 96.4 | 100.0 | 100.0 | 100.0 |
N = number of observations present in gold standard; sensitivity and positive predictive value before (BP) and after (AP) post-processing; HbA1c = glycosylated hemoglobin; HDL = high-density lipoprotein cholesterol; LDL = low-density lipoprotein cholesterol.
Time to conduct data retrieval from EPR systems operating in actual practice ranged from 1.8 to 8.2 minutes per 100 patients. For the data interpretation, the software required 0.3–7.7 minutes per 100 patients. Total data extraction (retrieval plus interpretation), on average took 7.8 (s.d. 4.0) minutes per 100 patients. Effort to perform post-processing procedures varied between 42 and 184 minutes per 100 patients (mean 119). The total number of numeric measurements of interest extracted per practice ranged from 2,216 to 14,090 per 100 patients. The proportion of extracted data considered valid before post-processing was 72%–97% (▶). Deletion was the most common corrective action, followed by addition of data that had not been recognized by the computerized data extraction method (▶). An average of 87% (range per practice 56%–99%) of deletions removed erroneously identified values. For example, numeric values with “t” sometimes labeled other things than the wanted blood pressure (“tension”) results. Remaining deletions removed either incorrect values due to general database problems, or addressed data entry errors, e.g., tagging urine creatinine values as serum creatinines, or typing errors. Most often (49%), value modifications addressed unit conversions for height (m vs. cm). Overall, the proportion of corrections required to address data entry errors by GPs was 11%, ranging between 2% and 41%; the proportion due to general database errors was 14% (▶).
Table 2.
Table 2 Numbers of Valid Data Extractions and Corrective Actions Needed During Post-processing in the Field Test (10 GP practices), Standardized to a Population of 100 Patients per Practice
| Min | Max | Mean (SD) | |
|---|---|---|---|
| Valid extractions (n) | 2,216 | 14,090 | 9,156 (4,362) |
| Corrective actions (n) | 172 | 1,894 | 815 (504) |
| % Valid extractions | 72.4 | 97.4 | 89.8 (8.1) |
| Ratio Valid/Actions | 2.6 | 37.1 | 15.3 (11.3) |
Table 3.
Table 3 Type and Underlying Reasons for Corrective Actions Needed During Post-processing in the Field Test (10 GP practices), Standardized to a Population of 100 Patients per Practice
| General Database Error Mean (SD) | Data Entry Error GP Mean (SD) | Extraction Error Mean (SD) | Total Mean (SD) | |
|---|---|---|---|---|
| Deleted data (n) | 45.1 (110.5) | 28.1 (27.8) | 455.1 (406.3) | 528.3 (500.6) |
| Modified value (n) | 71.9 (63.2) | 32.7 (36.7) | 20.8 (25.0) | 125.3 (96.7) |
| Modified variable (n) | 0.0 (0.0) | 0.0 (0.0) | 20.6 (22.2) | 20.6 (22.2) |
| Added data (n) | 0.0 (0.0) | 21.9 (43.6) | 118.9 (123.4) | 140.8 (128.2) |
| % Corrective actions | 13.6 (10.8) | 11.4 (11.9) | 75.0 (16.6) |
Discussion
While the study EPRs could store data in structured tables, in practice, 80% of the numeric data for 13 diabetes-related clinical measurements occurred in free text segments of EPRs. The study data extraction method performed with a generally high sensitivity and positive predictive value, despite large variations in relevant-data-identifying labels both within and between practices. Results depended upon combination of a highly customizable text recognition algorithm, a number-oriented extraction method, and a semi-automated post-processing procedure.
Study data extraction procedures demonstrated feasibility of improved data collection without additional data registration or verification work for participating clinicians. Most of the study workload occurred at the central level during post-processing, with a maximum of 1.8 minutes needed per patient. Post-processing helps to maximize rates of correct observations extracted, 27 and was considered necessary to eliminate GP data entry errors. Information obtained during post-processing can improve text recognition definitions, increase accuracy of the extraction methods, and decrease workload of post-processing.
The alternative to the approach used in this study is to manually collect and review patient records for data extraction. In a Dutch project focusing on diabetes care in primary care, 28 manually collecting relevant clinical data from patient records required at least ten minutes per patient.
Recently, a JAMIA study reported abstracting blood pressure measurements from text notes using a label-oriented approach with similar accuracy to the current study. 29 Such label-oriented approaches apply lists of regular expressions for extraction, and are only feasible in situations where labeling styles are invariate. Otherwise, label-oriented approaches can result in poorer accuracy than number-oriented approaches.
Some researchers view structured data registration and standardization as the ultimate solution to achieve full benefits from EPRs. However, a combination of free text and structured data registration must exist for adequate patient record-keeping. 30 All record systems must accommodate clinicians’ workflows, and efforts for correct EPR use should be minimal. 31 For most GPs, the quickest way to record data involves generating a few lines of text. 10,32 When computer-based decision support systems require onerous efforts to achieve better data registration during routine practice, rates of system usage are often suboptimal. 33–35 Inflexible data collection forms with predefined items may fit the workflow of a specialized environment, but in general practice anticipation of data registration is difficult.
