Abstract
This review focuses on the monoterpene, sesquiterpene, and diterpene synthases of plant origin that use the corresponding C10, C15, and C20 prenyl diphosphates as substrates to generate the enormous diversity of carbon skeletons characteristic of the terpenoid family of natural products. A description of the enzymology and mechanism of terpenoid cyclization is followed by a discussion of molecular cloning and heterologous expression of terpenoid synthases. Sequence relatedness and phylogenetic reconstruction, based on 33 members of the Tps gene family, are delineated, and comparison of important structural features of these enzymes is provided. The review concludes with an overview of the organization and regulation of terpenoid metabolism, and of the biotechnological applications of terpenoid synthase genes.
Keywords: terpene cyclase, isoprenoids, plant defense, genetic engineering, secondary metabolism
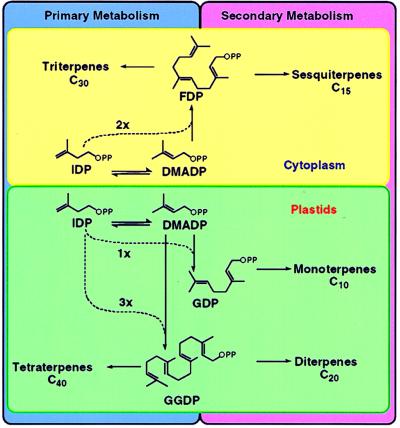
The pathways of monoterpene, sesquiterpene, and diterpene biosynthesis are conveniently divided into several stages. The first encompasses the synthesis of isopentenyl diphosphate, isomerization to dimethylallyl diphosphate, prenyltransferase-catalyzed condensation of these two C5-units to geranyl diphosphate (GDP), and the subsequent 1′-4 additions of isopentenyl diphosphate to generate farnesyl (FDP) and geranylgeranyl (GGDP) diphosphate (Fig. 1) (1). In the second stage, the prenyl diphosphates undergo a range of cyclizations based on variations on the same mechanistic theme to produce the parent skeletons of each class. Thus, GDP (C10) gives rise to monoterpenes (2), FDP (C15) to sesquiterpenes (3), and GGDP (C20) to diterpenes (4). These transformations catalyzed by the terpenoid synthases (cyclases) may be followed by a variety of redox modifications of the parent skeletal types to produce the many thousands of different terpenoid metabolites of the essential oils, turpentines, and resins of plant origin (5).
Figure 1.
Organization of terpene biosynthesis in plants. DMADP, dimethylallyl diphosphate. IDP, isopentenyl diphosphate.
This review focuses on the synthases that use prenyl diphosphate substrates to generate the enormous diversity of carbon skeletons characteristic of terpenoids. Most of these natural products are cyclic, and many contain multiple ring systems, the basic structures of which are determined by the highly specific terpenoid synthases; examples of synthases that produce acyclic products are also known. The terpenoid synthases may be involved in the regulation of pathway flux because they operate at metabolic branch points and catalyze the first committed steps leading to the various terpene classes (6). The synthases responsible for generating the parent compounds of the various types are quite similar in properties (7), and all operate by electrophilic reaction mechanisms, as do the prenyltransferases (8, 9). Comprehensive treatment of the topic, especially enzymological and mechanistic aspects, has been provided recently (2–4), and the field is periodically surveyed (10, 11). After brief coverage of the three types of terpene synthases from higher plants, with emphasis on common features of structure and function, we focus here on molecular cloning and sequence analysis of these important and fascinating catalysts.
Enzymology and Mechanism of Terpenoid Cyclization
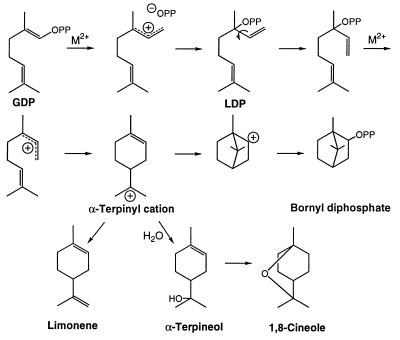
GDP is considered to be the natural substrate for monoterpene synthases, because all enzymes of this class efficiently utilize this precursor without the formation of free intermediates (12). Since GDP cannot be cyclized directly because of the C2-C3 trans-double bond, the reaction mechanism necessarily involves both isomerization and cyclization steps (12, 13). Thus, GDP ionizes with the assistance of a divalent metal ion (Fig. 2), as in the first step of the prenyltransferase reaction (8). The resulting allylic cation-diphosphate anion pair then rearranges to form the enzyme-bound, tertiary allylic isomer, 3R- or 3S-linalyl diphosphate (LDP, depending on the initial folding of the geranyl substrate). After rotation to the cisoid conformer, LDP ionizes and is cyclized in anti,endo-form to the corresponding 4R- or 4S-α-terpinyl cation. From this universal intermediate, the reaction may take one of several routes involving internal additions to the remaining double bond, hydride shifts, or rearrangements before the terminal carbocation is deprotonated to an olefin or captured by water or the diphosphate anion. In the simplest of all terpenoid cyclizations, the α-terpinyl cation is deprotonated to yield limonene (14) (Fig. 2). Alternatively, the α-terpinyl cation may undergo further cyclization, via the remaining double bond, to afford the pinyl cation (and then α- or β-pinene after deprotonation) (15) or the bornyl cation (to form bornyl diphosphate by capture of the diphosphate) (16). Hydride shifts in the α-terpinyl cation yield the terpinen-3-yl or terpinen-4-yl cations, providing access to the phellandrenes (17) and thujanes (18), respectively. Water capture of the α-terpinyl cation yields α-terpineol, which upon heterocyclization affords 1,8-cineole (19). All monoterpene cyclases are capable of catalyzing both the isomerization and cyclization reactions, and these steps occur via a series of ion pairs at the same active site (12). A few monoterpene synthases produce acyclic products such as myrcene (20) and linalool (21). Additional variations on the electrophilic isomerization–cyclization sequences illustrated (Fig. 2) account for essentially all monoterpene skeletal types, and their stereoisomers and derivatives.
Figure 2.
Biosynthesis of representative monoterpenes from GDP.
A number of monoterpene synthases from angiosperms, gymnosperms, and bryophytes have been partially purified and characterized (7, 22, 23), and all have similar properties; native molecular mass in the 50- to 100-kDa range (either monomers or homodimers), a requirement for a divalent metal ion (usually Mg2+ or Mn2+), a pI value near 5.0, and a pH optimum within a unit of neutrality. Monoterpene synthases are operationally soluble, although most are associated with plastids in vivo (6, 20, 24). The synthases of gymnosperms are distinguishable from their angiosperm counterparts by the requirement for a monovalent cation (K+ preferred), a preference for Mn2+ (or Fe2+) over Mg2+ as cofactor, and a generally higher pH optimum. An interesting feature of several monoterpene synthases is their ability to produce multiple products (14, 25). For example, the (−)-pinene synthases from sage and grand fir produce both (−)-α- and (−)-β-pinene (20, 25–28).
