Abstract
The 3.0-Å structure of a 190-residue fragment of intercellular adhesion molecule-1 (ICAM-1, CD54) reveals two tandem Ig-superfamily (IgSF) domains. Each of two independent molecules dimerizes identically with a symmetry-related molecule over a hydrophobic interface on the BED sheet of domain 1, in agreement with dimerization of ICAM-1 on the cell surface. The residues that bind to the integrin LFA-1 are well oriented for bivalent binding in the dimer, with the critical Glu-34 residues pointing away from each other on the periphery. Residues that bind to rhinovirus are in the flexible BC and FG loops at the tip of domain 1, and these and the upper half of domain 1 are well exposed in the dimer for docking to virus. By contrast, a residue important for binding to Plasmodium falciparum-infected erythrocytes is in the dimer interface. The presence of A′ strands in both domains 1 and 2, conserved hydrogen bonds at domain junctions, and elaborate hydrogen bond networks around the key integrin binding residues in domain 1 make these domains suited to resist tensile forces during adhesive interactions. A subdivision of the intermediate (I) set of IgSF domains is proposed in which domain 1 of ICAM-1 and previously described I set domains belong to the I1 set and domain 2 of ICAM-1, ICAM-2, and vascular cell adhesion molecule-1 belong to the I2 set.
Intercellular adhesion molecule-1 (ICAM-1, CD54) is a membrane protein with five Ig-like domains, a hydrophobic transmembrane domain, and a short cytoplasmic domain. ICAM-1 functions as a ligand for the leukocyte integrins LFA-1 (CD11a/CD18) and Mac-1 (CD11b/CD18) (1, 2). ICAM-1 is expressed on many cells important in immune and inflammatory responses, and is inducible on others. Increased ICAM-1 expression augments immune responses and leukocyte accumulation in inflamed tissues. In addition, ICAM-1 is subverted as the cell surface receptor for the major group of human rhinoviruses (HRV) and for sequestration of Plasmodium falciparum-infected red blood cells (IRBCs) in the peripheral vasculature. LFA-1, HRV, and IRBCs bind to the N-terminal Ig-like domain of ICAM-1 (3–7). Recombinant soluble forms of ICAM-1 have been shown to inhibit rhinovirus infection (8). Rhinoviruses are small, RNA-containing viruses with a protein capsid. ICAM-1 not only acts as a receptor, but also can induce irreversible disruption of the capsid, with release of the viral RNA (9–11). The kinetics of ICAM-1 binding measured in BIAcore under reversible conditions reveal two distinct, slow association rate constants and a single dissociation rate constant (12). Structural and mutational studies of rhinovirus and cryoelectron microscopy of a complex with a two-domain fragment of ICAM-1 suggest binding in or near a “canyon” or depression in the capsid that surrounds the virus 5-fold axis (13, 14).
At least four closely related Ig-superfamily (IgSF) members, ICAM-1, -2, -3, and -4, have been found to bind to the integrin LFA-1 (αLβ2) (2, 15). The more distant relatives, mucosal addressin cell adhesion molecule-1 (MAdCAM-1) and vascular cell adhesion molecule-1 (VCAM-1), bind to the α4 integrins, α4β7 and α4β1 (2). The crystal structures of ICAM-2 (16) and VCAM-1 (17, 18) reveal that domain 1 belongs to the I set of the IgSF, and domain 2 has been classified in the C2 set. In VCAM-1 the aspartic acid critical for integrin recognition lies in a protruding loop. That loop is absent in ICAM-2, and the functionally equivalent glutamic acid is in a β-strand surrounded by a flat recognition surface. The homologous Glu-34 in ICAM-1 is the most important residue for binding to LFA-1 (3). Binding of ICAM-1 and ICAM-2 to LFA-1 is mediated by an inserted or I-domain present in the α-chain of the integrin, which is structurally defined (19, 20). A Mg2+ in the I-domain is proposed to coordinate with the critical Glu in ICAMs and is surrounded by residues that provide ligand specificity (19, 21). By contrast, VCAM-1 and MAdCAM-1 bind to α4 integrins that lack an I-domain but like all integrins are predicted to contain a β-propeller domain in the α chain that is important in ligand binding (22).
ICAM-1 is more active than other ICAMs in binding to LFA-1. When present on the same cell in comparable amounts, ICAM-1 supports the preponderance of LFA-1-dependent binding, with lesser contributions from ICAM-2 and ICAM-3 (23). ICAM-1 exists predominantly as a dimer on the cell surface, whereas ICAM-2 does not (24, 25). Soluble dimeric ICAM-1 binds to LFA-1 with much greater affinity than monomeric ICAM-1. Dimerization also increases ICAM-1 affinity for rhinovirus (11, 12).