Limitations and Generalizability
This study focused on the extraction of numeric clinical measurements relevant for diabetes care. By including a wide range of measurements coming from physical examination and laboratory results, the study demonstrated applicability for collecting data from EPRs involving numeric values. The study applied data extraction software to two of the seven EPR systems used in Dutch general practice, but could easily be applied in other settings. No differences in sensitivity of data collection occurred between the two EPR systems. The authors have no reason to expect that this method will perform differently within other EPR systems. Conversely, a higher (adequate) use of structured tables for clinical data entry would improve upon observed PPVs before post-processing.
Conclusions
The study extraction method identifies and converts to structured data selected numeric clinical information stored anywhere within tested EPR systems. This method offers considerable advantages over existing methods that rely on structured data registration or manual data extraction. The method, through its generality, appears to hold potential value for conducting health services research and quality of care assessments in general practice.
Footnotes
The GIANTT project is funded by grants from the University Medical Center Groningen, The Netherlands.
The support of the physicians at the practices where data were collected is greatly appreciated. We thank Ineke van de Ven for double coding the gold standard dataset. We thank Promedico ICT B.V., The Netherlands, and iSoft, The Netherlands, for providing their EPR Information Systems for our test environment. Flora Haaijer-Ruskamp and Hans Hillege provided helpful comments on the draft of this paper, as did the anonymous reviewers of this journal.
The Groningen Initiative to Analyse Type 2 Diabetes Treatment (GIANTT) group are D. de Zeeuw, F.M. Haaijer-Ruskamp, P. Denig (Department of Clinical Pharmacology, University Medical Center Groningen), R.O.B. Gans (Department of Internal Medicine, University Medical Center Groningen), B.H.R. Wolffenbuttel (Department of Endocrinology, University Medical Center Groningen), F.W. Beltman (Department of General Practice, University Medical Center Groningen), K. Hoogenberg (Department of Internal Medicine, Martini Hospital Groningen), P. Bijster (Regional Diabetes Facility, General Practice Laboratory LabNoord, Groningen), J. Bolt (District Association of General Practitioners, Groningen), L.T.W. de Jong-van den Berg (Department of Social Pharmacy and Pharmacoepidemiology, University of Groningen), J.G.W. Kosterink (Hospital Pharmacy, University Medical Center Groningen), J.L. Hillege (Trial Coordination Center, Department of Clinical Epidemiology, University Medical Center Groningen).
References
- 1.Treweek S. The potential of electronic medical record systems to support quality improvement work and research in Norwegian general practice BMC Health Serv Res 2003;3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mansson J, Nilsson G, Bjorkelund C, Strender LE. Collection and retrieval of structured clinical data from electronic patient records in general practiceA first-phase study to create a health care database for research and quality assessment. Scand J Prim Health Care 2004;22:6-10. [DOI] [PubMed] [Google Scholar]
- 3.Lagasse R, Desmet M, Jamoulle M, et al. European situation of the routine medical data collection and their utilisation for health monitoring Euro-med-data, final report 2001.
- 4.Metsemakers JFM. Huisartsgeneeskundigen registraties in Nederland Onderzoeksinstituut voor ExTramurale en Transmurale Gezondheidszorg, editie 3, April 1999.
- 5.Jick SS, Kaye JA, Vasilakis-Scaramozza C, et al. Validity of the general practice research database Pharmacotherapy 2003;23:686-689. [DOI] [PubMed] [Google Scholar]
- 6.Carey IM, Cook DG, De Wilde S, et al. Developing a large electronic primary care database (Doctors’ Independent Network) for research Int J Med Inform 2004;73:443-453. [DOI] [PubMed] [Google Scholar]
- 7.Mosis G, Vlug AE, Mosseveld M, et al. A technical infrastructure to conduct randomized database studies facilitated by a general practice research database J Am Med Inform Assoc 2005;12:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lusignan S, Valentin T, Chan T, et al. Problems with primary care data quality: osteoporosis as an exemplar Inform Prim Care 2004;12:147-156. [DOI] [PubMed] [Google Scholar]
- 9.Prins H, Kruisinga FH, Buller HA, Zwetsloot-Schonk JH. Availability and usability of data for medical practice assessment Int J Qual Health Care 2002;14:127-137. [DOI] [PubMed] [Google Scholar]
- 10.McDonald CJ. The barriers to electronic medical record systems and how to overcome them J Am Med Inform Assoc 1997;4:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lusignan S, Chan T, Wells S, et al. Can patients with osteoporosis, who should benefit from implementation of the national service framework for older people, be identified from general practice computer records?A pilot study that illustrates the variability of computerized medical records and problems with searching them. Public Health 2003;117:438-445. [DOI] [PubMed] [Google Scholar]