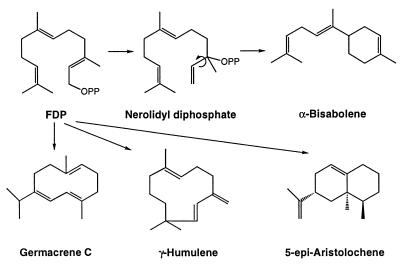
All sesquiterpenes are derived from FDP (29), and the structural diversity of this class is greater than that of the monoterpenes because of the increased number of cyclizations possible from a precursor with five additional carbon atoms [the monoterpenes number approximately 1,000, the sesquiterpenes more than 7,000 (5)]. As with the monoterpenes, the formation of cyclohexanoid compounds, such as α-bisabolene, requires preliminary isomerization of the trans-farnesyl precursor to the sesquiterpene analog of LDP, i.e., 3R- or 3S- nerolidyl diphosphate, followed by ionization-dependent cyclization (Fig. 3). The increased size of the farnesyl chain also permits cyclization to 10- and 11-membered macrocycles such as germacrene C (30) and γ-humulene (31). Internal additions to the remaining double bonds of the initially formed cyclic carbocations also occur that, along with hydride shifts, methyl migrations, and Wagner–Meerwein rearrangements, permit generation of a broad range of structures, including δ-cadinene (32), δ-selinene (31), epi-aristolochene (33), vetispiradiene (31), and longifolene (31) (Fig. 3). Acyclic sesquiterpenes, such as β-farnesene, are also derived from FDP by related synthases (35, 36). The similarities in reaction mechanisms between the plastidial monoterpene synthases and the cytosolic sesquiterpene synthases are paralleled by similarities in properties (7).
Figure 3.
Biosynthesis of representative sesquiterpenes from FDP.
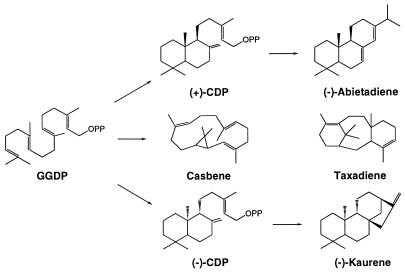
The diterpenoids are also widespread in the plant kingdom, and they often are encountered in the resins of conifers, woody legumes, composites, and members of the Euphorbiaceae. More than 3,000 different diterpenoid structures have been defined, all of which appear to be derived from GGDP (37). The most common diterpene is phytol, the acyclic alcohol side chain of chlorophyll. However, most diterpenoids are cyclic, and there appear to be two major, and fundamentally different, modes of cyclization in this class. The macrocyclic diterpenes, such as casbene (38), cembrene (39), and taxadiene (40, 41), are formed by cyclizations analogous to those of the monoterpene and sesquiterpene series (Fig. 4).
Figure 4.
Biosynthesis of representative diterpenes involving ionization and cyclization of GGDP directly, and preliminary cyclization to CDP by protonation of the terminal double bond.
The second mode of cyclization involves generation of copalyl diphosphate (CDP) as the initial intermediate (37) (Fig. 4). This reaction cascade is initiated by protonation of the terminal double bond of GGDP followed by two internal additions and proton elimination, in a sequence similar to that catalyzed by squalene cyclase in the triterpene series (42). CDP then can be transformed into a variety of tricyclic and tetracyclic diterpenoids by ionization of the diphosphate ester and subsequent internal additions, rearrangements, and terminations. These reactions proceed along parallel routes leading to different stereoisomers of CDP. In the biosynthesis of (−)-kaurene, the precursor of the gibberellin plant hormones, (−)-CDP synthase (kaurene synthase A) catalyzes the protonation-initiated cyclization of GGDP to (−)-CDP (43, 44), whereas a separate enzyme, kaurene synthase B (45), transforms this intermediate to (−)-kaurene via a more typical, ionization-dependent cyclization (46, 47) (Fig. 4). In the biosynthesis of (−)-abieta-7(8),13(14)-diene, the precursor of abietic acid of conifer resin (48, 49), a single enzyme, abietadiene synthase, catalyzes the conversion of GGDP to (+)-CDP and the cyclization of this bound intermediate to the olefin (50).
In spite of the different modes of cyclization in the diterpene series, the properties of the responsible enzymes are quite similar, and they resemble those of the other terpenoid synthases. The remarkably similar characteristics of the monoterpene, sesquiterpene, and diterpene synthases are certainly related to the fact that these enzymes carry out similar electrophilic cyclizations involving common steps, i.e., the generation, transformation, and stabilization of highly reactive carbocations and their ultimate quenching by deprotonation or nucleophile capture. The unique features of each individual cyclization relate to the precise means by which each synthase enforces conformation on the substrate and intermediates to effect a particular reaction channel while protecting such reactive species from premature termination. Although the size and shape of the active sites must differ (51), the basic means by which these enzymes enforce regiochemistry and stereochemistry of product formation are also probably very similar. This similarity in function is reflected in similarity in primary structure of all of the terpene synthases of plant origin (20, 24, 30–34, 36, 50, 52–55).
Molecular Cloning of Terpenoid Synthase Genes
More than 30 plant terpenoid synthases have now been cloned as cDNAs, many of which encode enzymes of secondary metabolism. Because expression of terpene synthase genes is highly up-regulated in specialized cells such as those of glandular trichomes in mint (36), or restricted to certain developmental stages (56) or short periods of transient defense reactions (20, 32–34, 53), most molecular cloning efforts have relied on enriched plant materials for mRNA isolation. Sesquiterpene synthases, such as tobacco epi-aristolochene synthase (33), cotton δ-cadinene synthase (32), and Hyoscyamus muticus vetispiradiene synthase (34), catalyze cyclizations involved in phytoalexin biosynthesis, and expression of these enzymes is induced in cell cultures by elicitation. Thus, cDNA libraries enriched for terpenoid biosynthetic genes were constructed from mRNA isolated from induced cells. A similar approach was used to clone casbene synthase, an elicitor-responsive diterpene synthase from castor bean (53). Grand fir responds to mechanical wounding of stems, which mimics bark beetle attack, by the induced biosynthesis of defense-related monoterpenes, sesquiterpenes, and diterpenes (20, 31, 57). Therefore, an induced grand fir stem cDNA library (50), in which monoterpene synthases are represented at a frequency of approximately 1% (20), was employed for the cloning of 11 defense-related terpene synthases (20, 31, 50) representing the largest group of synthases available from any plant source.
Another means of cloning terpenoid synthases exploits their often tissue- or cell-specific pattern of expression. Thus, enriched libraries from tomato leaf epidermis and from flower petals of Clarkia breweri were employed for cloning of germacrene C synthase (30) and linalool synthase (56), respectively. Highly enriched, tissue-specific libraries are also useful for cloning of terpenoid synthases in cases where hybridization probes are not easily accessible. Random cloning from such libraries relies on diagnostic sequences of terpenoid synthases for selection, with confirmation by functional expression of target activities. Random sequencing of a peppermint oil gland cDNA library, in which limonene synthase is represented at a frequency of approximately 4%, was successfully employed for isolation of a (E)-β-farnesene synthase clone (36).