Here we present the x-ray crystal structure at 3.0 Å resolution of domains 1 and 2 of ICAM-1 with four N-linked glycans. The structure helps define a new set of IgSF domains and has implications for the dimerization of ICAM-1 and ligand recognition.
METHODS
A cDNA encoding the N-terminal 190-aa residues of ICAM-1 (IC1-P191*) was subcloned into the PBJ5-GS vector. CHO Lec3.2.8.1 cell transfectants were selected and ICAM-1 was purified as described (12). Crystals were grown at 4°C by using the hanging-drop vapor diffusion method. Each drop (2 μl) contained a 1:1 ratio of protein (16 mg/ml in 10 mM Tris, pH 8.0) and the crystallization solution (17% PEG 4K, 10 mM Tris⋅HCl, pH 8.0, and 100 mM β-octyl-glucopyranoside). Crystals were harvested with 25% PEG 4K in Tris buffer and dialyzed at 4°C overnight against 28% PEG 4K/15% ethylene glycol (EG) in Tris buffer and then against 32.5% PEG 4K/30% EG in Tris buffer for 24 hr. Alternatively, crystals were obtained with the same method except 10 mM Na cacodylate, pH 6.5 was substituted for Tris buffer. Crystals belong to the monoclinic P2 space group with two molecules per asymmetric unit and about 60% solvent. The unit cell is a = 88.0 Å, b = 42.2 Å, c = 93.0 Å, β = 109.3 Å.
Each data set was collected from a single flash-frozen crystal by using a MAR-Research 180-mm area detector (MAR Research, Hamburg, Germany) equipped with a Rigaku RU200 rotating anode x-ray generator (Rigaku, Tokyo). Data were reduced and scaled by using the HKL package (HKL Research Inc., Charlottesville, VA). A 95% complete native data set with an average Rmerge of 10% for resolution ranging from 20 to 2.9 Å was collected for crystals grown at pH 6.5. The Rmerge for data from 3.25 to 3.0 Å was 28%, and therefore data beyond 3 Å were not used for structure determination.
Data collected from a crystal soaked for 30 min in 0.5 mM uranyl nitrate was isomorphous (Rder = 16.8) to native data from 15 to 3.8 Å, and the corresponding difference Patterson map was readily interpreted. Correct orientations and positions for the four Ig-like domains per asymmetric unit were determined by molecular replacement (26) using the program amore (27) and search models of ICAM-2 domain 1 (16) for domain 1 and a composite of ICAM-1 and VCAM-1 domains 2 (18) for domain 2. Models contained the buried and conserved residues of the β-framework, and nonconserved residues were truncated at Cβ. The first and second domain of each molecule were subjected to rigid body refinement with x-plor (4–8 Å) and used to calculate initial molecular replacement phases. These initial phases were used to locate the heavy atom positions in difference Fourier maps as a cross-check. Heavy atom positions were refined with heavy and phases were calculated with mlphare. These phases were combined with molecular replacement phases using sigmaa (CCP4 suite, Science and Engineering Research Council collaborative computer project 4; Daresbury Laboratory, Daresbury, U.K.). Finally, density modification (DM, CCP4) with noncrystallographic 2-fold averaging, solvent flattening, and histogram matching was applied to the combined phases for improvement and generation of the electron density map. An initial ICAM-1 model was built into a 3.5-Å DM map. The model was subjected to positional and B factor refinement with x-plor (28), and an improved map was obtained. About 90% of the protein residues were traced and built using this map, and a considerable R factor improvement was obtained after manual rebuilding and refinement using data from 15 to 3.3 Å. Several cycles of manual rebuilding and refinement with x-plor using solvent-corrected data gave a final model with free and working R factors of 27.9 and 22.2%, respectively, for the resolution range of 15 to 3.0 Å. About 10% of 11,300 unique reflections with F > 2σ were used to determine R free. The rms deviations from ideality for bond length and angles were 0.018 Å and 2.8°, respectively.
The main chain is well defined for molecule A in the current electron density map, with the exception of the FG and BC loops in domain 1. The electron density of molecule B was more difficult to interpret compared with A. The final average B factors are 28 Å2 for A and 69 Å2 for B. Some residues in the BC and DE loops in domain 1 and the top of domain 2 are not well defined in molecule B. The three C-terminal residues are not well defined in either molecule. The first N-acetyl glucosamine of each N-linked glycan is clearly seen in the map, except the site at Asn-118.