- 12.Wiesman F, Hasman A, van den Herik HJ. Information retrieval: an overview of system characteristics Int J Med Inf 1997;47:5-26. [DOI] [PubMed] [Google Scholar]
- 13.Friedman C, Hripcsak G. Natural language processing and its future in medicine Acad Med 1999;74:890-895. [DOI] [PubMed] [Google Scholar]
- 14.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing J Am Med Inform Assoc 2004;11:392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meystre S, Haug PJ. Automation of a problem list using natural language processing BMC Med Inform Decis Mak 2005;5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman WW, Christensen LM, Wagner MM, et al. Classifying free-text triage chief complaints into syndromic categories with natural language processing Artif Intell Med 2005;33:31-40. [DOI] [PubMed] [Google Scholar]
- 17.Xu H, Anderson K, Grann VR, Friedman C. Facilitating cancer research using natural language processing of pathology reports Medinfo 2004;11:565-572. [PubMed] [Google Scholar]
- 18.Mendonça EA, Haas J, Shagina L, Larson E, Friedman C. Extracting information on pneumonia in infants using natural language processing of radiology reports J Biomed Inform 2005;38:314-321. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Friedman C. CliniViewer: A tool for viewing electronic medical records based on Natural Language Processing and XML Medinfo 2004;11:639-643. [PubMed] [Google Scholar]
- 20.Grant RW, Buse JB, Meigs JBCN - University HealthSystem Consortium (UHC) Diabetes Benchmarking Project Team Quality of diabetes care in U.S. academic medical centers: low rates of medical regimen change Diabetes Care 2005;28:337-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Berardis G, Pellegrini F, Franciosi M, et al.; for the QuED (Quality of Care and Outcomes in Type 2 Diabetes) Study Group. Quality of diabetes care predicts the development of cardiovascular events: Results of the QuED study. Nutr Metab Cardiovasc Dis (e-pub ahead of print). 2006; doi: 10.1016/j.numecd.2006.04.009. [DOI] [PubMed]
- 22.Ackermann RT, Thompson TJ, Selby JV, et al. Is the number of documented diabetes process-of-care indicators associated with cardiometabolic risk factor levels, patient satisfaction, or self-rated quality of diabetes care?The Translating Research into Action for Diabetes (TRIAD) study. Diabetes Care 2006;29:2108-2113. [DOI] [PubMed] [Google Scholar]
- 23.UK Department of Health National service framework for diabetes—the delivery strategy. 2001. London.
- 24.Anonymous Diabeteszorg Beter. Rapport van de Taakgroep Programma Diabeteszorg. The Netherlands: Ministry of Health; 2005.
- 25. The Health Plan Employer Data and Information Set (HEDIS). Washington DC: National Committee for Quality Assurance; 2006. Accessed at www.ncqa.org/programs/hedis on Oct 10.
- 26.Kerr EA, Smith DM, Hogan MM, et al. Comparing clinical automated, medical record, and hybrid data sources for diabetes quality measures Jt Comm J Qual Improv 2002;28:555-565. [DOI] [PubMed] [Google Scholar]
- 27.Andersen M. Is it possible to measure prescribing quality using only prescription data? Basic Clin Pharmacol Toxicol 2006;98:314-319. [DOI] [PubMed] [Google Scholar]
- 28.Schaars CF, Denig P, Kasje WN, Stewart RE, Wolffenbuttel BH, Haaijer-Ruskamp FM. Physician, organizational, and patient factors associated with suboptimal blood pressure management in type 2 diabetic patients in primary care Diabetes Care 2004;27:123-128. [DOI] [PubMed] [Google Scholar]
- 29.Turchin A, Kolatkar NS, Grant RW, Makhni EC, Pendergrass ML, Einbinder JS. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes J Am Med Inform Assoc 2006;13:691-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Ginneken AM. The computerized patient record: balancing effort and benefit Int J Med Inf 2002;65:97-119. [DOI] [PubMed] [Google Scholar]
- 31.Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using CDSS: a systematic review of trials to identify features critical to success. BMJ. doi: 10.1136/bmj.38398.500764.8F (published 14 March 2005). [DOI] [PMC free article] [PubMed]
- 32.Los RK, van Ginneken AM, van der Lei J. OpenSDE: a strategy for expressive and flexible structured data entry Int J Med Inform 2005;74:481-490. [DOI] [PubMed] [Google Scholar]
- 33.Hetlevik I, Holmen J, Kruger O. Implementing clinical guidelines in the treatment of hypertension in general practiceEvaluation of patient outcome related to implementation of a computer-based clinical decision support system. Scand J Prim Health Care 1999;17:35-40. [DOI] [PubMed] [Google Scholar]
- 34.Montgomery AA, Fahey T, Peters TJ, MacIntosh C, Sharp DJ. Evaluation of computer based clinical decision support system and risk chart for management of hypertension in primary care: randomised controlled trial BMJ 2000;320:686-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balas EA, Krishna S, Kretschmer RA, Cheek TR, Lobach DF, Boren SA. Computerized knowledge management in diabetes care Med Care 2004;42:610-621. [DOI] [PubMed] [Google Scholar]