Probes for targeted library screening have been acquired by three basic strategies: reverse genetic approaches based on purified enzymes, similarity-based PCR methods, and mutant-based techniques. By applying reverse genetics, epi-aristolochene synthase (33) and casbene synthase (53) were obtained with antibody probes, and limonene synthase (24), abietadiene synthase (50), linalool synthase (56), and kaurene synthase B (45) were cloned by using protein sequence information. Subsequently, closely related genes were cloned by using heterologous hybridization probes or heterologous, nondegenerate PCR primers. Thus, Hyoscyamus vetispiradiene synthase (34) and cotton δ-cadinene synthase (32) sequences were amplified by PCR using primers designed from tobacco epi-aristolochene synthase, and the spearmint limonene synthase cDNA retrieved a Perilla limonene synthase cDNA (54).
Because purification of plant terpenoid synthases often is difficult, a general, similarity-based cloning technique was developed (20, 52, 57) that was founded on the use of consensus sequence elements for design of degenerate primers for PCR amplification. This strategy has been employed to amplify specific probes for terpenoid synthases from angiosperms and gymnosperms, and these probes were used to obtain cDNAs encoding taxadiene synthase from Pacific yew (52), a range of monoterpene and sesquiterpene synthases from grand fir (20, 31), three different monoterpene synthases from common sage, and germacrene C synthase from tomato (30). Degenerate oligonucleotides based on consensus sequences for (−)-CDP synthase from Arabidopsis thaliana (43) and maize (58) were employed to isolate a pea (−)-CDP synthase cDNA (44) and a bifunctional kaurene synthase cDNA from the fungus Phaeosphaeria (69).
Mutant-based cloning has been used only for the isolation of (−)-CDP synthase from Arabidopsis (43) and maize (58). The target synthase catalyzes the initial cyclization in gibberellin biosynthesis and is encoded by single-copy genes in Arabidopsis and maize. Mutations at the Arabidopsis GA1 locus (60) and at the maize An1 locus (58) were identified based on gibberellin-responsive dwarf phenotypes, and the isolated genes were confirmed by complementation and functional expression in E. coli. Only the terpenoid synthases of primary metabolism have been observed to affect plant growth and development directly, thereby allowing, by mutation, an alteration in morphological phenotype. By contrast, mutated terpenoid synthases of secondary metabolism result in altered chemotypes that are difficult to screen. Terpenoid synthases of secondary metabolism are also often represented by multiple-copy gene families that encode functionally identical or highly similar enzymes (20, 32–34); such redundancy drastically hampers mutant-based cloning.
In addition to directed cDNA cloning of terpenoid synthases, plant genome sequencing will inadvertently reveal new terpenoid synthase-like genes (61). Until now, most terpenoid synthases have been cloned as cDNAs, and the structures of only a few terpene synthase genes have been described. Comparison of the genes for tobacco epi-aristolochene synthase, castor bean casbene synthase, Hyoscyamus vetispiradiene synthase, mint limonene synthase, and a putative terpene synthase from Arabidopsis reveals a similar overall structure with six positionally conserved introns (33, 53, 61). This conservation pattern guided a domain-swapping experiment with epi-aristolochene synthase and vetispiradiene synthase, which demonstrated that exon domains can be interchanged among different synthases to produce functional chimeric proteins, some of which produce mixtures of epi-aristolochene and vetispiradiene (62).
Heterologous Expression of Terpenoid Synthases
Sequence comparison with extant terpenoid synthases may allow placement of a new sequence into one of the six subdivisions of the plant Tps gene family (Fig. 5) (20) but does not allow functional characterization of a new synthase. Heterologous expression as an active recombinant protein and identification of the enzymatic reaction products formed with the possible C10, C15, or C20 prenyl diphosphate substrates is required. Because most terpenoid synthases are present in plants at low levels and are not easily purified, heterologous expression of cloned synthases has played an important role in obtaining high yields of pure proteins for antibody preparation or crystallization for structural analysis. Terpenoid synthases are operationally soluble enzymes localized to the cytosol (sesquiterpene synthases) or plastids (monoterpene synthases and diterpene synthases), and they often can be expressed functionally in E. coli using standard plasmid expression vectors. However, in some cases, the N-terminal transit peptides of these synthases, which target the nuclear-encoded preproteins to the plastids for proteolytic processing to the mature forms, promote formation of inclusion bodies when expressed in E. coli. High-yield expression of soluble monoterpene synthases can be achieved by truncation of the cDNAs to remove the targeting sequences. Another commonly encountered problem with expression of plant terpenoid synthases in E. coli relates to the frequency of arginine residues that use rare tRNAs in the prokaryotic host. Coexpression of the terpenoid synthase cDNA with the required tRNA can eliminate translational difficulty and yield high-level expression of active recombinant enzymes (31, 65).
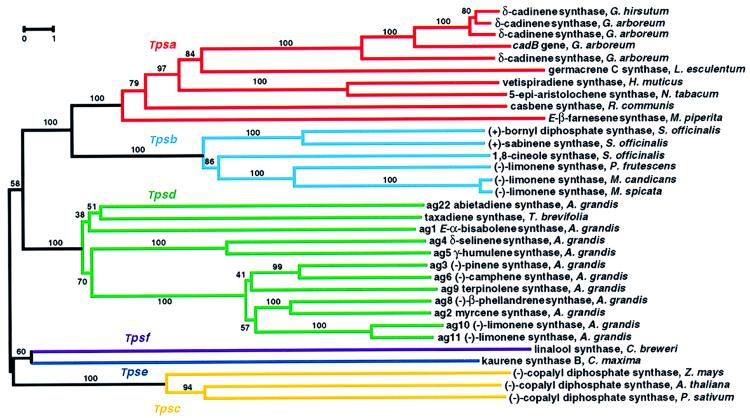
Figure 5.
Phylogenetic tree of plant terpenoid synthases reconstructed by using Dayhoff’s (63) distances between proteins and the neighbor-joining method (64). Scale bar indicates 1% sequence divergence. Numbers are the actual bootstrap values of branches. Tpsa through Tpsf designate Tps gene subfamilies defined by a minimum of 40% identity between members at the amino acid level.
cDNA cloning and functional expression has provided unambiguous proof that many terpenoid synthases form multiple products in fixed ratios. For example, recombinant limonene synthase converts GDP to (−)-limonene, α- and β-pinene, and myrcene in a ratio identical to the native enzyme (14, 24), and grand fir (−)-pinene synthase, in both native and recombinant form, produces (−)-α- and (−)-β-pinene at a ratio of 2:3 (20, 27). Expression of the cDNAs for several grand fir monoterpene synthases also has provided evidence that the complex turpentine mixture produced by this conifer (66) is formed by a family of single-product and multiproduct enzymes encoded by closely related genes (20). Single-product and multiproduct sesquiterpene synthases also exist. Thus, δ-selinene synthase and γ-humulene synthase of grand fir form, respectively, 34 and 52 different sesquiterpenes (31), whereas a third synthase from grand fir produces only (E)-α-bisabolene.