RESULTS
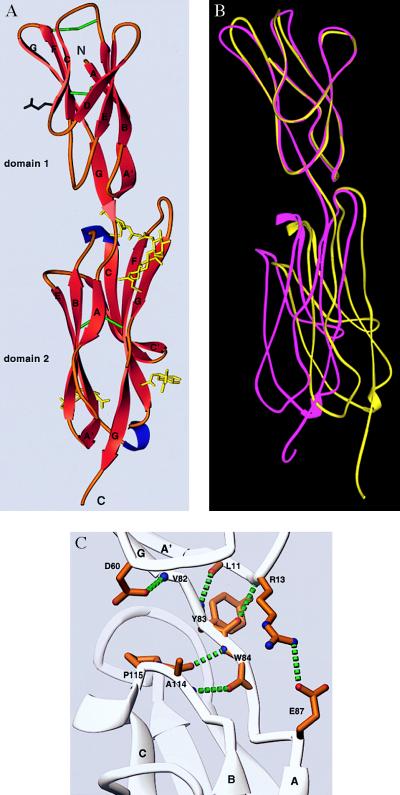
We expressed fragments of ICAM-1 with high mannose carbohydrates at all four N-linked sites and terminated after amino acid residue 185 or 190. The 185-residue fragment bound and neutralized rhinovirus markedly less well than the five-domain fragment, as previously reported (9). By contrast, the 190-residue fragment bound to rhinovirus and a panel of mAb with kinetics identical to the five-domain fragment as measured with BIAcore (29). The 190-residue fragment readily crystallized, with two molecules per asymmetric unit, and revealed two tandem IgSF domains (Fig. 1A).
Figure 1.
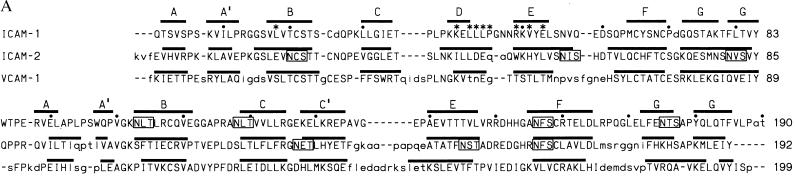
The crystal structure of the N-terminal two domains of ICAM-1. (A) Ribbon diagram (39) with β-strands in red, α-helix in blue, and coil in orange. The last residue of domain 1 (Tyr-83) is rose. N-linked sugars (yellow), Glu-37 in strand C of domain 1 (black), and disulfide bonds (green) are included. The last two residues of the structure are omitted. (B) Movement between domains 1 and 2 and comparison of domain 1 of molecules A and B. The Cα backbones of ICAM-1 molecules A (magenta) and B (yellow) are shown after superposition on domain 1. (C) Hydrogen bonds at the boundary between domains 1 and 2.
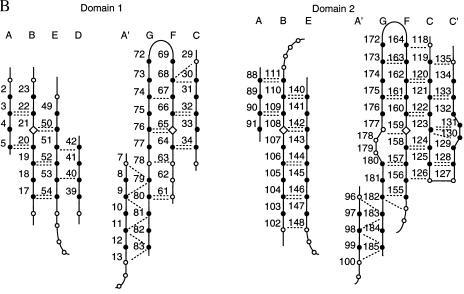
Domain 1 belongs to the I set of IgSF domains (30). This set is intermediate between the V and C sets, and is usually defined as a β-sandwich domain with one sheet containing ABED strands and another containing A′GFCC′ strands. Domain 1 of ICAM-1, ICAM-2 (16), VCAM-1 (17, 18), and MAdCAM-1 (K. Tan, J.M.C., J.-H.L., M. J. Briskin, T.A.S., and J.-H.W., unpublished data) are classified as I set domains although they lack C′ strands. A kink in place of a C′ strand is present in VCAM-1 and MAdCAM-1 and contains the Asp residue critical for recognition of α4 integrins. This kink is absent in ICAM-1 and ICAM-2. The Glu-34 residue in ICAM-1 that is critical for binding to LFA-1 is the last residue of β-strand C in domain 1 (Figs. 1A and 2). Two β-sheet hydrogen bonds link Glu-34 to Met-64 in strand F (Fig. 2B). The side chains of Thr-35 and Lys-39 hydrogen-bond to the backbone amide of Leu-37 and carbonyl of Ile-33, respectively. This hydrogen-bond network surrounding Glu-34 is conserved in ICAM-2 and maintains a rigid orientation for the integrin-binding site. The greatest structural difference with domain 1 of ICAM-2 is in the DE loop, which widens the top of domain 1 in ICAM-1 (Fig. 2A).
Figure 2.
Domain structure. (A) Structural alignment of ICAM-1 with ICAM-2 and VCAM-1. β-strands defined by DSSP (31) for ICAM-1 molecules A or B, for ICAM-2 (16), and the longer form of VCAM-1 (17) are overlined. The α carbons of molecules A and B of ICAM-1, ICAM-2, and molecule A of VCAM-1 were structurally aligned with 3dmalign of modeller (http://guitar.rockefeller.edu/modeller/modeller.html) with a gap penalty of 1.75 Å, separately for domain 1 and 2. Residues are in lowercase in ICAM-1 if they are not within 3.5 Å in molecules A and B, and in ICAM-2 and VCAM-1 if they are not within 1.75 Å of the framework (the average position of capitalized residues including both molecules A and B of ICAM-1). ICAM-1 residues in the dimer interface in domain 1 are asterisked, and a dot marks every 10th residue. N-linked glycosylation sites are boxed. (B) β-sheet frameworks. β-sheet framework residues in ICAM-1 are solid circles if defined as β-strand in molecules A or B by DSSP (31). Open circles represent conserved loop structures. Backbone hydrogen bonds are dashed lines and are shown if ≥1.0 kcal/mol with DSSP or found with look (Molecular Applications Group, Palo Alto, CA).
The loops in the tip of domain 1 in ICAM-1 are flexible as shown by the high B factors. The FG loop and BC loops appear flexible only one residue away from the “extra” cysteines that covalently link these loops. Conformational differences between molecules A and B (Fig. 1B) are demonstrated by residues 72 and 73, which are well defined in the electron density.
The last residue of domain 1, Tyr-83 in the G strand, is only four residues away from the beginning of the regular β-strand A in domain 2 (Figs. 1 A and C and 2A). The first residue of domain 2, Trp-84, has two main-chain hydrogen bonds to Ala-114 in the BC loop of domain 2 (Fig. 1C). A cis proline follows Ala-114. The hydrogen bonds and the conformation of the BC loop with its cis-proline are conserved in domain 2 of molecules A and B as well as in ICAM-2, VCAM-1, and MAdCAM-1. The angle between the planes of the β-sheets in the two domains is about 170° in both molecules of ICAM-1. The interdomain orientation difference between the two molecules is about 17° (Fig. 1B). The interface between domains 1 and 2 buries only 350 Å2 per domain (31) and is largely hydrophobic, consistent with movement between domains in ICAM-1. By contrast, a larger number of hydrogen bonds that link domains 1 and 2 in ICAM-2 may limit movement more (16). The FG loop of domain 2 of ICAM-1 extends the furthest toward domain 1 and makes the most contacts with domain 1 (Fig. 1A). The C′E loop in domain 2 of ICAM-1 is markedly shorter than that in ICAM-2 or VCAM-1 (Fig. 2A).
Domain 2 of ICAM-1 is structurally similar to domain 2 of ICAM-2 and VCAM-1, and these domains clearly belong in the same IgSF subset. Previously recognized IgSF fold subsets are the V, C1, C2, and I sets (30, 32). These differ from one another with respect to edge strands on each beta sheet, in how far strands extend toward the “top” of the domain relative to the cysteines in the B and F strands, and with respect to certain framework residues. The second domains of VCAM-1 and ICAM-2 have been placed in the C2 set, because they lack a D strand. However, their β-sheets more resemble the I set in other respects, including extension toward the “top” of the domain and the presence of an A′ strand (Fig. 2). All other C set domains lack A′ strands. The A′ strand in domain 2 of ICAM-1 forms four hydrogen bonds to the G strand (Fig. 2B). The A′G hydrogen-bond network is identical to that in domain 1, and the last residue in domain 2, Phe-185, is at the same position in this network as the last residue in domain 1, Tyr-83 (Fig. 2B). A′ strands are also present but not previously reported in domain 2 of ICAM-2 (16), and in VCAM-1 when the cognate residues in the G strand are intact (17) (Fig. 2A).
Domain 2 displays four N-linked glycans; either one or two N-acetylglucosamine residues are visualized at each site (Fig. 1A). The glycans attached to Asn-118 at the top of strand C and N175 in strand G are near the top of domain 2. The chitobiose moiety at the latter site points up and packs close to Tyr-83 and Phe-173 at the domain junction. These two N-linked glycans lie on either side of the FG loop and may limit interdomain flexion. The glycans N-linked to Asn-101 and Asn-156 lie near the bottom of domain 2 and lack protein side-chain contacts.
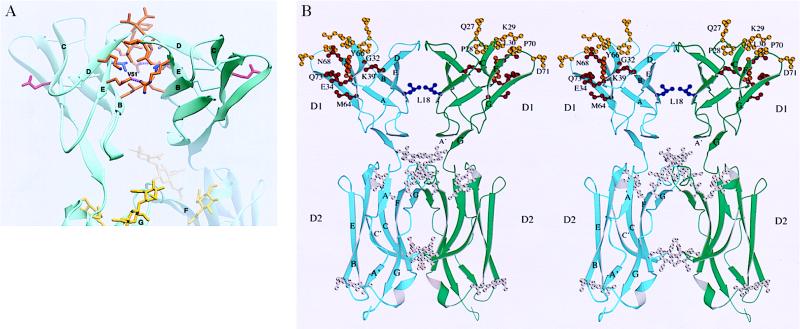
Two types of dimers are found in the crystal. The AB dimer has polar intermolecular interactions and appears to be a packing artifact. By contrast, the symmetry-related AA and BB dimers bury 550–580 Å2 of surface area per monomer (31) (Fig. 3). Of this, 430–460 Å2 come from contacts across the BED face of domain 1, which are identical in the AA and BB dimers. These contacts have 2-fold symmetry and involve hydrophobic residues, with Val-51 at the center (Figs. 2A and 3A). The aliphatic portions of Lys-40, Arg-49, and Glu-53 chains pack against this hydrophobic core, with their charged groups forming salt bridges at its periphery. Compared with nonspecific contacts, as exemplified by translation and screw-axis contacts, the contact area is unusually large (33). These observations suggest that dimerization at the BED interface may be relevant for formation of ICAM-1 dimers on the cell surface. It is striking that the ligand-binding surfaces for LFA-1 are on opposite sides of the dimer, with the Glu-34 side-chains 42 Å apart and pointing away from one another (Fig. 3). The centers of the rhinovirus-binding interfaces on the BC and FG loops of domain 1 are 50 Å away and are oriented more toward the top of the dimer (Fig. 3B).