Testing with different prenyl diphosphate substrates of heterologously expressed enzymes indicates that substrate specificities have evolved differently in the three classes of terpenoid synthases. All monoterpene synthases and diterpene synthases reveal strict substrate specificity in accepting only GDP or GGDP, respectively, as might be expected for the purpose of controlling flux through these plastidial pathways for isoprenoid metabolism (Fig. 1). Selectivity in the case of monoterpene synthases can be explained as a result of size exclusion for FDP or GGDP, but other refinements in active-site structure are required to exclude GDP and FDP as substrates for diterpene synthases. Interestingly, several sesquiterpene synthases will accept GDP (but not GGDP) as an alternate, but inefficient, substrate for the formation of the simple olefin limonene (30, 31, 36). Under physiological conditions, the cytosolic sesquiterpenoid synthases are unlikely to encounter GDP, which arises in plastids (6), and thus there is no evolutionary pressure for discrimination against this adventitious substrate.
Sequence Relatedness and Phylogenetic Reconstruction
Amino acid sequence relatedness of plant terpenoid synthases allows subdivision of the Tps gene family into six subfamilies (Fig. 5) (20), designated Tpsa through Tpsf, each distinguished by sharing a minimum of 40% identity among members. The terpenoid synthases of primary metabolism, (−)-CDP synthase (43, 44, 58) (Tpsc) and kaurene synthase B (45) (Tpse), are only distantly related to those of secondary metabolism, including members of subfamilies Tpsa, Tpsb, and Tpsd. However, all plant terpene synthases share a common evolutionary origin (Fig. 5), and it appears that the bifurcation of terpenoid synthases of primary and secondary metabolism occurred before separation of angiosperms and gymnosperms. Terpenoid synthases of secondary metabolism constitute the most extensively studied Tps subfamilies, including Tpsa, Tpsb, Tpsd, and the distant and possibly ancient Tpsf branch containing linalool synthase. The Tpsa subfamily consists of angiosperm sesquiterpene synthases (32–34, 36, 55) and a diterpene synthase gene encoding casbene synthase (53). The alignment of casbene synthase with sesquiterpene synthases, as opposed to (−)-CDP synthase and kaurene synthase B of the diterpene series, is consistent with the similarity in reaction mechanism of casbene synthase to those of the sesquiterpene synthases. The angiosperm monoterpene synthase subfamily Tpsb, representing members of the Lamiaceae (30, 54), is clearly distinct from angiosperm sesquiterpene synthases (Tpsa) and from gymnosperm monoterpene synthases (Tpsd). Sequence comparison (20) and phylogenetic reconstruction (Fig. 5) consistently reveal that gymnosperm monoterpene, sesquiterpene, and diterpene synthases represented by genes from grand fir (20, 31, 50) and Pacific yew (52) are more closely related to each other than they are to their counterparts of angiosperm origin. Thus, the gymnosperm terpenoid synthases form a separate branch designated as the Tpsd group (20). The pattern of bifurcation of gymnosperm and angiosperm terpenoid synthases from a common ancestor implies independent functional specialization after separation of the angiosperm and gymnosperm lineages (Fig. 5). For example, functional limonene synthases (sharing only 30–35% identity) have evolved separately in the Lamiaceae and in grand fir. Substitution rates are significantly lower in the Tpsd group than among the angiosperm terpene synthases, consistent with the evolution of 18S rRNA in gymnosperms (67).
Terpenoid synthases of subfamilies Tpsa, Tpsb, and Tpsd show much greater functional diversity than do members of the Tpsc group of (−)-CDP synthases. The observation that genes of secondary metabolism evolve functional diversity with little change in primary structure may be explained in two ways that relate to gene copy number and to the physiological tolerance of functional mutation. Many terpenoid synthases of secondary metabolism are encoded by multiple-gene copies (32–34) that arose by duplication and then provided the basis for diversification. In grand fir, at least seven very closely related genes (minimum of 65% identity) encode monoterpene synthases with distinct product patterns (20). By contrast, (−)-CDP synthases are encoded by single-copy genes in Arabidopsis (43) and maize (58). Genes of gibberellin biosynthesis are less likely to tolerate mutations leading to alteration in product outcome. Genes of secondary metabolism, however, are not essential for growth and development (68) and therefore may tolerate functional mutations leading to product diversity. Moreover, increased diversity in terpenoid chemistry may prove beneficial in ecological interactions with competing plants, as well as pathogens, herbivores, and pollinators.
The rather limited sequence similarity of plant terpenoid synthases to their microbial counterparts does not provide clear indication of a common evolutionary origin. Unclear also is the phylogenetic relationship between the terpenoid synthases and the mechanistically related prenyltransferases (1). The suggestion that these enzymes, which catalyze related steps in isoprenoid biosynthesis, evolved from a common ancestor is intriguing (42, 69), and recent crystal structure analyses reveal significant three-dimensional similarities between FDP synthase (70) and sesquiterpene cyclases (51, 69), in spite of very limited sequence similarity (33, 71). Although these similarities in tertiary structure may have evolved convergently as a consequence of common reaction mechanisms, they nevertheless allow three-dimensional rationalization of these mechanisms and provide confirmation of previous, but indirect, evidence concerning active-site organization (72–75). To address this question of relatedness in detail will require structural evaluation of terpene synthases and prenyltransferases from the same species.
General Structural Features
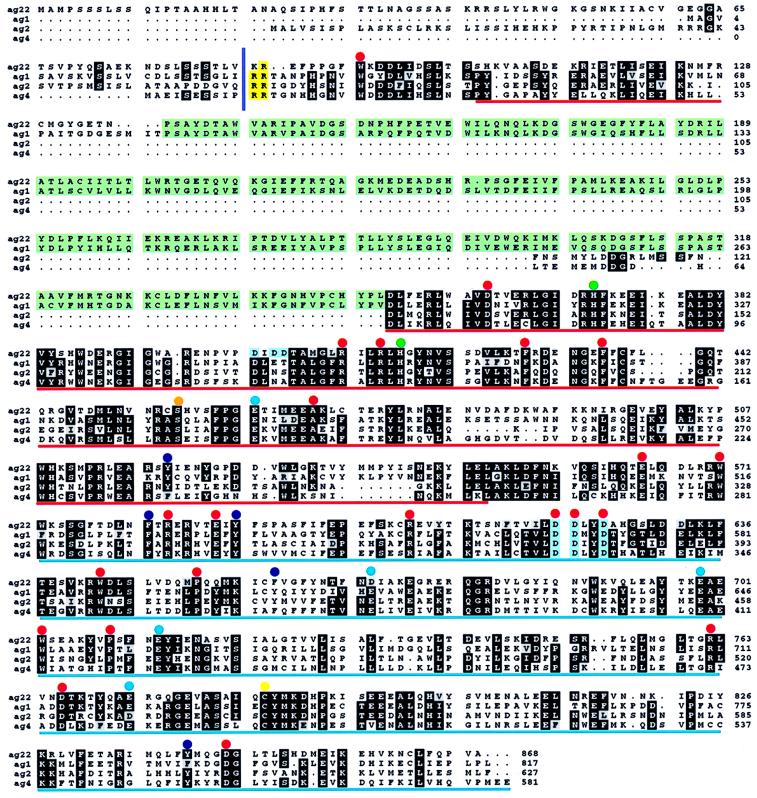
Terpenoid synthase cDNAs encode proteins of 550–850 aa (Fig. 6), in agreement with observed native molecular masses of 50–100 kDa. Based on the crystal structure of epi-aristolochene synthase (51), it appears that terpene synthases of subfamilies Tpsa, Tpsb, and Tpsd are composed of two distinct structural domains, a C-terminal active site domain, and an N-terminal domain that structurally resembles catalytic cores of glycosyl hydrolysases. In general, monoterpene synthases are between 600 and 650 aa in length (20, 24, 54) and are larger than sesquiterpene synthases by 50–70 aa (30–34, 36, 55). This difference largely is a result of the N-terminal transit peptides required for plastidial targeting of monoterpene synthases. Although lacking similarity in primary structure, the targeting sequences are characterized by a high content of serine and threonine and a low number of acidic residues (20). N-terminal deletion has demonstrated that residues upstream of a highly conserved RR-motif (Fig. 6) are not required for monoterpene synthase activity. Because sequence similarity among these synthases is significant only downstream of the RR-motif, this element may define the approximate N terminus of mature processed proteins.