Figure 3.
The dimer interface and ligand-binding residues. (A) Interacting residues in domain 1. Side chains are shown for residues that interact across the dimer interface in domain 1 (Fig. 2A). The conserved central Val-51 residue is blue, and Glu-34 is red. Salt bridges between residues at the periphery of the interface are dashed lines. (B) Stereoview (40) of the dimer. Side chains and α carbons are shown for residues important in binding to LFA-1 (red and orange) (3, 37), human rhinoviruses 3, 14, 15, 36, and 41 (yellow and orange) (3–5), and P. falciparum (blue) (6). Only single amino acid substitutions that reduced binding 50% or 2 SD below control are shown.
DISCUSSION
ICAM-1 is perhaps the most intensively studied adhesion molecule of the immune system, and the x-ray crystal structure of its two N-terminal IgSF domains has important implications for its function both in the immune response and as a receptor for pathogens. The interface between domains 1 and 2 is small and hydrophobic, allowing interdomain movement. Movement is limited by hydrogen bonding of the last residue of domain 1 to the A′ strand of domain 1, and of the first residue of domain 2 to the BC loop of domain 2. Flexion may also be limited by the long FG loop of domain 2 and by the two N-linked carbohydrates at the top of domain 2.
The proper folding of domain 1 of ICAM-1 depends on domain 2 (3). The two most disruptive amino acid substitutions for domain 1 occur at its bottom boundary. The side chain of Asp-60 accepts a hydrogen bond from the backbone of Val-82, the penultimate residue of the G strand (Fig 1C). This hydrogen bond is required, as shown by the deleterious effect of mutation of Asp-60 to Lys and Gln but not to Asn. Similarly, the requirement for the salt bridge between the side chains of Arg-13 and Glu-87 near the beginning of domain 2 (Fig. 1C) is demonstrated by the deleterious effect of mutating Arg-13 to Glu and Gln but not to Lys (3). It is remarkable that although Phe-185 is at the end of the G strand and appears to be the last residue of domain 2, truncations here diminish affinity for rhinovirus by almost 10-fold and decrease affinity for one mAb that binds to domain 1 (29). The presence of a free carboxyl group on Phe-185 may abolish its two hydrogen bonds to the A′ strand (Fig. 2B), as occurs in VCAM-1 when the homologous Tyr-196 residue is mutated to Asp and is C-terminal (18). The sensitivity of this region is further highlighted by the A-178 → G mutation in the bulge of the G strand in domain 2 of ICAM-1 (Fig. 2B), which reduces LFA-1 and mAb binding to domain 1 (3).
Domains 1 and 2 of ICAM-1, ICAM-2, and VCAM-1 resemble one another except by the presence of a D strand in domain 1 and a C′ strand in domain 2. The presence of an A′ strand in domain 2, shown here in ICAM-1 and present but previously unremarked in ICAM-2 and VCAM-1, is another similarity between domains 1 and 2, and distinguishes domain 2 from the C2 set of the IgSF (30, 32). The A′ strand is common to the V and I sets but is lacking in the C1 and C2 sets. Therefore, we propose a division of the I set, with currently acknowledged members possessing D strands belonging to the I1 set, and domain 2 of ICAM-1, ICAM-2, VCAM-1, and similar domains that contain A′ strands and lack D strands belonging to the I2 set. This division is analogous to that of the C set into the C1 and C2 sets, which also possess and lack D strands, respectively. The I1 and I2 sets are readily interconverted in evolution, as shown by domain 2 of MAdCAM-1 and VCAM-1, which are the closest relatives in the IgSF as shown by sequence, but belong to the I1 and I2 sets, respectively (K. Tan, et al., unpublished data).