Figure 6.
Alignment of representative deduced amino acid sequences of monoterpene, sesquiterpene, and diterpene synthases of grand fir. ag1, (E)-α-bisabolene synthase; ag2, myrcene synthase; ag4, δ-selinene synthase; and ag22, abietadiene synthase (20, 31, 50). Conserved tandem arginines are shaded yellow with the minimum mature protein indicated by a vertical bar. A conserved insert sequence is shaded green. Aspartate-rich motifs are shaded blue. Dots mark absolutely conserved residues (red), highly conserved histidines (green), a highly conserved serine (orange), conserved acidic residues (blue), conserved aromatic residues (purple), and a highly conserved cysteine (yellow). The glycosyl hydrolysase-like domain is underlined in red. The active-site domain is underlined in blue.
Most diterpene synthases are approximately 210 aa longer than even monoterpene synthases. The difference in length is accounted for by an additional internal element (Fig. 6) that is conserved in sequence and position among all diterpene synthases regardless of cyclization mechanism (52, 43–45, 50, 58), with the exception of casbene synthase (53). Interestingly, this sequence is also conserved in the grand fir sesquiterpene synthase (E)-α-bisabolene synthase and in Clarkia linalool synthase (56), a monoterpene synthase. These terpene synthases represent members of four distantly related subfamilies (Tpsc, Tpsd, Tpse, Tpsf), implying that this structural element existed in a common ancestor (Fig. 5). However, not all members of the Tpsd group carry the insertion, nor do enzymes that contain the insertion share any other common property. Location of the insertion N-terminal to the active site domain (51) suggests a function other than catalysis. Until a function in stability, targeting, or regulation is established, it will remain unclear why this conserved sequence had been lost in some synthases.
Specific Structural Elements
Comparison of 28 terpene synthases of the tpsa, tpsb, and tpsd subfamilies reveals several conserved amino acid residues (see Fig. 6 for representative sequences). These include absolutely conserved positions corresponding to grand fir (E)-α-bisabolene synthase ag1 Trp35, 516, 588, 647, Asp306, 566, 567, 570, 714, 794, Arg356, 359, 529, 552, 710, Phe373, 380, Ala414, Glu509, 534, and Pro597, 654. Two His-residues (His316, 361), Cys733, and Ser401 are highly conserved. Furthermore, six additional positions (residues 465, 527, 536, 604, 641, 790 in ag1) are absolutely conserved for aromatic amino acids, and four positions (residues 408, 613, 644, 658) are absolutely conserved for acidic amino acids. Very few of the residues that are absolutely conserved among synthases of the tpsa, tpsb, and tpsd subfamilies are also conserved in distantly related synthases of subfamilies tpsf, tpse, and tpsc. Residues that are absolutely conserved in all 33 known sequences are Trp35, 516, 647, Arg359, 552, Phe373, and Glu509, with the numbering corresponding to ag1.
The first crystal structure of a plant terpene synthase recently was solved for tobacco epi-aristolochene synthase (51) and revealed the protein to be composed entirely of α-helices with short, connecting loops and turns, forming a two-layer α-barrel active site. This structure resembles that of microbial synthases (42, 69), and modeling studies suggest that all terpenoid synthases bear the same overall fold structure. This common three-dimensional framework, when coupled to directed mutagenesis, provides the means of evaluating the functional relevance of conserved amino acids.
The hydrophobic, aromatic-rich active-site pocket of epi-aristolochene synthase is formed by α-helices of the C-terminal domain to accommodate the olefin chain of the substrate, whereas the diphosphate moiety is complexed by two Mg2+ ions at the entrance of the active site. One Mg2+ is coordinated by conserved residues corresponding with ag1 Asp566, Asp570, and Glu644. Both aspartate residues are part of an absolutely conserved aspartate-rich motif, DDxxD (Fig. 6). Mutagenesis of any of the three aspartates of the DDxxD motif of limonene synthase to either Ala or Glu reduces catalytic activity by 1,000-fold. The second Mg2+ is coordinated by an Asp residue corresponding to highly conserved ag1 Asp714, a nonconserved Thr-residue, and by a well conserved Glu/Asp-residue corresponding to ag1 Glu721. The side chain of an absolutely conserved Arg-residue (Arg529 in ag1) also interacts with substrate analogues in epi-aristolochene synthase. This residue, together with the Mg2+ ions and a second absolutely conserved Arg (Arg710 in ag1), is thought to stabilize negative charge of the diphosphate after ionization and to direct this species away from the active site to prevent reaction with carbocationic intermediates of the catalytic cycle. Aromatic residues, many of which are conserved, may stabilize carbocationic intermediates via π–cation interaction (2). The carboxyl of an Asp-residue (conserved as ag1 Asp794) could be involved in proton translocation (deprotonation and allotopic reprotonation) in reaction intermediates, as modeled for epi-aristolochene synthase, and a tryptophan residue has been suggested as an unusual terminating proton acceptor in this case (51), in part because of the absence of a suitable His-residue that has been implicated as the general base in other terpenoid synthases (69, 72). Active-site chemical modification studies implicate essential Cys, His, and Arg residues in monoterpene synthases, with differential placement (or roles) in angiosperm and gymnosperm synthases (22, 72, 73).
Although the evolutionary relationship of plant and microbial terpene synthases and prenyltransferases is unclear, all of these enzymes contain the aspartate-rich DDxxD motif involved in coordination of divalent metal ions for substrate binding (69, 70). The DDxxD motif is absent in (−)-CDP synthase (43, 44, 58), which initiates ionization of the substrate by protonation rather than by diphosphate ester scission (Fig. 4). (−)-CDP synthase, however, contains another aspartate-rich motif, DxDD, which also is conserved in the mechanistically related bacterial squalene-hopene cyclase (42). Only two enzymes contain both DxDD and DDxxD elements, the bifunctional abietadiene synthase (Fig. 6) (50) and the bifunctional kaurene synthase of fungal origin (59), both of which catalyze cyclization reactions by protonation-initiated and diphosphate ester cleavage steps (Fig. 4).
Gene Expression and Regulation
Terpenoid secondary metabolites are recognized as signal molecules in many interactions of plants with other organisms, including competitors, beneficial insects, herbivores, and microbial pathogens (76–78). Terpenoid phytoalexins of the sesquiterpene and diterpene series serve as induced defenses against fungal pathogens (79). Volatile terpenes serve as attractants for pollinators (80) and mediate tritrophic interactions (81). Plant terpenoids can influence insect communication as pheromones (36) or pheromone precursors (82) and can interfere with insect development as hormone analogues (83, 84). Terpenoid-mediated chemical communication between plants and insects reflects eons of coevolution leading, in part, to both the diversity and specificity of terpene synthase catalysts. Plants also coordinately evolved mechanisms to control gene expression of terpene synthases, for example, in the elicitor-inducible transcriptional activation of synthase genes involved in phytoalexin production (32–34, 85). For epi-aristolochene synthase, an elicitor-dependent promoter and several responsive cis-elements were recently identified (86). Tissue-specific and developmentally controlled gene expression of linalool synthase in flower organs of C. breweri were interpreted as early steps in floral scent evolution for attraction of pollinators (56).
Grand fir has been developed as a model system to study conifer defense responses to bark beetles and their associated fungal pathogens, a process that involves induced production of monoterpenes, sesquiterpenes, and diterpenes (57, 87). Monoterpenes are toxic to invaders and furnish the volatile solvent for the diterpene resin acids, which upon monoterpene evaporation harden to a mechanical barrier that seals the wound site (88). Inducible resin sesquiterpenoids could function as phytoalexins, and derivatives of bisabolene may serve as hormone analogues that interfere with insect reproduction and development (83, 84). Genes for all three classes of resin terpene synthases have been identified (20, 31, 50), and they are transcriptionally activated by stem wounding (20, 87). Monoterpene synthase mRNA accumulation occurs within 2 hr of wounding and reaches maximum by 2–4 days (87). Diterpene synthase gene expression is similarly induced, but the time course is slightly delayed (87). By contrast, mRNA levels of the inducible (E)-α-bisabolene synthase, thought to be involved in production of sesquiterpenoid juvenile hormone analogues, increases to a maximum about 12 days after wounding. These differences in the timing of induced gene expression are consistent with the different roles of terpenoid defense compounds. Thus, toxic monoterpenes are required immediately after beetle attack, diterpene resin acids are mobilized somewhat later, whereas sesquiterpenoid juvenile hormone analogues are produced still later, presumably to interfere with insect reproduction should infestation of the host succeed. Expression of both single-product and multiproduct terpenoid synthases in grand fir has consequences for the regulation of resin composition that underlie both chemical adaptation of individual trees as well as chemical diversity within a population (89). δ-Selinene synthase and γ-humulene synthase allow the species to produce a highly complex mixture of terpenoid secondary metabolites by means of expression of a few genes. On the other hand, variations in the expression pattern of a terpenoid synthase multigene family provide for physiological adaptations in resin composition.
Perspective
Recent progress in the molecular biology of terpenoid synthases has opened new avenues of research by providing immunological tools and nucleic acid probes for examining the organization and both developmental and inducible regulation of terpenoid metabolism. The first crystal structures of two sesquiterpenoid synthases recently have been reported (51, 69). These advances certainly will be followed by structural investigation of other terpenoid synthase types from which comparative study will reveal finer details of reaction mechanism and the determinants of substrate specificity and product outcome, and permit the rational design of these catalysts. Although highly conserved structural elements likely are involved in general cyclization reaction chemistry (ionization, charge stabilization, deprotonation), it is the differences in active-site size and shape that enforce conformation on substrates and intermediates to direct the selectivity of this group of fascinating catalysts. Comparative investigations of closely related synthases should target those structural features that determine the basic modes of cyclization and that underlie regiochemical and stereochemical diversity so characteristic of this enzyme family. Comparison of highly specific synthases with those that produce an assortment of products can identify structural determinants of fidelity, or the lack thereof.
Biotechnological applications made possible by recent molecular advances include the engineering of terpenoid-based defenses in crop plants and the alteration of these pathways in foodstuffs to impart desirable flavor properties. Modifying the aroma profiles of ornamental species offers another useful possibility, as does the transgenic improvement of slow biosynthetic steps to increase the production yields of essential oils, phytopharmaceuticals, insecticides, and a wide range of industrial intermediates that are economically inaccessible by traditional chemical synthesis. The genetic engineering of terpenoid-based plant defenses is particularly appealing, given the storehouse of possibilities and the high probability that these substances are active against pests not adapted to them. In addition, the many ecological concepts that posit terpenoids as mediators of plant–pathogen and plant–insect interactions can now be experimentally evaluated by recombinant approaches, which may in turn lead to new strategies for crop protection.
Acknowledgments
J.B. is a Feodor Lynen Fellow of the Alexander-von-Humboldt-Foundation.