Stress forces acting on IgSF and integrin molecules as they function in adhesion in the vasculature or to provide traction during cell migration will strain, i.e. lengthen, distort, and pull apart, the receptor-ligand binding interface and the domains that lie between this interface and anchorage points in the cell membrane and cytoskeleton. The integrin-binding site on ICAM-1 at Glu-34 and surrounding residues is reinforced with an elaborate backbone and side-chain-backbone hydrogen bond network that will diminish distortion of the binding interface by applied force. The I set structure seen in domains 1 and 2 may also be important in its ability to resist tensile forces. Within a domain, tensile forces will be distributed over multiple β-strands; however, at domain junctions, forces will be borne primarily by the G strand of the domain on the N-terminal side and the A strand of the domain on the C-terminal side. The presence of the A′ strand means that the G strand is not an edge strand, as it would be in a C2 domain, and thus should be less susceptible to being peeled from the β-sheet. In domain 2 on the C-terminal side of the domain junction, forces will be borne both by the A strand, which should be supported by the A′ strand, and by the hydrogen bonds of the first residue of domain 2 to the BC loop. These features of the domain 1 and 2 boundaries of IgSF integrin ligands are conserved and may function to kinetically resist the tendency of force to unfold and denature IgSF domains, while at the same time allowing, at least in the case of ICAM-1 and VCAM-1, interdomain flexibility. It is interesting that the IgSF domains of titin and twitchin, which are muscle proteins that bear tensile force, also belong to the I set (32).
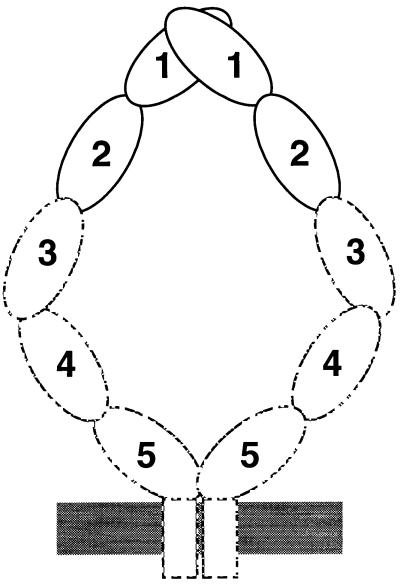
ICAM-1 is dimeric on the cell surface, suggesting that a dimerization interface on the BED face of domain 1 in the ICAM-1 crystal may be biologically important. The residue at the center of the contact is Val-51 in strand E, which is conserved as a Val or Ile in all known ICAM-1 molecules, but is hydrophilic in ICAM-2, which does not dimerize (24). The amount of buried interface in domain 1 of the ICAM-1 dimer of about 450 Å2 per monomer is similar to that in the CD4 domain 4 dimer interface (34) and less than that in CD2 dimers (650 Å2) (32) or antibody–antigen complexes (700 Å2) (33). In agreement with the observation that the two-domain fragment is monomeric at μM concentrations (25, 29, 35), the observed dimer interface is too small to be expected to drive dimerization on its own. The transmembrane domain of ICAM-1 contains a dimerization motif and contributes to dimer formation (24, 25). Electron micrographs show that ICAM-1 is bent (3), and antibody tags suggest that the bend occurs between domains 3 and 4 (36). In the crystal dimer, the C termini of domain 2 of each monomer are far apart; however, the bend could bring the more C-terminal domains closer together, so that the transmembrane domains could associate (Fig. 4). Binding of a mAb to domain 5 is hindered by dimerization, consistent with propinquity of domain 5 in the ICAM-1 dimer (25). In solution, ICAM-1 is monomeric unless the transmembrane domain or a flu peptide linked to domain 5 is present to help drive dimerization (1, 2). Dimerization of the N-terminal 190-residue fragment occurs after removal of all but one N-linked carbohydrate residue at each site, and the dimer antagonizes LFA-1 function (35). In our dimer structure the C termini of domain 2 are 80 Å apart, consistent with the observation that a flu peptide attached to a two-domain fragment is unable to assist dimerization (2).
Figure 4.
A model for the ICAM-1 dimer on the cell surface. Domains 1 and 2 and their orientation in the dimer are from the crystal structure. The rod-like shape of domains 1–5 in the monomer and the bend between domains 3 and 4 are from electron microscopy (3, 36). Dimerization or proximity between domain 5 is based on hindrance of antibody binding to this domain in the dimer (25), and association at the transmembrane domain is based on its role in dimerization (24, 25).
The putative dimerization interface on the BED face of domain 1 has important implications for interactions of ICAM-1 on the cell surface. The angle between domains of about 120° places each domain at an angle of about 60° with respect to a line perpendicular to the cell membrane. Constant region domains in antibodies also dimerize on the BED face, but with an angle between domains of about 60°. Domain 1 of CD2 dimerizes with an angle of about 110° between domains (32). This is similar to the angle between domains in ICAM-1 dimers; however, dimerization of CD2 is through the GFCC′C′′ sheet. CD2–CD2 interaction is thought to mimic interactions in trans between molecules on opposite cells, and thus would place domain 1 at an angle about 20° from perpendicular to the cell membrane. Dimerization in cis between ICAM-1 molecules on the same cell may have an important function in orienting ICAM-1 for interactions in trans with LFA-1. Furthermore, orientation may be important to prevent cis interactions between LFA-1 and ICAM-1, because LFA-1 and ICAM-1 are frequently expressed on the same cell (1).