ABBREVIATIONS
- CDP
copalyl diphosphate
- GDP
geranyl diphosphate
- GGDP
geranylgeranyl diphosphate
- FDP
farnesyl diphosphate
- LDP
linalyl diphosphate
References
- 1.Ogura K. In: Comprehensive Natural Products Chemistry: Isoprenoid Biosynthesis. Cane D E, editor. Oxford: Pergamon; 1998. , in press. [Google Scholar]
- 2.Wise M L, Croteau R. In: Comprehensive Natural Products Chemistry: Isoprenoid Biosynthesis. Cane D E, editor. Oxford: Pergamon; 1998. , in press. [Google Scholar]
- 3.Cane D E. In: Comprehensive Natural Products Chemistry: Isoprenoid Biosynthesis. Cane D E, editor. Oxford: Pergamon; 1998. , in press. [Google Scholar]
- 4.MacMillan J. In: Comprehensive Natural Products Chemistry: Isoprenoid Biosynthesis. Cane D E, editor. Oxford: Pergamon; 1998. , in press. [Google Scholar]
- 5.Connolly J D, Hill R A. Dictionary of Terpenoids. London: Chapman & Hall; 1991. [Google Scholar]
- 6.Gershenzon J, Croteau R. In: Lipid Metabolism in Plants. Moore T S Jr, editor. Boca Raton, FL: CRC Press; 1993. pp. 339–388. [Google Scholar]
- 7.Alonso W R, Croteau R. Methods Plant Biochem. 1993;9:239–260. [Google Scholar]
- 8.Poulter C D, Rilling H C. Acc Chem Res. 1978;11:307–313. [Google Scholar]
- 9.Poulter C D, Rilling H C. In: Biosynthesis of Isoprenoid Compounds. Porter J W, Spurgeon S L, editors. Vol. 1. New York: Wiley; 1981. pp. 162–224. [Google Scholar]
- 10.Dewick P M. Nat Prod Rep. 1995;12:507–534. doi: 10.1039/np9951200507. [DOI] [PubMed] [Google Scholar]
- 11.Dewick P M. Nat Prod Rep. 1997;14:111–144. doi: 10.1039/np9971400111. [DOI] [PubMed] [Google Scholar]
- 12.Croteau R. Chem Rev. 1987;87:929–954. [Google Scholar]
- 13.Croteau R. In: Models in Plant Physiology and Biochemistry. Newman D W, Wilson K G, editors. Boca Raton, FL: CRC Press; 1987. pp. 55–63. [Google Scholar]
- 14.Rajaonarivony J I M, Gershenzon J, Croteau R. Arch Biochem Biophys. 1992;296:49–57. doi: 10.1016/0003-9861(92)90543-6. [DOI] [PubMed] [Google Scholar]
- 15.Gambliel H, Croteau R. J Biol Chem. 1984;259:740–748. [PubMed] [Google Scholar]
- 16.Croteau R, Shaskus J J, Renstrøm B, Felton N M, Cane D E, Saito A, Chang C. Biochemistry. 1985;24:7077–7085. doi: 10.1021/bi00346a009. [DOI] [PubMed] [Google Scholar]
- 17.LaFever R E, Croteau R. Arch Biochem Biophys. 1993;301:361–366. doi: 10.1006/abbi.1993.1156. [DOI] [PubMed] [Google Scholar]
- 18.Croteau R. In: Recent Developments in Flavor and Fragrance Chemistry. Hopp R, Mori K, editors. Weinheim: VCH; 1992. pp. 263–273. [Google Scholar]
- 19.Croteau R, Alonso W R, Koepp A E, Johnson M A. Arch Biochem Biophys. 1994;309:184–192. doi: 10.1006/abbi.1994.1101. [DOI] [PubMed] [Google Scholar]
- 20.Bohlmann J, Steele C L, Croteau R. J Biol Chem. 1997;272:21784–21792. doi: 10.1074/jbc.272.35.21784. [DOI] [PubMed] [Google Scholar]
- 21.Pichersky E, Lewinsohn E, Croteau R. Arch Biochem Biophys. 1995;316:803–807. doi: 10.1006/abbi.1995.1107. [DOI] [PubMed] [Google Scholar]
- 22.Savage T J, Hatch M W, Croteau R. J Biol Chem. 1994;269:4012–4020. [PubMed] [Google Scholar]
- 23.Adam K-P, Crock J E, Croteau R. Arch Biochem Biophys. 1996;332:352–356. doi: 10.1006/abbi.1996.0352. [DOI] [PubMed] [Google Scholar]
- 24.Colby S M, Alonso W R, Katahira E, McGarvey D J, Croteau R. J Biol Chem. 1993;268:23016–23024. [PubMed] [Google Scholar]
- 25.Wagschal K, Savage T J, Croteau R. Tetrahedron. 1991;47:5933–5944. [Google Scholar]
- 26.Croteau R, Wheeler C J, Cane D E, Ebert R, Ha H J. Biochemistry. 1987;26:5383–5389. doi: 10.1021/bi00391a025. [DOI] [PubMed] [Google Scholar]
- 27.Lewinsohn E, Gijzen M, Croteau R. Arch Biochem Biophys. 1992;293:167–173. doi: 10.1016/0003-9861(92)90380-f. [DOI] [PubMed] [Google Scholar]
- 28.Wagschal K C, Pyun H J, Coates R M, Croteau R. Arch Biochem Biophys. 1994;308:477–487. doi: 10.1006/abbi.1994.1068. [DOI] [PubMed] [Google Scholar]
- 29.Cane D E. Chem Rev. 1990;90:1089–1103. [Google Scholar]
- 30.Colby S M, Crock J, Dowdle-Rizzo B, Lemaux P G, Croteau R. Proc Natl Acad Sci USA. 1998;95:2216–2221. doi: 10.1073/pnas.95.5.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steele C L, Crock J, Bohlmann J, Croteau R. J Biol Chem. 1998;273:2078–2089. doi: 10.1074/jbc.273.4.2078. [DOI] [PubMed] [Google Scholar]
- 32.Chen X-Y, Chen Y, Heinstein P, Davisson V J. Arch Biochem Biophys. 1995;324:255–266. doi: 10.1006/abbi.1995.0038. [DOI] [PubMed] [Google Scholar]
- 33.Facchini P J, Chappell J. Proc Natl Acad Sci USA. 1992;89:11088–11092. doi: 10.1073/pnas.89.22.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Back K, Chappell J. J Biol Chem. 1995;270:7375–7381. doi: 10.1074/jbc.270.13.7375. [DOI] [PubMed] [Google Scholar]
- 35.Salin F, Pauly G, Charon J, Gleizes M. J Plant Physiol. 1995;146:203–209. [Google Scholar]
- 36.Crock J, Wildung M R, Croteau R. Proc Natl Acad Sci USA. 1997;94:12833–12838. doi: 10.1073/pnas.94.24.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West C A. In: Biosynthesis of Isoprenoid Compounds. Porter J W, Spurgeon S L, editors. Vol. 1. New York: Wiley; 1981. pp. 376–411. [Google Scholar]
- 38.Dueber M T, Adolf W, West C A. Plant Physiol. 1978;62:598–603. doi: 10.1104/pp.62.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Z, Wagner G J. Plant Sci. 1995;110:1–10. [Google Scholar]
- 40.Koepp A E, Hezari M, Zajicek J, Stofer Vogel B, LaFever R E, Lewis N G, Croteau R. J Biol Chem. 1995;270:8686–8690. doi: 10.1074/jbc.270.15.8686. [DOI] [PubMed] [Google Scholar]
- 41.Lin X, Hezari M, Koepp A E, Floss H G, Croteau R. Biochemistry. 1996;35:2968–2977. doi: 10.1021/bi9526239. [DOI] [PubMed] [Google Scholar]
- 42.Wendt K U, Poralla K, Schulz G E. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 43.Sun T P, Kamiya Y. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ait-Ali T, Swain S M, Reid J B, Sun T P, Kamiya Y. Plant J. 1997;11:443–454. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi S, Saito T, Abe H, Yamane H, Murofushi N, Kamiya Y. Plant J. 1996;10:203–213. doi: 10.1046/j.1365-313x.1996.10020203.x. [DOI] [PubMed] [Google Scholar]
- 46.Duncan J D, West C A. Plant Physiol. 1981;68:1128–1134. doi: 10.1104/pp.68.5.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Railton I D, Fellows B, West C A. Phytochemistry. 1984;23:1261–1267. [Google Scholar]
- 48.Funk C, Croteau R. Arch Biochem Biophys. 1994;308:258–266. doi: 10.1006/abbi.1994.1036. [DOI] [PubMed] [Google Scholar]
- 49.LaFever R, Stofer Vogel B, Croteau R. Arch Biochem Biophys. 1994;313:139–149. doi: 10.1006/abbi.1994.1370. [DOI] [PubMed] [Google Scholar]
- 50.Stofer Vogel B, Wildung M R, Vogel G, Croteau R. J Biol Chem. 1996;271:23262–23268. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- 51.Starks C M, Back K, Chappell J, Noel J P. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 52.Wildung M R, Croteau R. J Biol Chem. 1996;271:9201–9204. doi: 10.1074/jbc.271.16.9201. [DOI] [PubMed] [Google Scholar]
- 53.Mau C J D, West C A. Proc Natl Acad Sci USA. 1994;91:8497–8501. doi: 10.1073/pnas.91.18.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuba A, Yazaki K, Tabata M, Honda G, Croteau R. Arch Biochem Biophys. 1996;332:280–287. doi: 10.1006/abbi.1996.0343. [DOI] [PubMed] [Google Scholar]
- 55.Chen X-Y, Wang M, Chen Y, Davisson J, Heinstein P. J Nat Prod. 1996;59:944–951. doi: 10.1021/np960344w. [DOI] [PubMed] [Google Scholar]
- 56.Dudareva N, Cseke L, Blanc V M, Pichersky E. Plant Cell. 1996;8:1137–1148. doi: 10.1105/tpc.8.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steele C L, Lewinsohn E, Croteau R. Proc Natl Acad Sci USA. 1995;92:4164–4168. doi: 10.1073/pnas.92.10.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bensen R J, Johal G S, Crane V C, Tossberg J T, Schnable P S, Meeley R B, Briggs S P. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawaide H, Imai R, Sassa T, Kamiya Y. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- 60.Sun T P, Goodman H M, Ausubel F M. Plant Cell. 1992;4:119–128. doi: 10.1105/tpc.4.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aubourg S, Takvorian A, Cheron A, Kreis M, Lecharny A. Gene. 1997;199:241–253. doi: 10.1016/s0378-1119(97)00374-0. [DOI] [PubMed] [Google Scholar]
- 62.Back K, Chappell J. Proc Natl Acad Sci USA. 1996;93:6841–6845. doi: 10.1073/pnas.93.13.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dayhoff M O. Atlas of Protein Sequence and Structure. 5/3. Washington, DC: Natl. Biomed. Res. Found.; 1987. pp. 351–352. [Google Scholar]
- 64.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–420. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 65.Hill A M, Cane D E, Mau C J D, West C A. Arch Biochem Biophys. 1996;336:383–289. doi: 10.1006/abbi.1996.0559. [DOI] [PubMed] [Google Scholar]
- 66.Lewinsohn E, Savage T J, Gijzen M, Croteau R. Phytochem Anal. 1993;4:220–225. [Google Scholar]
- 67.Chaw S-M, Zharkikh A, Sung H-M, Lau T-C, Li W-H. Mol Biol Evol. 1997;14:56–68. doi: 10.1093/oxfordjournals.molbev.a025702. [DOI] [PubMed] [Google Scholar]
- 68.Hartmann T. Entomol Ex Appl. 1996;80:177–188. [Google Scholar]
- 69.Lesburg C A, Zhai G, Cane D E, Christianson D W. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 70.Tarshis L C, Yan M, Poulter C D, Sacchettini J C. Biochemistry. 1994;33:10871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 71.Cane D E, Sohng J, Lamberson C R, Rudnicki S M, Wu Z, Lloyd M D, Oliver J S, Hubbard B R. Biochemistry. 1994;33:5846–5857. doi: 10.1021/bi00185a024. [DOI] [PubMed] [Google Scholar]
- 72.Rajaonarivony J I M, Gershenzon J, Miyazaki J, Croteau R. Arch Biochem Biophys. 1992;299:77–82. doi: 10.1016/0003-9861(92)90246-s. [DOI] [PubMed] [Google Scholar]
- 73.Savage T J, Ichii H, Hume S D, Little D B, Croteau R. Arch Biochem Biophys. 1995;320:257–265. doi: 10.1016/0003-9861(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 74.Cane D E, Shim J H, Xue Q, Fitzsimons B C, Hohn T M. Biochemistry. 1995;34:2480–2488. doi: 10.1021/bi00008a011. [DOI] [PubMed] [Google Scholar]
- 75.Cane D E, Xue Q. J Am Chem Soc. 1996;118:1563–1564. [Google Scholar]
- 76.Harborne J B. In: Ecological Chemistry and Biochemistry of Plant Terpenoids. Harborne J B, Tomas-Barberan F A, editors. Oxford: Clarendon; 1991. pp. 399–426. [Google Scholar]
- 77.Gershenzon J, Croteau R. In: Herbivores: Their Interaction with Secondary Metabolites. Rosenthal G A, Berenbaum M, editors. New York: Academic; 1991. pp. 165–219. [Google Scholar]
- 78.Langenheim J H. J Chem Ecol. 1994;20:1223–1280. doi: 10.1007/BF02059809. [DOI] [PubMed] [Google Scholar]
- 79.Ku’c J. Annu Rev Phytopathol. 1995;33:275–297. doi: 10.1146/annurev.py.33.090195.001423. [DOI] [PubMed] [Google Scholar]
- 80.Dobson H E M. In: Insect–Plant Interactions. Bernays E, editor. Vol. 5. Boca Raton, FL: CRC Press; 1993. pp. 47–81. [Google Scholar]
- 81.Turlings T C J, Loughrin J H, McCall P J, Röse U S R, Lewis W J, Tumlinson J H. Proc Natl Acad Sci USA. 1995;92:4169–4174. doi: 10.1073/pnas.92.10.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pickett J A. In: Ecological Chemistry and Biochemistry of Plant Terpenoids. Harborne J B, Tomas-Barberan F A, editors. Oxford: Clarendon; 1991. pp. 297–313. [Google Scholar]
- 83.Bowers W S, Tomihisha O, Cleere J S, Marsella P A. Science. 1976;193:542–547. doi: 10.1126/science.986685. [DOI] [PubMed] [Google Scholar]
- 84.Bowers W S. In: Herbivores: Their Interaction with Secondary Plant Metabolites. Rosenthal G A, Berenbaum M R, editors. Vol. 1. San Diego: Academic; 1991. pp. 431–456. [Google Scholar]
- 85.Lois A F, West C A. Arch Biochem Biophys. 1990;276:270–277. doi: 10.1016/0003-9861(90)90038-z. [DOI] [PubMed] [Google Scholar]
- 86.Yin S, Leng M, Newman J, Back K, Chappell J. Plant Physiol. 1997;115:437–451. doi: 10.1104/pp.115.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Steele, C., Katoh, S., Bohlmann, J. & Croteau, R. (1998) Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- 88.Johnson M A, Croteau R. Am Chem Soc Symp Ser. 1987;325:67–91. [Google Scholar]
- 89.Katoh S, Croteau R. Phytochemistry. 1998;47:577–582. [Google Scholar]