The binding surface for LFA-1 on ICAM-1 (3, 37) is completely exposed in the dimer (Fig. 3B). Interacting residues are located primarily in the C and F strands, with contributions of one residue each from the G strand and D strand. The C′E loop of domain 2 is also exposed, but makes no contribution to binding to LFA-1 (3), in contrast to the contribution of the C′E loop in VCAM-1 to binding to α4 integrins (38). The 60° angle between domain 1 in the dimer exposes the interaction surface for binding by an LFA-1 molecule on another cell. The proposed orientation on the cell surface, with the transmembrane domain toward the bottom in Figs. 3 and 4, is similar to that proposed for the bent ICAM-2 molecule (16). The critical Glu-34 residues point in opposite directions, and the geometry appears ideal for simultaneous binding of two LFA-1 molecules to the dimer. The finding that soluble ICAM-1 dimers are far more avid for LFA-1 than monomers implies either bivalent binding to LFA-1 or that each monomer contributes to binding to a single LFA-1 molecule (24, 25). Dimerization of ICAM-1 on the cell surface thus far has been measured with a mAb to domain 5, but whether the putative interaction through domain 1 is affected is not known (25). Disruption of the domain 5 interaction results in 2-fold loss of cell–cell adhesion; whether dimerization per se or the orientation of the ligand-binding site on the cell surface is most important is currently unknown.
The only single amino acid substitution known to affect binding of P. falciparum-infected erythrocytes to ICAM-1 is Leu-18 to Gln, in the dimer interface. Whether binding involves dimeric or monomeric ICAM-1 is not known. This mutation had no effect on LFA-1 or rhinovirus14 binding (6).
The residues important for binding to rhinovirus have been mapped with five different rhinovirus serotypes (3–5). Six of these residues map to the BC and FG loops on the tip of domain 1, and one maps partway down the side of domain 1 in the F strand (Fig. 3B). The flexibility of the BC and FG loops to which rhinovirus binds is remarkable (Fig. 1B). The particular ICAM-1 residues that are important for binding vary depending on the rhinovirus serotype (5), and it is possible that variation in the ICAM-1-binding surface between serotypes could be accommodated by changes in loop conformation. It has been proposed that ICAM-1 docks in a depression in the capsid surface (13), and localization of interacting residues to the tip and side of domain 1 as well as cryoelectron microscopy of a two-domain fragment of ICAM-1 bound to rhinovirus are consistent with this (14). The bound fragment appears monomeric. A dimer could dock to the proposed binding site in the canyon through one of its monomers, because the upper half of domain 1 is unobscured in the dimer. However, the geometry of the dimer is inappropriate for bivalent docking to symmetry-related sites on the virus. ICAM-1 binding triggers rhinovirus disruption (9–11). It will be interesting to learn whether the flexible tip of ICAM-1 adapts to conformational changes in the virus capsid during uncoating and whether ICAM-1 dimerization plays any role in this process.
Acknowledgments
We thank Linda Chee for assistance with protein crystallization. We are especially thankful to Dr. Stephen C. Harrison for his support during many years of work on this project. We thank Dan Leahy and Cyrus Chothia for refereeing the manuscript. This work was supported by National Institutes of Health Grants AI31921 and HL48675.
ABBREVIATIONS
- ICAM-1
intercellular adhesion molecule-1
- IgSF
Ig-superfamily
- MAdCAM-1
mucosal addressin cell adhesion molecule-1
- VCAM-1
vascular cell adhesion molecule-1
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1ic1).
References
- 1.Springer T A. Nature (London) 1990;346:425–433. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- 2.Springer T A. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 3.Staunton D E, Dustin M L, Erickson H P, Springer T A. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 4.McClelland A, DeBear J, Yost S C, Meyer A M, Marlor C W, Greve J M. Proc Natl Acad Sci USA. 1991;88:7993–7997. doi: 10.1073/pnas.88.18.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Register R B, Uncapher C R, Naylor A M, Lineberger D W, Colonno R J. J Virol. 1991;65:6589–6596. doi: 10.1128/jvi.65.12.6589-6596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ockenhouse C F, Betageri R, Springer T A, Staunton D E. Cell. 1992;68:63–69. doi: 10.1016/0092-8674(92)90206-r. [DOI] [PubMed] [Google Scholar]
- 7.Berendt A R, McDowall A, Craig A G, Bates P A, Sternberg M J E, Marsh K, Newbold C I, Hogg K. Cell. 1992;68:71–81. doi: 10.1016/0092-8674(92)90207-s. [DOI] [PubMed] [Google Scholar]
- 8.Marlin S D, Staunton D E, Springer T A, Stratowa C, Sommergruber W, Merluzzi V. Nature (London) 1990;344:70–72. doi: 10.1038/344070a0. [DOI] [PubMed] [Google Scholar]
- 9.Greve J M, Forte C P, Marlor C W, Meyer A M, Hoover-Litty H, Wunderlich D, McClelland A. J Virol. 1991;65:6015–6023. doi: 10.1128/jvi.65.11.6015-6023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoover-Litty H, Greve J M. J Virol. 1993;67:390–397. doi: 10.1128/jvi.67.1.390-397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin S, Casasnovas J M, Staunton D E, Springer T A. J Virol. 1993;67:3561–3568. doi: 10.1128/jvi.67.6.3561-3568.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casasnovas J M, Springer T A. J Biol Chem. 1995;270:13216–13224. doi: 10.1074/jbc.270.22.13216. [DOI] [PubMed] [Google Scholar]
- 13.Rossmann M G. J Biol Chem. 1989;264:14587–14590. [PubMed] [Google Scholar]
- 14.Olson N H, Kolatkar P R, Oliveira M A, Cheng R H, Greve J M, McClelland A, Baker T S, Rossmann M G. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian L, Yoshihara Y, Mizuno T, Mori K, Gahmberg C G. J Immunol. 1997;158:928–936. [PubMed] [Google Scholar]
- 16.Casasnovas J M, Springer T A, Liu J-h, Harrison S C, Wang J-h. Nature (London) 1997;387:312–315. doi: 10.1038/387312a0. [DOI] [PubMed] [Google Scholar]
- 17.Jones E Y, Harlos K, Bottomley M J, Robinson R C, Driscoll P C, Edwards R M, Clements J M, Dudgeon T J, Stuart D I. Nature (London) 1995;373:539–544. doi: 10.1038/373539a0. [DOI] [PubMed] [Google Scholar]
- 18.Wang J-h, Pepinsky R B, Stehle T, Liu J-h, Karpusas M, Browning B, Osborn L. Proc Natl Acad Sci USA. 1995;92:5714–5718. doi: 10.1073/pnas.92.12.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J-O, Rieu P, Arnaout M A, Liddington R. Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 20.Qu A, Leahy D J. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C, Springer T A. J Biol Chem. 1995;270:19008–19016. doi: 10.1074/jbc.270.32.19008. [DOI] [PubMed] [Google Scholar]
- 22.Springer T A. Proc Natl Acad Sci USA. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Fougerolles A R, Springer T A. J Exp Med. 1992;175:185–190. doi: 10.1084/jem.175.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reilly P L, Woska J R, Jr, Jeanfavre D D, McNally E, Rothlein R, Bormann B-J. J Immunol. 1995;155:529–532. [PubMed] [Google Scholar]
- 25.Miller J, Knorr R, Ferrone M, Houdei R, Carron C P, Dustin M L. J Exp Med. 1995;182:1231–1241. doi: 10.1084/jem.182.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossmann M G, Blow D M. Acta Crystallogr. 1962;15:24–31. [Google Scholar]
- 27.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 28.Brunger A T. x-plor: A System for Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1990. [Google Scholar]
- 29.Casasnovas, J. M., Bickford, J. K. & Springer, T. A. (1998) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 30.Harpaz Y, Chothia C. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 32.Chothia C, Jones E Y. Annu Rev Biochem. 1997;66:823–862. doi: 10.1146/annurev.biochem.66.1.823. [DOI] [PubMed] [Google Scholar]
- 33.Janin J. Nat Struct Biol. 1997;4:973–974. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- 34.Wu H, Kwong P D, Hendrickson W A. Nature (London) 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 35.Weber C, Lu C-F, Casasnovas J M, Springer T A. J Immunol. 1997;159:3968–3975. [PubMed] [Google Scholar]
- 36.Kirchhausen T, Staunton D E, Springer T A. J Leukocyte Biol. 1993;53:342–346. doi: 10.1002/jlb.53.3.342. [DOI] [PubMed] [Google Scholar]
- 37.Fisher K L, Lu J, Riddle L, Kim K J, Presta L G, Bodary S C. Mol Biol Cell. 1997;8:501–515. doi: 10.1091/mbc.8.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newham P, Craig S E, Seddon G N, Schofield N R, Rees A, Edwards R M, Jones E Y, Humphries M J. J Biol Chem. 1997;272:19429–19440. doi: 10.1074/jbc.272.31.19429. [DOI] [PubMed] [Google Scholar]
- 39.Carson M. J Mol Graph. 1987;5:103–106. [Google Scholar]
- 40.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]